Abstract
The balance between carbon and nitrogen is a key determinant of seed storage components, and thus, is of great importance to rice and other seed-based food crops. To clarify the influence of the rhizosphere carbon/nitrogen balance during the maturation stage of several seed components, transcriptome analysis was performed on the seeds from rice plants that were provided additional nitrogen fertilization at heading time. As a result, it was assessed that genes associated with molecular processes such as photosynthesis, trehalose metabolism, carbon fixation, amino acid metabolism, and cell wall metabolism were differentially expressed. Moreover, cellulose and sucrose synthases, which are involved in cellulose synthesis, were down-regulated. Therefore, we compared cellulose content of mature seeds that were treated with additional nitrogen fertilization with those from control plants using calcofluor staining. In these experiments, cellulose content in endosperm from plants receiving additional nitrogen fertilization was less than that in control endosperm. Other starch synthesis-related genes such as starch synthase 1, starch phosphorylase 2, and branching enzyme 3 were also down-regulated, whereas some α-amylase and β-amylase genes were up-regulated. On the other hand, mRNA expression of amino acid biosynthesis-related molecules was up-regulated. Moreover, additional nitrogen fertilization caused accumulation of storage proteins and up-regulated Cys-poor prolamin mRNA expression. These data suggest that additional nitrogen fertilization at heading time changes the expression of some storage substance-related genes and reduces cellulose levels in endosperm.
Introduction
Regulation of carbon (C) and nitrogen (N) metabolism is indispensable for plant growth and development. Carbon and nitrogen species are essential constituents of both macronutrients and signaling metabolites, which influence several cellular processes and gene expression [1]–[6]. In crop plants, starch and protein content in seed are determinants of yield and quality. These are synthesized using sugars and amino acids from the plant body, and share photosynthetic carbon sources for their synthesis. Thus, control of C/N balance during the reproductive stage is critical for high-yield and high-quality crop production.
The amount and timing of nitrogen fertilization are the most important factors for beneficial control of C/N balance, because the distribution of carbon sources from photosynthesis is generally influenced by plant nitrogen conditions [7], [8]. For example, the expression of photosynthetic and carbon fixation-related genes rapidly decreases in rice roots and leaves under low nitrogen conditions [9]. Under these conditions, rice leaves turn to pale green, carbon fixation is reduced, and remobilized nitrogen is used for other metabolic processes. In contrast, additional nitrogen fertilization facilitates maturation and elevates grain yields in commercial rice cultivation [10]. A high-nitrogen condition retards leaf senescence by maintaining nitrogen-containing compounds such as chlorophyll and photosynthetic proteins, thus elevating photosynthetic activity and transport of photosynthetic materials to the seed throughout the seed-maturation period. Such conditions may alter accumulation of seed components. In fact, protein content of rice seeds is elevated under conditions of high nitrogen fertilization [11].
Above results suggest that rhizospheric nitrogen influences global gene expression in the plant body, thereby implying the existence of gene networks that control C/N balance. In addition, gene expression in seeds is strongly influenced by the condition of the plant body. However, no studies have examined the influence of nitrogen on gene expression, metabolic processes, and accumulation of components in rice seeds. In this study, we examined the effects of nitrogen fertilization at heading time of rice, because plant body sizes and numbers of spikelets are fixed before fertilization. Rhizospheric nitrogen may directly affect metabolic processes during seed maturation. Thus, we examined changes in gene expression that correspond with nitrogen fertilization at heading time using DNA microarray analysis. Subsequently, we examined whether these changes in gene expression are correlated with seed components. The present data indicate that nitrogen fertilization at heading time decreases cellulose synthesis.
Materials and Methods
Rice Cultivation under Field Conditions
Oryza sativa L. cv. Nipponbare was used in all experiments. Rice plants were grown in a paddy field at the NARO Agricultural Research Center, Niigata, Japan. Field trials were conducted for three years.
At heading time, the experimental field was divided into a control plot and a nitrogen-fertilized (N-fertilized) plot using a plastic board. Ammonium chloride was sprayed on the soil surface of the N-fertilized section at a rate of 8 kg/1,000 m2. The sampling area contained 10×10 plants in each section, and all mature grains in the sampling area were harvested and prepared for analysis of nitrogen and amino acid content.
Rice Cultivation in a Plant Incubator
To improve reproducibility, a plant incubator with fluorescent lamps on inner walls (model FLI2000A, Tokyo Rikakikai, Tokyo, Japan) was used to simulate paddy field conditions.
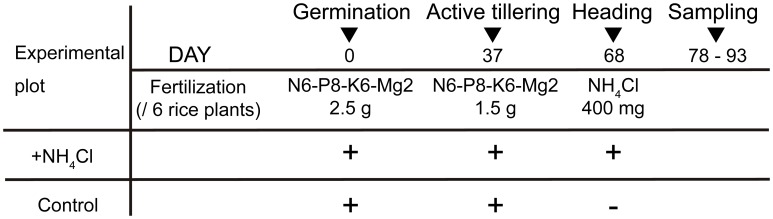
The schedule of cultivation is shown in Fig. 1. Plastic containers (C-AP fruit 500-1; 173×123×70 mm; Chuo Kagaku, Saitama, Japan) were filled with 500 mL rice nursery soil (Honen Agri, Niigata, Japan) and were supplied with 2.5 g fertilizer containing 0.15 g nitrogen, 0.2 g phosphate, 0.15 g potassium, and 0.05 g magnesium. Six plants were cultivated in each plastic container. Each of the four containers was placed on stainless trays (TRAY SUS No. 9, 367×257×92 mm; AZ ONE, Osaka, Japan); two trays, one for the control plot and the other for the N-fertilized plot, were placed in an incubator. Each plot contained 24 plants and tap water was provided to the depth such that the container was submerged. These plants were cultivated under short-daytime conditions with a 12-h maximum illumination (28°C)/12-h dark (22°C) cycle. Water depth was recovered every 2 days using tap water. The growth of each plant was restricted to the main culm by removing tillers. After 5 weeks, 1.5 g fertilizer was supplied to each container. At heading stage, ammonium chloride was sprayed on the soil surface of the N-fertilized plot tray at a rate of 400 mg/container.
Figure 1. Rice cultivation schedule.
Samples were grown in a growth chamber at 28°C/22°C over a 12-h light/12-h dark cycle. Fertilizer was supplied at planting and 37 days after germination. In addition, 400 mg ammonium chloride (NH4Cl) was supplied at heading time to the “+NH4Cl” group.
To examine the synthesis of storage materials in more detail, samples were taken at 10, 15, 20, and 25 DAF during incubator cultivation. Spikelets were marked as for field cultivation and were subsequently sampled. Developing grains were collected randomly from 24 plants in each experimental plot. All dehusked grains from the same sampling time were mixed, frozen in liquid nitrogen, and stored at −80°C until use. For microarray analysis, RNAs from 15-DAF grains were used, because storage materials are synthesized actively during this stage [12]. Finally, mature grains were harvested from each plant and their nitrogen content was determined.
Analysis of Nitrogen Content, Amino Acid Composition, and Protein Composition
Nitrogen content was determined using the Kjeldahl method in field trials. Amino acid analysis was performed at the Japan Food Research Laboratories (http://www.jfrl.or.jp/e/index.html; Tokyo, Japan) using an automated amino acid analyzer and HPLC according to their standard protocol. Total amino acid content was analyzed after hydrolysis of rice samples using hydrogen chloride. Free amino acid content was analyzed using sulfosalicylic acid- extractable fraction of rice samples. After harvest of all three trials, polished rice seeds were ground to powder in a cyclone sample mill (Shizuoka Seiki, Shizuoka, Japan), and nitrogen and amino acid content were determined for each trial. The sum of individual amino acid content was calculated, and differences between means were used for statistical analysis.
Nitrogen content was determined using a rapid-N-III automated nitrogen analyzer (Elementar Analysensysteme GmbH, Hanau, Germany) in incubator cultivation. Three independent plants from each fertilization division were selected, and nitrogen content of six polished grains from each plant was determined.
Protein composition of polished rice was analyzed using SDS-PAGE as shown previously with some modifications [13]. Subsequently, six seeds from each plant were ground to a powder, and soluble protein was extracted by adding 2.4 mL globulin extraction buffer containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 0.5 M NaCl. Insoluble protein was then extracted by adding 2.4 mL of the total protein extraction buffer containing 50 mM Tris-HCl, (pH 6.8), 8 M urea, 4% SDS, and 5% 2-mercaptoethanol. Subsequently, a 10 µL aliquot of extracted protein was used for SDS-PAGE.
Total RNA Isolation and DNA Microarray Preparation
Seeds at 15 DAF were collected from 24 control plants and 24 N-fertilized plants, immediately frozen in liquid nitrogen and pooled. Three sets of five seeds were selected from each seed pool. Samples were then ground to a fine powder, and RNA was extracted using an RNeasy Plant Mini kit (Qiagen, Hilden, Germany).
DNA microarray analysis was performed using a One-Cycle cDNA Synthesis kit (Affymetrix, Santa Clara, CA), a Sample Cleanup Module (Affymetrix), and a GeneChip IVT Labeling kit (Affymetrix). Experimental procedures were performed according to the manufacturer’s protocols. Briefly, after synthesizing cDNA from purified RNA, biotinylated cRNA was transcribed from cDNA using T7 RNA polymerase, fragmented, and was then added to a GeneChip Rice Genome Array containing over 50,000 rice genes (Affymetrix). Fragmented RNA was hybridized with the array at 45°C for 16 h. The array was then washed, labeled with phycoerythrin, and scanned for fluorescence using the Affymetrix GeneChip System. All microarray data were submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/; GEO Series ID GSE49818). Further, Affymetrix GCOS software was used to convert array images and probe intensities to CEL files. CEL files were then transferred to a personal computer and were analyzed using R statistical language and Bioconductor software. Files were normalized using the distribution-free weighted (DFW) method [14]. The pvclust function was then used to perform clustering analysis of the samples.
Gene expression patterns in rice grown under +NH4/−NH4 conditions were compared using the “rank products” function [15]. Genes with false discovery rates (FDR) of <0.05 were extracted, analyzed with the “pvclust” function [16], and divided according to their expression patterns. Identified genes were analyzed using the annotation file for the Rice Genome Array, which was downloaded from the NetAffx database on the Affymetrix website.
Gene-annotation enrichment analysis of differentially expressed genes (DEGs) was performed using the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/) [17] and Quick GO (http://www.ebi.ac.uk/QuickGO/) [18]. EASE scores from modified Fisher’s exact test P-values [19] were used to extract statistically over represented GO terms from DEGs. When annotations could not be obtained using the NetAffx database, we consulted the following databases: Rice TOGO (http://agri-trait.dna.affrc.go.jp/index.html), RiceXPro (http://agri-trait.dna.affrc.go.jp/index.html), MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), RAP-DB (http://rapdblegacy.dna.affrc.go.jp/), and UniProt (http://www.uniprot.org/).
β-glucan Staining
β-glucan staining was performed on 30 mature seeds that were randomly selected from each fertilization plot in the third field trial. Fresh-frozen rice grain sections were prepared for fluorescence microscopy as described in Saito et al., (2008). Frozen sections (5 µm thick) were cut from each seed, soaked in a 10% (w/v) potassium hydroxide solution, and were then stained with 0.05% (w/v) calcofluor white for 1–2 min. Further, sections were observed under a microscope using UV illumination. ImageJ software (http://rsbweb.nih.gov/ij/) was used to quantify fluorescence intensities. Fluorescence intensity per unit area in the part of endosperm that was surrounded by the dotted lines was measured was adjusted by subtraction of background fluorescence intensity.
Results and Discussion
In this study, the timing of additional nitrogen fertilization is the key factor. At heading time, plant body growth ceases and number of spikelets are fixed. Therefore, the effects of additional nitrogen (ammonium) are likely to be reflected in seed maturation and storage material synthesis.
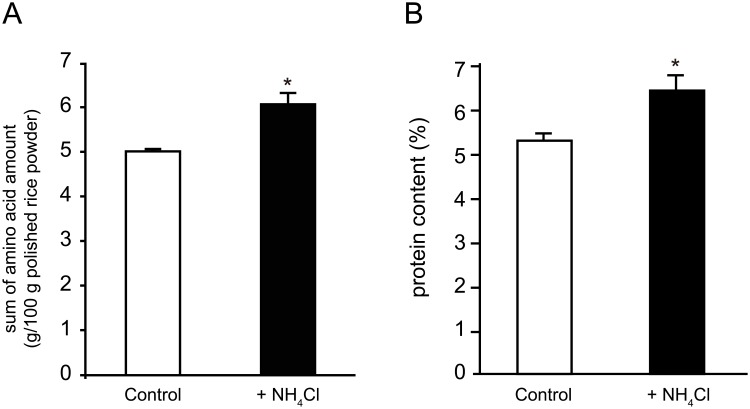
In general, enriched nitrogen fertilization elevates protein content, as determined using the Kjeldahl method [10]. In agreement, the sum of amino acid and protein content, were elevated in mature rice treated with additional fertilization in all trials (Fig. 2 and Table S1). As shown in Table S1, amino acid content was also higher in individuals from the N-fertilized plot in all trials. However, free amino acid levels remained low or undetectable in all trials (Table S1). These results indicate that nitrogen fertilization at heading time was mainly utilized for protein synthesis and not for the accumulation of free amino acids. At the third field trial, we took some preliminary results about developing grains. Grain weight did not differ significantly between plants from the N-fertilized and control plot at every sampling time (data not shown). Although differences were not statistically confirmed, seed nitrogen content was higher in the N-fertilized plot than that in the control plot at every sampling time. It is a reasonable speculation that gene expression in developing grain also changes in response to additional nitrogen fertilization.
Figure 2. Amino acid and protein contents of mature polished rice cultivated in a field.
Field trials were conducted for three years and data are expressed as means of the three trials. Filled bars correspond with samples from the N-fertilized plot and unfilled bars correspond with samples from the control plot. Error bars represent standard deviation (SD; *P<0.01) (A) Total amino acid content was analyzed after hydrolysis using hydrogen chloride. (B) Protein content was estimated using the Kjeldahl method.
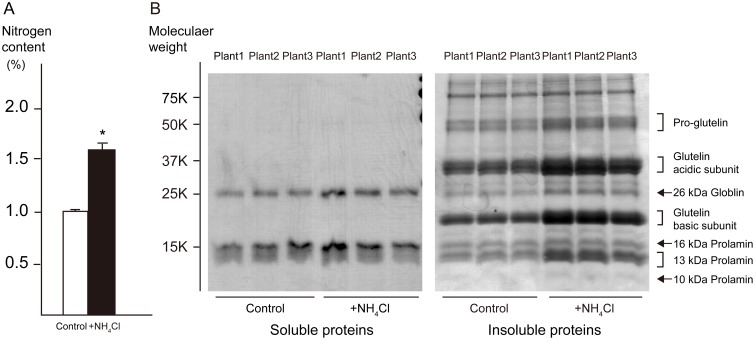
To investigate the genomic response to nitrogen fertilization, a plant incubator was used to control growth conditions between replicate experiments. Rice plants from the two experimental plots were morphologically similar at heading stage, although the leaf blades of N-fertilized rice clearly showed a darker green color than those of the control group (Fig. S1). Mature seeds were obtained from all but one control plant. Moreover, the average maturation rate was calculated from the number of mature seeds divided by the number of spikelets, and was found to be 88.0% in the N-fertilization plot and 92.2% in the control plot. Nitrogen content of mature polished grains was much higher in plants from the N-fertilized plot (Fig. 3A), and this difference of seed nitrogen content between two plots was greater in incubator cultivation than in field trial. In incubator cultivation, water was pooled in trays and plant sizes were reduced by removing tillers. Thus, all fertilizer remained in incubator trays, allowing for higher quantities of nitrogen to be utilized for grain development. Accordingly, higher nitrogen content of seeds from the N-fertilized plot was reflected by band density of insoluble proteins in SDS-PAGE analysis (Fig. 3B). Each density corresponding to glutelin and a 13 kDa prolamin was higher in the case of N-fertilized plot.
Figure 3. Analysis of proteins in polished rice from incubator cultivated plants.
Three independent plants were selected from the N-fertilized and control plots. (A) Nitrogen content of polished rice was determined using a Rapid-N analyzer. Error bars represent standard deviation (SD; *P<0.001) (B) Protein composition of polished rice was analyzed using SDS-PAGE. Soluble proteins and insoluble proteins were extracted sequentially from the same sample. Major storage proteins in rice seed were extracted as insoluble proteins, as indicated at the right side of the gel.
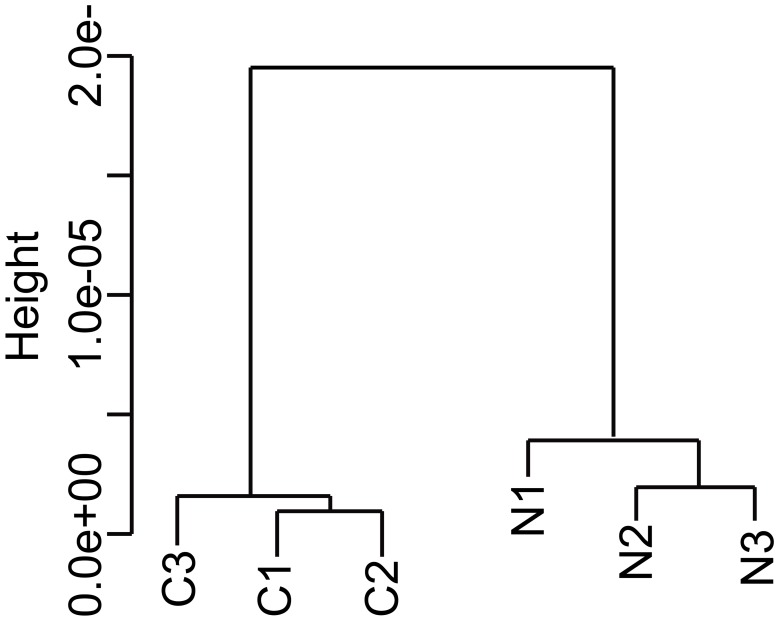
Dehusked 15-DAF developing seeds from rice plants were subjected to DNA microarray analysis, and data from six samples (three per experimental plot) were normalized using the DFW method and subjected to hierarchical clustering analysis. As shown in Fig. 4, a distinct separation was observed between the N-fertilized and control samples.
Figure 4. Cluster dendrogram.
A cluster dendrogram was generated using rice seed gene expression data from six samples and the “pvclust” function. Each sample was prepared at 15-DAF seeds from 24 control or 24 N-fertilized plants and were pooled. Three sets of five seeds were picked from each seed pool. C, control samples; N, N-fertilized samples.
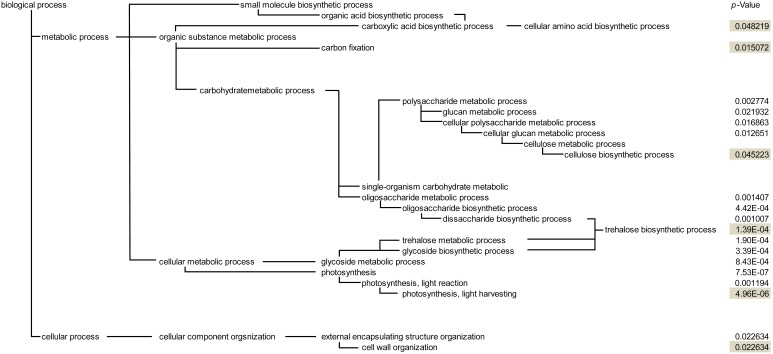
Furthermore, DEGs were identified between the control and N-fertilized samples using the rank products method. DEGs with FDR<0.05 were extracted, revealing 678 significantly up-regulated genes and 687 significantly down-regulated genes due to additional fertilization (Tables S2 and S3). The extracted DEGs were classified into functional categories according to gene ontology (GO) terms. Significantly enriched categories of DEGs (FDR<0.05) are shown in Fig. 5. The hierarchical GO structure indicated a more specific category with a deeper hierarchy. Therefore, most important categories appeared at the lower end of the tree (depth of hierarchy is shown in shadowed areas; Fig. 5). Furthermore, six functional categories were significantly enriched in DEGs. These included “photosynthesis, light harvesting,” “trehalose biosynthetic process,” “carbon fixation,” “cell wall organization,” “cellulose biosynthetic process,” and “cellular amino acid biosynthetic process.” Genes that were predicted to be involved in the synthesis of storage substances were also identified from the list of DEGs (Table 1). The “photosynthesis, light harvesting” category included genes that encode chlorophyll-binding proteins, and Rubisco was identified from the “carbon fixation” category (Table 2). Consistent with these results, the leaf blades of the N-fertilized samples were clearly darker in color than those of the control blades (Fig. S1). Because DNA microarray samples were collected from 15-DAF seeds with green pericarps, the molecules mentioned above are likely localized to the pericarp.
Figure 5. Significantly enriched categories were identified using QuickGO.
In response to additional fertilization, 1,365 genes were up- or down-regulated; FDR-corrected P-values of categories at the deepest hierarchical level are shaded; *P<0.05.
Table 1. Genes related to synthesis of storage substances in seeds with altered mRNA expression in response to additional fertilizer.
| Probe set ID | RAP-DB ID | RAP-DB Annotation | Gene Expression |
| Os.26109.1.S1_x_at | Os05g0329100, Os05g0329200, Os05g0329400, Os05g0330600 | Cysteine-poor 13 kDa prolamin | Up |
| Os.25998.1.S1_at | Os05g0329100, Os05g0329200, Os05g0329400, Os05g0330600 | Cysteine-poor 13 kDa prolamin | Up |
| Os.8502.4.S1_at | Os05g0329300 | Cysteine-poor 13 kDa prolamin | Up |
| Os.8502.5.S1_x_at | Os05g0328333, Os05g0329100, Os05g0329200, Os05g0329400, Os05g0330600, Os07g0219300, Os07g0219400, Os07g0220050 | Cysteine-poor 13 kDa prolamin | Up |
| Os.20396.1.A1_at | Os02g0456150 | 11-S plant seed storage protein family protein. | Up |
| Os.20396.1.A1_s_at | Os02g0456150 | 11-S plant seed storage protein family protein. | Up |
| OsAffx.2749.1.S1_at | Os02g0456150 | 11-S plant seed storage protein family protein. | Up |
| Os.17979.1.S1_at | Os02g0244100 | Grain weight 2 (OsGW2). | Up |
| OsAffx.16823.1.S1_at | Os08g0137250 | Fertilization-Independent endosperm(Protein Fertilization-Independent Seed 3) (OsFIE1). | Up |
| Os.12593.1.S1_s_at | Os08g0473600 | Alpha-amylase isozyme 3E precursor (EC 3.2.1.1). | Up |
| Os.10339.1.S1_at | Os03g0141200 | Similar to Beta-amylase PCT-BMYI (EC 3.2.1.2). | Up |
| Os.46618.1.S1_at | Os10g0565200 | Similar to Beta-amylase PCT-BMYI (EC 3.2.1.2). | Up |
| Os.13907.1.S1_at | Os02g0248800 | Similar to Glutelin type-B 2 precursor. | Up |
| Os.29800.1.S1_x_at | Os01g0702900, Os02g0771500 | Sucrose-phosphate synthase (EC 2.4.1.14). | Up |
| Os.12725.1.S1_at | Os06g0160700 | Similar to Starch synthase I, chloroplast precursor (EC 2.4.1.21)(Soluble starch synthase 1) (SSS 1). | Down |
| Os.2623.1.S1_at | Os01g0851700 | Similar to Cytosolic starch phosphorylase (Fragment)/Starch phosphorylase 2. | Down |
| Os.4179.1.S1_at | Os02g0528200 | Branching enzyme-3 precursor (EC 2.4.1.18). | Down |
| OsAffx.13550.1.S1_s_at | Os03g0808200 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. | Down |
| Os.33722.1.S1_at | Os01g0736100 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. | Down |
| Os.9127.1.S1_a_at | Os06g0194900 | Sucrose synthase 2 (EC 2.4.1.13). | Down |
| Os.9860.1.S1_at | Os07g0616800 | Sucrose synthase 3 (EC 2.4.1.13). | Down |
Table 2. Differentially expressed genes in significantly enriched GO categories (*P<0.05).
| RAP-DB ID | RAP-DB annotation | Gene Expression | |
| Photosynthesis, light harvesting | Os08g0435900, Os07g0562700, Os07g0558400, Os01g0600900, Os03g0592500, Os04g0457000 | Chlorophyll a/b-binding protein | Up |
| Trehalose biosynthetic process | Os03g0386500 | Trehalose-6-phosphate phosphatase 9 | Up |
| Os02g0661100 | Trehalose-6-phosphate phosphatase 1 | Down | |
| Os09g0369400 | Trehalose-6-phosphate phosphatase 7 | Down | |
| Os01g0730300 | Trehalose-6-phosphate synthase 3 | Down | |
| Os02g0790500 | Trehalose-6-phosphate synthase 5 | Up | |
| Os05g0517200 | Trehalose-6-phosphate synthase 6 | Up | |
| Carbon fixation | Os01g0791033, Os05g0427800, Os12g0207600 | Ribulose bisphosphate carboxylase large chain precursor (EC 4.1.1.39) (RuBisCO large subunit). | Up |
| Os12g0292400, Os12g0291400 | Ribulose bisphosphate carboxylase small chain | Up | |
| Cell wall organization | Os10g0555900, Os10g0548600 | Beta-expansin precursor | Up |
| Os08g0160500 | Cellulose synthase-like protein F6 (OsCslF6) | Down | |
| Os10g0450900 | Glycine-rich cell wall structural protein 2 precursor | Up | |
| Os07g0208500 | Cellulose synthase A8 (OsCESA8) | Down | |
| Os02g0130200 | Virulence factor, pectin lyase fold family protein | Up | |
| Os07g0252400 | Cellulose synthase A6 (OsCESA6) | Down | |
| Os02g0738600 | Endoglucanase 7 | Down | |
| Os03g0377700 | Cellulose synthase-like A5 (CSLA5) | Down | |
| Os03g0808100 | Cellulose synthase A2 (OsCESA2) | Down | |
| Os03g0837100 | Cellulose synthase A5 (OsCESA5) | Down | |
| Os08g0237000 | Xyloglucan endotransglycosylase/hydrolase protein 8 precursor(End-xyloglucan transferase) (OsXTH8) | Down | |
| Cellulose biosynthetic process | Os07g0252400 | Cellulose synthase A6 (OsCESA6) | Down |
| Os08g0160500 | Cellulose synthase-like protein F6 (OsCslF6) | Down | |
| Os07g0208500 | Cellulose synthase A8 (OsCESA8) | Down | |
| Os03g0808100 | Cellulose synthase A2 (OsCESA2) | Down | |
| Os03g0837100 | Cellulose synthase A5 (OsCESA5) | Down | |
| Cellular amino acid biosynthetic process | Os04g0669800 | Methylthioribose kinase | Up |
| Os01g0720700 | Serine acetyltransferase 1 | Up | |
| Os11g0256000 | Acetolactate synthase, small subunit family protein | Up | |
| Os09g0565700 | Prephenate dehydratase domain containing protein | Up | |
| Os12g0578200 | Chorismate mutase, chloroplast precursor (CM-1) | Up | |
| Os03g0291500 | Asparagine synthase domain containing protein | Up | |
| Os01g0681900 | NADH - Glutamate Synthase 1 | Down | |
| Os03g0279400 | Arginine biosynthesis bifunctional protein ArgJ, chloroplastic | Down | |
| Os02g0510200 | Acetohydroxyacid synthase | Up | |
| Os03g0826500 | Anthranilate synthase alpha 1 subunit | Up | |
| Os03g0389700 | Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplastic | Up |
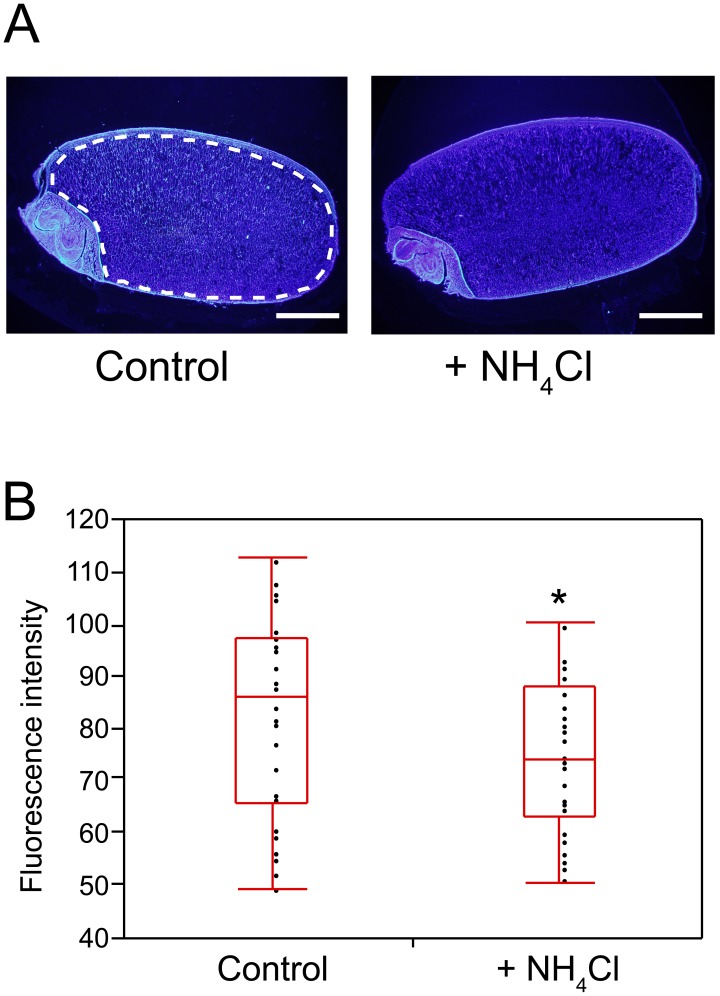
Cell wall-related genes such as those encoding cellulose synthase A catalytic subunit (CESA) and xyloglucan endotransglucosylase/hydrolase, were down-regulated following additional fertilization (Table 2). Genes encoding OsCESA2, OsCESA5, OsCESA6, OsCESA8, CESA-like protein A5 (OsCslA5), and CESA-like protein F6 (OsCslF6) are all involved in cell wall synthesis, and were down-regulated in the presence of additional fertilization. Thus, seeds were examined to determine whether β-glucan constituents of cellulose and (1, 3; 1, 4)-β-D-glucan actually decreased. Staining with calcofluor white demonstrated weaker fluorescence intensity in seed endosperms from the N-fertilized plot than from the control plot (Fig. 6). Cell walls of rice endosperm comprise cellulose and hemicellulose [21]. Rice hemicellulose comprises arabinoxylans and (1, 3; 1, 4)-β-D-glucan, also known as mixed-linkage glucan (MLG). Although calcofluor white stains both cellulose and MLG, rice does not accumulate significant amount of MLG in its grains [21], [22]. Thus the present data suggest decreased cellulose content of endosperm. In general, cellulose is synthesized from UDP-glucose by CESA, which comprises a cellulose synthase complex of six subunits [23]–[25]. Although at least 11 genes have been annotated as CESA in the MSU rice genome annotation project, the functions of only three genes, OsCESA4, OsCESA7, and OsCESA9, have been examined [26]. Sucrose synthase (SUS), which is involved in UDP-glucose metabolism, was down-regulated by additional fertilization, and plays a direct role in cell wall biosynthesis by forming a complex with CESA [27]–[30]. Six SUS homologs are present in the rice genome [31]. Among these, SUS2 and SUS3 were extracted from DEGs as genes that were down-regulated by additional fertilization (Table 1). In particular, SUS3 is reportedly localized primarily in the endosperm and in the aleurone layer [31], [32], indicating that CESA2, CESA5, CESA6, and CESA8 may form a complex with SUS3 to synthesize cellulose in the endosperm. The relationship between rice quality and cellulose content remains poorly understood. However, it is accepted that rice with a high nitrogen content has less stickiness, greater hardness after cooking, less palatability, and less processing quality [33]–[35]. Accordingly, cell walls may become harder with increased cellulose content. However, in the present experiments, rice with high nitrogen content contained less cellulose. Thus, the complexities of relationship between cooked-rice properties and rice-seed components require further detailed study.
Figure 6. β-Glucan content of rice grains.
(A) Histochemical staining of mature rice seed. Cross-sections of grains from the control plot (left) and from the N-fertilized plot (+NH4Cl; right) are shown. Sections were stained for β-glucan using calcofluor white. The white-dotted line indicates the area analyzed for fluorescence intensity. The fluorescence intensity of the endosperm was calculated by adjusting the background intensity. Scale bars, 1 mm. (B) Endosperm fluorescence intensity was analyzed using ImageJ. Sections were cut from 30 randomly selected grains from each plot. The horizontal line inside the box plot indicates the median value. The inner box indicates the interquartile range and runs between 25th and 75th percentiles. The upper line extending from the box indicates the largest value between the 75th percentile and the point at 1.5 times the interquartile range. The lower line extending from the box indicates the smallest point between the 25th percentile and the point at 1.5 times the interquartile range; *P<0.05.
Other polysaccharide-related genes were identified from DEGs with FDR<0.05 (Table 1). The genes encoding starch synthase 1, starch branching enzyme 3, and starch phosphorylase 2 were down-regulated. These enzymes participate in starch synthesis from UDP-glucose. However, α-amylase and β-amylase, which further process starch into smaller sugars, were up-regulated in plants receiving additional fertilization (Table 1). Although various α-amylases are present in rice, only RAmy3E (Os08g0473600) was identified in this study, and is reportedly localized in seeds [36]–[38]. Because genes associated with starch synthesis were down-regulated concomitantly with increases in mRNA expression of starch-degrading enzymes, the starch content of rice may have decreased. This phenomenon was previously reported in rice kept under high temperature conditions during maturation [39]. Besides, starch synthesis is reportedly suppressed when plant bodies are subjected to carbon starvation, favoring monosaccharide production [36], [40].
Some genes of cellular amino acid biosynthetic processes, expect for NADH-glutamate synthase 1 and arginine biosynthesis bifunctional protein, were up-regulated, (Table 2 and Fig. S2). This result indicates that amino acid metabolism was activated to meet the demands of storage protein synthesis. In fact, amino acid and storage protein content were increased in polished rice from the N-fertilized plot (Figs. 2 and 3). Also, DNA microarray analysis showed remarkable increase in mRNA expression of storage proteins, in particular that of Cys-poor prolamins. Glutelin B2 and Os02g0456150 mRNA, annotated as “11 S-plant seed storage protein family protein”, were also increased. The latter gene was found to be a homologous pseudogene of glutelin C. Other storage proteins were not extracted in our analysis, suggesting that mRNA response to additional nitrogen fertilization varies between types of storage protein.
Prolamin comprises several molecular species of various sizes, including 10-kDa, Cys-poor 13-kDa, Cys-rich 13-kDa, and 16-kDa prolamins [20], [41], [42]. Cys-poor prolamins are accumulated at late stages of grain filling [43], and their expression is increased during suppression of glutelins, which are the most abundant storage protein in rice [44]. Both the present data and previous reports suggest that the expression of Cys-poor prolamin is controlled at the mRNA level, and effectively stores excess nitrogen during additional fertilization [43], [44]. This may be because Cys-poor prolamin has the simple primary structure with the absence of cysteine residues, and is directly accumulated in the endoplasmic reticulum where protein synthesis occurs. Rice with a high protein content shows poor cooking quality, and prolamins are suggested to associate such phenomena [33]–[35], [45]. Thus, Cys-poor prolamins may be an important gene target in rice breeding, and may reflect cooking properties of rice.
Conclusion
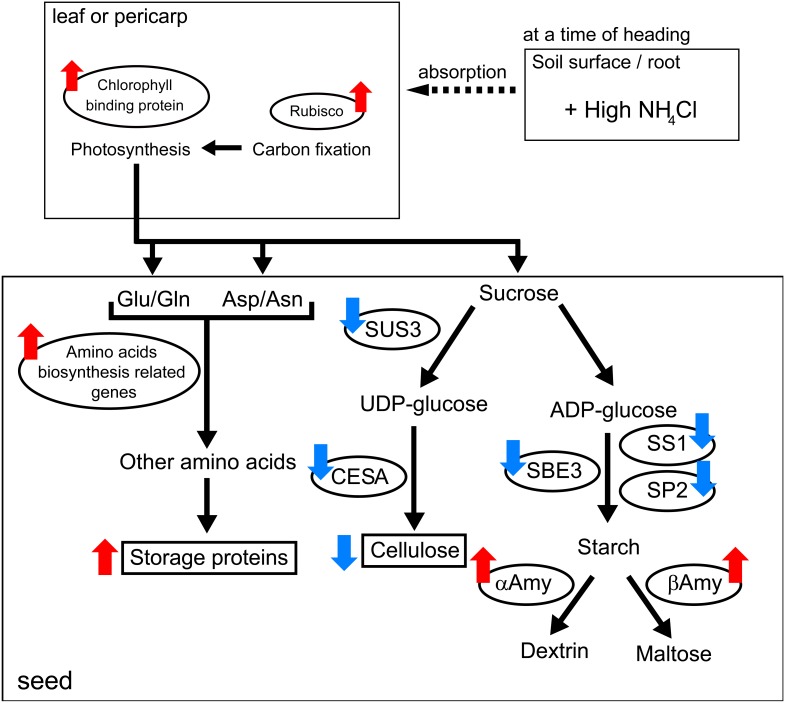
The present data reveal rapid changes in C/N balance in response to rhizosphere nitrogen fertilization at heading time. Notable changes in mRNA expression and seed compounds are summarized in Figure 7. Leaf color showed a darker green and the mRNA expression of Rubisco and chlorophyll-binding protein in seed pericarp was greater in the N-fertilized plot. In general, such situation contributes to increased total photosynthesis, carbon fixation, and nitrogen absorption in the rice plant body, and can improve final yield of grains per unit area [10]. More amounts of sucrose and glutamine/glutamate and asparagine/aspartic acid may be transported into seeds in N-fertilized plot. However, compounds of seeds were not always increased in the N-fertilized plot. The accumulation of nitrogen compounds such as storage proteins increased, whereas that of polysaccharides such as cellulose decreased, correlating with changes in mRNA expression for synthetic processes of these compounds. The mRNA expression profile for starch biosynthesis and starch degradation suggested that levels of other polysaccharides such as starch may also change. The possible mechanism underlying our result is that the amount of carbon backbones may be a limiting factor in seed and it may be primarily used for nitrogen accumulation in endosperm under high nitrogen fertilization (Fig. 7). This study provides new insights into the relationship between fertilization and grain maturation and contributes to the understanding of nutrient distribution during rice production.
Figure 7. Changes in gene expression and rice seed compounds with additional fertilization.
Notable change in mRNA expression and seed compounds are summarized. Molecules in the open oval are DEGs from DNA microarray experiments. Rectangles show the compound whose content was measured in this study. Red and blue arrows indicate up- and down-regulation by additional nitrogen fertilization, respectively. SUS3, sucrose synthase 3; CESA, cellulose synthase A catalytic subunit; SBE3, starch-branching enzyme 3; SS1, starch synthase 1; SP2, starch phosphorylase 2; αAmy, α-amylase; βAmy, β-amylase.
Supporting Information
Rice of 10 days after additional nitrogen fertilization.
(PDF)
Heat map of differentially expressed genes in additional nitrogen fertilization. Each column represents results from an independent samples. C1, C2 and C3 indicate control samples, and N1, N2 and N3 N-fertilized ones. Each line corresponds to a single probe. The heat map was prepared by obtaining the Z scores from the signal value of each probe after DFW normalization. More reddish and more greenish stand for higher and lower expression levels than the mean, relatively. Significantly enriched Gene ontology (GO) terms (P<0.05) and storage substances related genes are shown on the right side of the heat map.
(PPTX)
The amino acid content and the protein content of mature polished rice cultivated in a field.
(XLSX)
The gene in which expression significantly down-regulated with the additional fertilizer (FDR 0.05>).
(XLSX)
The gene in which expression significantly up-regulated with the additional fertilizer (FDR 0.05>).
(XLSX)
Acknowledgments
We thank Dr. Tamura (Tokyo University of Agriculture) for her helpful discussions and technical assistance.
Funding Statement
This work was supported by Nissin Food Holdings Co., LTD (http://www.nissinfoods-holdings.co.jp/). It was supported by grant in aid for food science research to T. A. and M. K. from the Tojuro Iijima Foundation for Food Science and Technology (http://www.iijima-kinenzaidan.or.jp/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stitt M (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970. [DOI] [PubMed] [Google Scholar]
- 4. Rolland F, Moore B, Sheen J (2002) Sugar Sensing and Signaling in Plants. Plant Cell 14: S185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price J, Laxmi A, St Martin SK, Jang J-C (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thum KE, Shin MJ, Palenchar PM, Kouranov A, Coruzzi GM (2004) Genome-wide investigation of light and carbon signaling interactions in Arabidopsis. Genome Biol 5: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593. [DOI] [PubMed] [Google Scholar]
- 8. Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, et al. (1997) Nitrate Acts as a Signal to Induce Organic Acid Metabolism and Repress Starch Metabolism in Tobacco. Plant Cell 9: 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lian X, Wang S, Zhang J, Feng Q, Zhang L, et al. (2006) Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol Biol 60: 617–631. [DOI] [PubMed] [Google Scholar]
- 10.Matsushima S (1995) 3. Phisiology of high-yielding rice and its cultivation. In: Matsuo T, Kumazawa K, Ishii R, Ishihara K, Hirata H, editors. Science of the rice plant. Tokyo: Food and Agriculture Plicy Resarch Center. 753–766.
- 11.Taira H (1995) 1. Physical properties. In: Matsuo T, Kumazawa K, Ishii R, Ishihara K, Hirata H, editors. Science of the rice plant. Tokyo: Food and Agriculture Plicy Resarch Center. pp. 1064–1078.
- 12. Suzuki K, Hattori A, Tanaka S, Masumura T, Abe M, et al. (2005) High-coverage profiling analysis of genes expressed during rice seed development, using an improved amplified fragment length polymorphism technique. Funct Integr Genomics 5: 117–127. [DOI] [PubMed] [Google Scholar]
- 13. Kuroda M, Kimizu M, Mikami C (2010) A simple set of plasmids for the production of transgenic plants. Biosci Biotechnol Biochem 74: 2348–2351. [DOI] [PubMed] [Google Scholar]
- 14. Chen Z, McGee M, Liu Q, Scheuermann RH (2007) A distribution free summarization method for Affymetrix GeneChip arrays. Bioinformatics 23: 321–327. [DOI] [PubMed] [Google Scholar]
- 15. Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92. [DOI] [PubMed] [Google Scholar]
- 16. Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542. [DOI] [PubMed] [Google Scholar]
- 17. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 18. Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, et al. (2009) QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 25: 3045–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA (2003) Identifying biological themes within lists of genes with EASE. Genome Biol 4: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saito Y, Nakatsuka N, Shigemitsu T, Tanaka K, Morita S, et al. (2008) Thin frozen film method for visualization of storage proteins in mature rice grains. Biosci Biotechnol Biochem 72: 2779–2781. [DOI] [PubMed] [Google Scholar]
- 21. Shibuya N (1985) Comparative Studies on Cell Wall Preparations from Rice Bran, Germ, and Endosperm. Cereal Chem 62: 252–258. [Google Scholar]
- 22. Burton RA, Fincher GB (2012) Current challenges in cell wall biology in the cereals and grasses. Front Plant Sci 3: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Atanassov II, Pittman JK, Turner SR (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284: 3833–3841. [DOI] [PubMed] [Google Scholar]
- 26. Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, et al. (2003) Three Distinct Rice Cellulose Synthase Catalytic Subunit Genes Required for Cellulose Synthesis in the Secondary Wall. Plant Physiol 133: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujii S, Hayashi T, Mizuno K (2010) Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol 51: 294–301. [DOI] [PubMed] [Google Scholar]
- 28. King SP, Lunn JE, Furbank RT (1997) Carbohydrate Content and Enzyme Metabolism in Developing Canola Siliques. Plant Physiol 114: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Usuda H, Demura T (1999) Development of sink capacity of the “storage root” in a radish cultivar with a high ratio of “storage root” to shoot. Plant Cell Physiol 40: 369–377. [Google Scholar]
- 30. Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92: 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang JW, Chen JT, Yu WP, Shyur LF, Wang a Y, et al. (1996) Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Biosci Biotechnol Biochem 60: 233–239. [DOI] [PubMed] [Google Scholar]
- 32. Hirose T, Scofield GN, Terao T (2008) An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci 174: 534–543. [Google Scholar]
- 33.Ken’ ichi O, Akiharu K, Hisashi S (1993) Quality Evaluation of Rice in Japan. Japan Agric Res Q: 95–101.
- 34. Okadome H (2005) Application of instrument-based multiple texture measurement of cooked milled-rice grains to rice quality evaluation. Japan Agric Res Q 39: 261–268. [Google Scholar]
- 35. Zhou Z, Robards K, Helliwell S, Blanchard C (2002) Composition and functional properties of rice. Int J Food Sci Technol 37: 849–868. [Google Scholar]
- 36. Yu SM, Kuo YH, Sheu G, Sheu YJ, Liu LF (1991) Metabolic derepression of alpha-amylase gene expression in suspension-cultured cells of rice. J Biol Chem 266: 21131–21137. [PubMed] [Google Scholar]
- 37. Karrer E, Rodriguez R (1992) Metabolic regulation of rice alpha-amylase and sucrose synthase genes in planta. Plant J 2: 517–523. [PubMed] [Google Scholar]
- 38. Umemura T, Perata P, Futsuhara Y, Yamaguchi J (1998) Sugar sensing and α-amylase gene repression in rice embryos. Planta 204: 420–428. [DOI] [PubMed] [Google Scholar]
- 39. Yamakawa H, Hakata M (2010) Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol 51: 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Akihiro T, Mizuno K, Fujimura T (2005) Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol 46: 937–946. [DOI] [PubMed] [Google Scholar]
- 41. Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, et al. (1987) Purification of Protein Body-I of Rice Seed and its Polypeptide Composition. Plant Cell Physiol 28: 1517–1527. [Google Scholar]
- 42. Fabian C, Ju Y-H (2011) A review on rice bran protein: its properties and extraction methods. Crit Rev Food Sci Nutr 51: 816–827. [DOI] [PubMed] [Google Scholar]
- 43. Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, et al. (2012) Formation mechanism of the internal structure of type I protein bodies in rice endosperm: relationship between the localization of prolamin species and the expression of individual genes. Plant J 70: 1043–1055. [DOI] [PubMed] [Google Scholar]
- 44. Shigemitsu T, Saito Y, Morita S (2012) Separation and Identification of Rice Prolamins by Two-Dimensional Gel Electrophoresis and Amino Acid Sequencing. Biosci Biotechnol Biochem 76: 594–597. [DOI] [PubMed] [Google Scholar]
- 45. Tanaka, Y Hayashida, S. Hongo M (1975) The relationship of the feces protein particles to rice protein bodies. Agric Biol Chem 39: 515–518. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rice of 10 days after additional nitrogen fertilization.
(PDF)
Heat map of differentially expressed genes in additional nitrogen fertilization. Each column represents results from an independent samples. C1, C2 and C3 indicate control samples, and N1, N2 and N3 N-fertilized ones. Each line corresponds to a single probe. The heat map was prepared by obtaining the Z scores from the signal value of each probe after DFW normalization. More reddish and more greenish stand for higher and lower expression levels than the mean, relatively. Significantly enriched Gene ontology (GO) terms (P<0.05) and storage substances related genes are shown on the right side of the heat map.
(PPTX)
The amino acid content and the protein content of mature polished rice cultivated in a field.
(XLSX)
The gene in which expression significantly down-regulated with the additional fertilizer (FDR 0.05>).
(XLSX)
The gene in which expression significantly up-regulated with the additional fertilizer (FDR 0.05>).
(XLSX)