Abstract
The pannexin family of channel-forming proteins is composed of 3 distinct but related members called Panx1, Panx2, and Panx3. Pannexins have been implicated in many physiological processes as well as pathological conditions, primarily through their function as ATP release channels. However, it is currently unclear if all pannexins are subject to similar or different post-translational modifications as most studies have focused primarily on Panx1. Using in vitro biochemical assays performed on ectopically expressed pannexins in HEK-293T cells, we confirmed that all 3 pannexins are N-glycosylated to different degrees, but they are not modified by sialylation or O-linked glycosylation in a manner that changes their apparent molecular weight. Using cell-free caspase assays, we also discovered that similar to Panx1, the C-terminus of Panx2 is a substrate for caspase cleavage. Panx3, on the other hand, is not subject to caspase digestion but an in vitro biotin switch assay revealed that it was S-nitrosylated by nitric oxide donors. Taken together, our findings uncover novel and diverse pannexin post-translational modifications suggesting that they may be differentially regulated for distinct or overlapping cellular and physiological functions.
Keywords: Pannexin, Panx1, Panx2, Panx3, post-translational modifications, caspase, nitrosylation, glycosylation
Introduction
The pannexin family constitutes as class of channel-forming proteins and is composed of 3 members: Panx1, Panx2, and Panx3.1,2 All 3 pannexins have been shown to form functional channels capable of dye uptake.3-6 Importantly, functional analysis of the channel properties of Panx1 and Panx3 have revealed a significant role for these channels in releasing ATP to extracellular spaces7,8 and facilitating Ca2+ influx into the cytosol.9,10 Panx1 has been the most studied as it is more ubiquitously expressed than Panx2 (predominantly found in the CNS) and Panx3 (found in skin, bone, cartilage, and mammary gland).1,11 An important physiological role for Panx1 is the regulation of vascular smooth muscle cell constriction in resistance arteries through an interaction with the α1-adrenergic receptor.12 Panx1 has also been implicated in ischemic neuronal cell death,13 epileptic seizures,14 and colitis15 raising its profile in a diverse array of pathologies. In apoptotic immune cells, the ATP released from caspase-cleaved Panx1 channels acts as “find-me” signals for monocyte recruitment and clearance of dead cells.16 Human PANX1 harbors 2 caspase cleavage sites, one residing in the intracellular loop (residues 164–167) and another located in the C-terminus of the protein (residues 376–379), with the C-terminal site being responsible for the activation of the channel during apoptosis.16,17 It is currently not known, however, whether the other 2 pannexin isoforms are also substrates for caspase cleavage.
One common modification for all 3 pannexins is N-linked glycosylation (or N-glycosylation), with Panx1 and Panx3 being modified to the high-mannose and complex glycosylation species associated with editing in the endoplasmic reticulum (ER) and Golgi apparatus. Panx2, on the other hand, appears to only be modified in the ER to a high mannose form, which remains sensitive to endoglycosidase H de-glycosylation.6 N-glycosylation has been reported to regulate the cellular localization of pannexins, their interactions with other pannexins, and potentially their function at the cell surface or in intracellular compartments.6,18,19
In addition to post-translational modification by caspase cleavage and glycosylation, we have recently reported that Panx1 can be targeted for S-nitrosylation. Targeted modification of cysteine thiols by the bioactive gas nitric oxide (NO) promotes reversible S-nitrosylation of a number of membrane proteins, having distinct effects on protein function.20-25 S-nitrosylation of Panx1 can occur at cysteines 40 and 346, with modification of both residues imparting inhibition of Panx1 channel currents and the ability of the channels to release ATP.25 To date, however, the ability of Panx2 and Panx3 to be modified by S-nitrosylation remains to be determined. Given the conserved sequence homology among the 3 pannexins, the fact that they are co-expressed in many cells and tissues and their shared function as large-pore membrane channels, we set out to investigate if the post-translational modifications that have been best reported for Panx1 are also involved in the regulation of Panx2 and Panx3. In parallel studies, we report that all 3 mouse pannexin isoforms are subject to variable N-glycosylation but do not change their apparent molecular weights in response to de-sialylation or O-deglycosylation. This is the first study to report that similar to Panx1, murine Panx2 is also a substrate for caspase cleavage, while the C-terminus of Panx3 is not cleaved by caspase 3 or 7. In addition, Panx3 can be S-nitrosylated like Panx1, while Panx2 appears to be resistant to NO-mediated cysteine modification.
Results
Glycosylation
Our research group and others have previously reported that all 3 pannexin members are N-glycosylated to different degrees, which affects the gel mobility of the proteins as assessed by SDS-PAGE.6,18 Utilizing an enzymatic deglycosylation kit (ProZyme/Glyko) we set out to investigate whether the 3 pannexin members are also modified by O-linked glycosylation or sialylation. Based on the assessment of SDS-PAGE gel mobility after sequential addition of the 3 enzymes: PNGase F, Sialidase A, and O-glycanase, we observed the greatest pannexin band shifts after PNGase F treatment (Fig. 1A, B, and C). Subsequent desialylation and O-glycanase digestion did not result in any further visible change in pannexin band mobility for any of the 3 pannexins, indicating N-glycosylation as the primary modification responsible for the band shift. As a positive control, Fetuin, a protein known to be N- and O- glycosylated as well as sialylated,26 exhibited increasing gel mobility following digestion with each enzyme, as visualized on a Sypro-stained gel (Fig. 1D). PNGase F digestion removes high-mannose as well as complex glycans present on glycoproteins, thus confirming that all 3 pannexins are N-glycosylated, with Panx1 and Panx3 exhibiting the largest gel mobility shift upon de-glycosylation (Fig. 1A and C). Panx2 presented a more subtle change in mobility after PNGase F treatment (Fig. 1B) consistent with previous reports indicating its existence as a high-mannose species.6
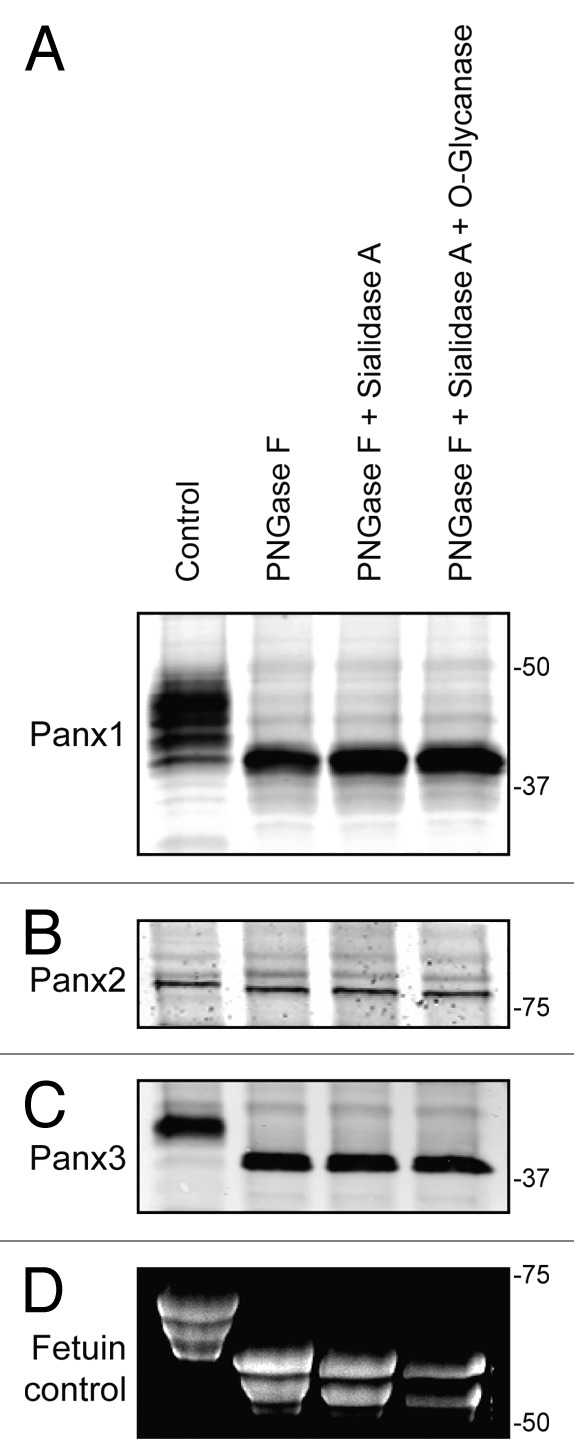
Figure 1. Pannexins are N-glycosylated but not O-glycosylated as defined by sensitivity to enzymatic digestion. Upon treatment of total protein lysates from HEK-293T cells ectopically expressing Panx1, 2, or 3 with PNGase F, a significant gel mobility shift in western blots was observed for Panx1 (A) and Panx3 (C) with a moderate change in mobility for Panx2 (B), indicating that all 3 pannexins are N-glycosylated. Sequential treatments with sialidase A and O-glycanase resulted in a change in the banding pattern of Fetuin in a Sypro-stained gel, used as a positive control (D), but did not generate visible banding shifts in any of the pannexin labeled blots (A, B, and C). The position of molecular weight standards are shown in kDa.
Caspase cleavage
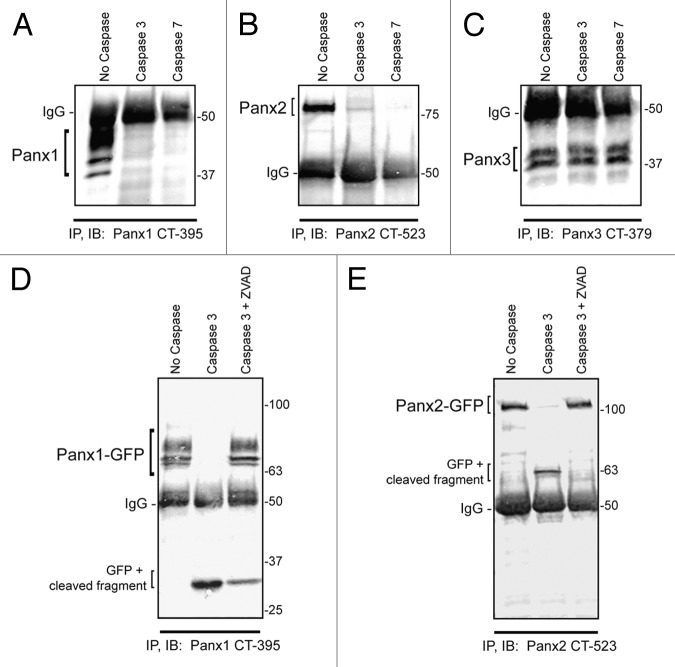
Human PANX1 has been shown to be a substrate for caspase-dependent cleavage in its C-terminal domain promoting ATP release and apoptotic cell clearance in immune cells.16 To determine whether the murine isoform of Panx1 (mPanx1) can be modified in the same manner, we utilized an in vitro cell-free caspase assay which demonstrated that mPanx1 is also susceptible to caspase 3 and caspase 7 cleavage. In these assays, ectopically-expressed Panx1 was immobilized on beads by immunoprecipitation with an antibody directed against the C-terminus, and subjected to caspase digestion. Following digestion, Panx1 was eluted from the beads and processed for immunoblotting for detection of modified Panx1. The loss in immunoreactivity to the Panx1 C-terminal antibody indicates cleavage of this epitope from the pannexin polypeptide. Only a small fragment (~5 kDa) of the protein C-terminus remained attached to the beads after caspase cleavage rendering it difficult to detect at the bottom of the gel after elution (Fig. 2A). We next tested whether the other 2 pannexins could be modified by caspase cleavage and found that Panx2, but not Panx3, is also a substrate for caspase 3 and caspase 7-dependent digestion (Fig. 2B, C). To better visualize the small C-terminal fragments left attached to the beads after caspase digestion, we utilized C-terminus GFP-tagged versions of Panx1 and 2 in the caspase 3 assay, which revealed C-terminal fragments for Panx1 and Panx2 following caspase digestion that were readily detected by immunoblotting (Fig. 2D and E). The predicted caspase-cleavage site in the Panx1 C-terminal resides at residue 378 (sequence DIID, according to CASVM server; http://www.casbase.org) and caspase cleavage of the GFP-tagged version of this protein yields a fragment of approximately 32 kDa. This fragment size is consistent with the predicted caspase cleavage site at amino acid 378, which is ~4.8 kDa from the end of the Panx1 polypeptide plus the molecular weight of GFP (26.9 kDa of GFP + ~5 kDa of mouse Panx1 C-terminus). Similarly, Panx2-GFP cleavage results in a ~63 kDa fragment (26.9 kDa of GFP + ~36 kDa of Panx2 C-terminus), indicating a caspase 3 cleavage site in the first part of the C-terminus of the 677 amino acid long Panx2 polypeptide. To verify that the absence of protein bands in these assays was not the result of non-specific protein degradation, addition of the pan-caspase inhibitor Z-VAD prevented caspase-dependent cleavage of both Panx1 and Panx2. (Fig. 2D and E). In summary, we report for the first time that Panx2, similar to Panx1, is a novel target for caspase-dependent modification.
Figure 2. Panx1 and Panx2 are substrates for caspase cleavage. In vitro caspase 3 and caspase 7 cleavage of Panx1, -2, and -3, ectopically expressed in HEK-293T cells. Panx-CT antibodies were used for pull down (IP) of all 3 pannexins, treated with: buffer only, exogenous caspase 3 or 7 (500 nM), or both caspase 3 (500 nM) and the pan-caspase inhibitor Z-VAD-OMe-FMK (50 µM). After immunoblotting (IB) with Panx-CT antibodies, the absence of protein bands indicated caspase cleavage of Panx1 (A) and Panx2 (B) as compared with controls. The presence of Panx3 protein bands despite the addition of activated exogenous caspase suggested that Panx3 is not a substrate for caspase-dependent cleavage in vitro. The IgG (50 kDa) antibody heavy chain is found in all 3 blots as expected. Cleaved protein bands detected at ~32 kDa for Panx1-GFP (D) and ~63 kDa for Panx2-GFP (E) suggested that the caspase cleavage sites are located in the C-terminal domains of both proteins. Cleavage is inhibited in the presence of the pan-caspase inhibitor Z-VAD. Protein sizes are noted in kDa.
S-Nitrosylation
While S-nitrosylation of Panx1 has previously been reported25 and shown to negatively regulate the activity of the channel, the ability of Panx2 and Panx3 to be modified in this manner has not been determined. To assess the potential for pannexins to be modified by S-nitrosylation, HEK-293T cells were engineered to express Panx1, Panx2, or Panx3 and subjected to the Biotin Switch assay. Following treatment of pannexin expressing cells with the NO donor molecule S-nitrosoglutathione (GSNO), S-nitrosylation of Panx1 was readily detected (Fig. 3A), in agreement with previous observations.25 In addition to Panx1, S-nitrosylation of Panx3 was also detected in response to GSNO treatment (Fig. 3C), with no observable NO-dependent modification of Panx2 (Fig. 3B). The ability of pannexins to be targeted for S-glutathionation, another cysteine post-translational modification, was assessed by treating pannexin-expressing cells with reduced glutathione (GSH), which did not result in cysteine modification. In addition, S-nitrosylation of Panx1 and Panx3 were reversible by reducing the modified cysteines with DTT. Taken together, these results indicate that nitric oxide can post-translationally modify both Panx1 and Panx3 by S-nitrosylation.
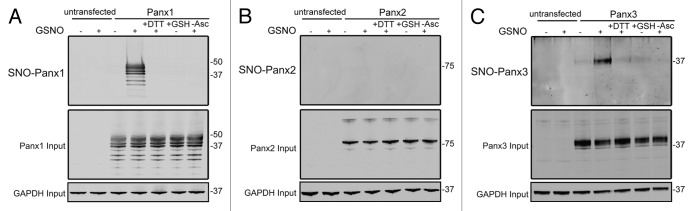
Figure 3. Panx1 and Panx3 are post-translationally modified by S-nitrosylation. Western blots for Panx1 (A), Panx2 (B), and Panx3 (C) following an assay for detection of S-nitrosylated proteins. HEK-293T cells ectopically expressing murine Panx1, Panx2, or Panx3 were treated with 100 µM GSNO to facilitate S-nitrosylation and samples were processed by the Biotin Switch assay. For controls, Panx expressing cells were stimulated with 100 µM reduced GSH, the backbone molecule in GSNO that lacks the capacity to release NO, or with the reducing agent DTT (1mM) following GSNO treatment to reduce S-nitrosylated protein thiols. As a negative control, ascorbate was omitted from the reducing step to prevent reduction of S-nitrosylated cysteines and subsequent biotinylation. GAPDH was used as input controls. Protein sizes are noted in kDa.
Discussion
Recent studies on the post-translational modifications of pannexins have focused on the most well characterized member, Panx1.27 With the notable exception of the S-palmitoylation of Panx2 in stem-like neural progenitor cells,28 our knowledge of the post-translational modifications of the other 2 pannexins is restricted to the evidence that they are N-linked glycoproteins. Protein glycosylation plays a crucial role in many key biological processes such as cell-cell interactions, adhesion, signal transduction, and recognition.29 Consequently, this and other post-translational modifications may be important for the regulation of pannexin channels as has been well documented for other ion channels and connexin channels modulated by phosphorylation, palmitoylation, S-nitrosylation, and SUMOylation.27 Most tissues express at least 2 pannexin isoforms that could, in some instances, functionally compensate for each other, particularly if they are differentially regulated by posttranslational modifications. Among the 3 pannexins, we have previously reported that sequence homology is highest between Panx1 and Panx3 (41% identical/59% conserved),6 particularly in the transmembrane domains of the 2 isoforms.30 Yet, N-glycosylation sites have been found in the second extracellular loop of Panx1 (N254,18,30) and in the first extracellular loop of Panx2 (predicted site at N86)6 and Panx3 (N71,30) indicating that these isoforms undergo distinct N-linked glycosylation targeted to very different polypeptide domains raising the possibility that this may have functional importance.
Panx1 and Panx3 are N-glycosylated to different levels going from an unglycosylated form (Gly0), to a high mannose modification (Gly1), and finally a complex glycosylated form (Gly2), while Panx2 seems to be present only in Gly0 and Gly1 forms.6 In our previous work, we used cell surface biotinylation of 293T cells ectopically expressing pannexins to detect the most prevalent glycosylation forms present at the surface of the cell.6 From those assays, it was apparent that the Gly2 forms (complex glycosylation) of Panx1 and Panx3, as well as a subpopulation of Panx2 (Gly1) are more abundant at the cell surface than their unglycosylated forms. However, as proof of principle in this overexpression system, a small subpopulation of the N-glycosylation deficient mutants (Gly0) of Panx1 and Panx3 were still able to traffic to the cell surface and form functional channels capable of dye uptake.6 Therefore, N-glycosylation may regulate cellular localization but does not seem to directly affect Panx1 or Panx3 single membrane channel function.
While all 3 pannexins are N-glycosylated, denaturing enzymatic assays with sequential addition of exoglycosidases to expose the core Galβ(1–3)GalNAc attached to a serine or threonine, followed by O-glycanase digestion, did not reveal any evidence for O-glycosylation. While O-linked deglycosylation did cause a band shift in the control fetuin used in this study, it is possible that pannexins have a minimal amount of O-linked glycosylation that fails to generate a band shift upon removal, while fetuin, containing 3 N-linked and 3 O-linked carbohydrate side chains significantly modifies its molecular weight.26 It is also possible that PNGase treatment and de-sialylation are not stringent enough to remove all the modifications of the core structure, which may block the action of the O-glycanase.31,32
Previously, Sandilos et al. reported a degree of sequence diversity between human PANX1 and murine Panx1 in a region of mismatched amino acids located between residues 371–391 in the predicted caspase cleavage site in the C-terminus.17 They also reported a constitutive basal current observed from mPanx1 channels which was significantly greater than the basal currents from the hPANX1 channel.17 However, generation of murine and human Panx1 constructs that harbor engineered Tobacco Etch Virus (TEV) protease sites replacing the endogenous caspase site revealed that both channels are capable of being activated by C-terminus cleavage.17 In the current study, we confirmed that murine Panx1 is indeed a substrate of both caspase 3 and caspase 7 with a predicted cleavage site in the C-terminus as reported for human PANX1.16 There is an additional predicted cleavage site in the intracellular loop of Panx1, but since our studies relied on immobilizing the proteins with the CT antibodies, it was not possible to detect other cleavage sites once the rest of the pannexin protein was cleaved off at the C-terminal site. Based on our analytical assay, the C-terminal domain of Panx3 does not appear to be a target for caspase digestion. However, since Panx3 has a predicted caspase site at amino acid 14 in the N-terminus (MLSD), it is possible that any cleavage that would remove only a small 1.4 kDa fragment may not be detected as a change in gel mobility using the C-terminus antibody for detection.
Importantly, murine Panx2 was shown for the first time to be a substrate for both effector caspases, and based on the size of the cleaved fragment, we predict that the caspase cleavage site resides in the first part of the carboxyl terminal domain. Based on the Panx2 sequence analysis on CASVM servers, this region has many predicted sites for caspase cleavage at residues 373 (NESD), 407 (QTVD), 416 (AEPD), and 479 (PLLD).
Although Panx2 tends to localize mostly to intracellular compartments,6 N-glycosylated Panx2 has the capacity to traffic to the cell surface, especially when co-expressed with Panx1.6 Swayne et al.28 also reported that while palmitoylated Panx2 is localized intra-cellularly in neural progenitor cells, the non-palmitoylated form is present at the cell surface of mature neurons. Bargiotas et al.33 reported the formation of Panx2 channels capable of dye release from calcein green-loaded cortical neurons even in the absence of Panx1. These authors also postulate that activation of pannexin channels during ischemic stroke may occur during apoptosis as a result of caspase 3 cleavage which is activated in cerebral ischemia.34 A double knockout (Panx1−/−, Panx2−/−) was required in that study to confer protection against ischemic brain damage resulting in reduced infarct size and better functional and neurological outcomes 24 h after ischemic stroke or 48 h after permanent occlusion of the distal middle cerebral artery.33,35 We hypothesize that Panx2 may play a similar function to that of Panx1, releasing ATP or other molecules during cell death, since both are potential substrates for caspase cleavage. However, it remains to be determined whether Panx2 channels can be activated by caspase cleavage in vivo, whether there is a role for caspase cleavage of Panx2 while a resident of the endoplasmic reticulum and the possible repercussions of Panx2 regulation on cellular physiology.33
The modification of cysteine thiols by nitric oxide has emerged as a key process in protein regulation and increasing evidence suggests that the pannexin family may serve as downstream targets for S-nitrosylation. Indeed, Panx1 was recently demonstrated to be modified by S-nitrosylation at 2 specific cysteine residues, resulting in functional inhibition of channel currents and ATP release into the extracellular milieu.25 Here we show that like Panx1, Panx3 can also be modified by S-nitrosylation, while Panx2 appears to be resistant to NO-mediated modification. Future studies will be instrumental in determining the specific functional impact of S-nitrosylation of Panx3 and the physiological context in which this modification may be relevant. Panx3 has been implicated in a number of cellular processes including its involvement in the regulation of keratinocyte and chondrocyte proliferation and differentiation, participation in Ca2+ leak from the endoplasmic reticulum and the promotion of bone development.5,8,9,36 The participation of NO signaling events in a number of these processes has also been well documented. For example, NO promotes bone and cartilage development by influencing osteoblast and chondrocyte differentiation, processes that are influenced by activation of Panx3 channels at the plasma membrane and ER.37,38 In this context, targeted S-nitrosylation of Panx3 in these cells may serve as an important regulatory element controlling cell growth and differentiation. In addition, keratinocytes are stimulated to proliferate in low NO while prompted to differentiate in high NO.39,40 In these same cells, Panx1 and Panx3 act to suppress proliferation and augment differentiation.5,41 Concurrently, the ability of Panx1 and Panx3 to be modified by S-nitrosylation may serve a role in regulating the activity or localization of these 2 pannexins during skin development and wound repair. These provocative observations may place the pannexin family at the heart of a number of NO-mediated responses, providing novel targets for clinical intervention.
In summary, our findings suggest that there is much diversity in the way all 3 pannexins are post-translationally modified, and these modifications may significantly modulate pannexin channel function and their roles in health and disease.
Materials and Methods
Ectopic expression
Transfections were completed as described by Penuela et al.6 Briefly, human embryonic kidney 293T (HEK-293T or 293T) cells were cultured in 100-mm petri dishes and 35-mm petri dishes. Cells were cultured in high-glucose Dulbecco’s modified eagle medium (DMEM), with 10% Fetal Bovine Serum (FBS), 2 mM L-glutamine, 100 units/ml penicillin G, and 100 μg/ml streptomycin. At 50–75% confluency, cells were transfected in Opti-MEM1 medium with Lipofectamine2000 (Invitrogen) and 5 µg of GFP tagged or untagged Panx1, 2, or 3 plasmid DNA for 4 h at 37 °C. Opti-MEM1 with Lipofectamine2000 was aspirated and replaced with culture media. After 48 h, proteins were extracted with Triton-based extraction buffer [1% Triton X-100, 150 mM NaCl, 10 mM Tris, 1 mM EDTA, 1 mM EGTA, 0.5% NP-40, 100 mM NaF, 100 mM Na3VO4, and ProteoGuard 1X EDTA-free proteinase inhibitor (Clonetech)]. Total protein concentrations were quantified by bicinchoninic acid (BCA) assay after protein extraction (Thermo Scientific Pierce BCA protein assay kit). Protein expression was assessed by western blot.
De-glycosylation
Total cell lysates were processed for sequential de-glycosylation using the “Enzymatic de-glycosylation kit” from ProZyme/Glyko following the manufacturer’s instructions. In brief, a denaturation protocol was performed using 100 µg of total protein heated in denaturation buffer solution for 5 min at 100 °C. PNGase, Sialidase A, and O-glycanase (1 µl of each enzyme) were added to different aliquots of total lysate in different combinations and incubated for 3 h at 37 °C, followed by western blot analysis (see below). Fetuin protein extract was used as positive control and visualized after enzymatic digestion using a Sypro-stained gel. Gels were scanned with a BIO-RAD UV scanner and images analyzed using QuantityOne software.
Western blot
Total protein was resolved by 8% sodium dodecyl sulfate PAGE (SDS-PAGE) and resolved proteins transferred to nitrocellulose membranes as described earlier.30 Membranes were blocked with 3% Bovine Serum Albumin (BSA) in 0.05% Tween-Phosphate Buffer Saline (T-PBS) for 45 min and probed overnight with affinity purified anti-Panx1, -Panx2, and -Panx3 carboxyl-terminal antibodies at 4 °C. Panx1-CT-395, and Panx3-CT-379 antibodies were previously reported by Penuela et al.6 Panx2-CT-523 antibody, from epitope GTKKAKTEAV PPALPASRS was custom-made by GeneMed Synthesis. Dilutions of antibodies in 3% BSA in 0.05% T-PBS were as follows: Panx1-CT (0.2 µg/ml), Panx2-CT (0.5 µg/ml), and Panx3-CT (0.2 µg/ml). After several washes with T-PBS, membranes were probed with anti-rabbit Alexa-Fluor-680 secondary antibody (1:10 000; Invitrogen) diluted in 3% BSA in 0.05% T-PBS for 45 min and scanned by an Odyssey imaging system (Li-Cor Biotechnology).
Immunoprecipitation
Equal amounts of protein lysates (500μg) were incubated overnight in 1X immunoprecipitation (IP) buffer [1% Triton X-100, 150 mM NaCl, 10 mM Tris, 1 mM EDTA, 1 mM EGTA, 0.5% NP-40,100 mM NaF, and 100 mM Na3VO4] at 4 °C with 5 µg of C-terminal (CT) anti-Panx1, -Panx2, and -Panx3 antibodies. Protein-antibody complexes were then incubated for 2 h at 4 °C with Protein A-agarose beads (Pierce) with gentle agitation. Beads were centrifuged at 4500 rpm for 2 min and washed 4 times with 1X IP buffer. Beads were then dried and resuspended in 2X Laemmli loading buffer, boiled for 5 min, and the eluates were processed by western blot as previously described.
Cell-free caspase assay
Caspase buffer was prepared as described by Stennicke et al.42 and caspase assays were conducted as described by Stennicke and Salvesen.43 Pannexin proteins were isolated by IP using C-terminus antibodies for all 3 pannexins as described above. Beads were washed twice with 500 μl of 1X IP buffer, and 2 times with 1X caspase assay buffer. The beads were resuspended in 250 μl of 2X caspase assay buffer containing 20 mM dithiothreitol (DTT) and preheated at 37 °C for 15 min to optimize enzymatic reactions. Activated caspase 3 and caspase 7 (500 nM) generated as described by Duncan et al.44 were incubated with the preheated mixture at 37 °C for 1 h. Samples were centrifuged (4500 rpm for 2 min) washed 2 times with 1X caspase assay buffer, mixed with 2X Laemmli loading buffer, boiled for 5 min, and western blotted. Blots were probed with C-terminal antibodies, washed, and probed with secondary antibodies to verify the presence of caspase cleavage as described above.
Cell-free exogenous caspase cleavage assay of GFP-tagged pannexin proteins with Z-VAD-OMe-FMK (Z-VAD) controls were done with proteins isolated by immunoprecipitation with Panx-CT antibodies and treated with buffer only, exogenous caspase 3 (500 nM), or both exogenous caspase 3 (500 nM) and pan-caspase inhibitor Z-VAD-OMe-FMK (50 µM, Millipore), and processed as described above.
Biotin switch assay
The biotin switch assay for assessment of pannexin S-nitrosylation was performed as described previously.25,29,45 In brief, HEK-293T cells were transfected with plasmids encoding murine Panx1, Panx2, or Panx3 using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s protocol. Following 48 h of culture, cells monolayers were treated with 100 µM S-nitrosoglutathione (GSNO) or vehicle for 10 min at 37 °C. Cells were then washed twice with 1x PBS and lysed in RIPA buffer containing protease inhibitors. Cell lysates were subjected to the Bradford assay for quantification of total protein. Three-hundred micrograms of total protein was precipitated for each sample by the addition of acetone and incubated at -20 °C for 30 min followed by centrifugation for 5 min at 10 000 x g to pellet precipitated proteins. Protein pellets were resuspended and non-modified cysteine residues were blocked with NEM for 20 min at 50 °C. Following an additional acetone precipitation to remove unreacted NEM, protein samples were treated with 1 mM ascorbate to reduce S-nitrosylated cysteine residues, and these reduced cysteines were biotinylated with EZ-link HPDP-Biotin (Thermo Scientific) for 1 h at room temperature. Biotinylated proteins were then precipitated by incubation with streptavidin-agarose beads (Sigma) for 1 h at room temperature and subjected to SDS PAGE and western blotting using primary antibodies against Panx1, Panx2, or Panx3.6 As a negative control, ascorbate was omitted from the assay, which prevents reduction of S-nitrosothiols and subsequent biotinylation. In some experiments, transfected cells were treated with GSNO followed by 1 mM DTT to reverse cysteine modifications.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors received funding from the Canadian Institutes of Health Research (CIHR) grant numbers MOP 37854 (Litchfield), MOP 130530 (Laird).
References
- 1.Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I, Shagin D, Nazarenko S, Geraymovych E, Litvin O, et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–16. doi: 10.1016/j.ygeno.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 2.Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol. 2000;10:R473–4. doi: 10.1016/S0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- 3.Seminario-Vidal L, Okada SF, Sesma JI, Kreda SM, van Heusden CA, Zhu Y, Jones LC, O’Neal WK, Penuela S, Laird DW, et al. Rho signaling regulates pannexin 1-mediated ATP release from airway epithelia. J Biol Chem. 2011;286:26277–86. doi: 10.1074/jbc.M111.260562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman W, Locovei S, Dahl G. Probenecid, a gout remedy, inhibits pannexin 1 channels. Am J Physiol Cell Physiol. 2008;295:C761–7. doi: 10.1152/ajpcell.00227.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celetti SJ, Cowan KN, Penuela S, Shao Q, Churko J, Laird DW. Implications of pannexin 1 and pannexin 3 for keratinocyte differentiation. J Cell Sci. 2010;123:1363–72. doi: 10.1242/jcs.056093. [DOI] [PubMed] [Google Scholar]
- 6.Penuela S, Bhalla R, Nag K, Laird DW. Glycosylation regulates pannexin intermixing and cellular localization. Mol Biol Cell. 2009;20:4313–23. doi: 10.1091/mbc.E09-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 2004;572:65–8. doi: 10.1016/j.febslet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto T, Nakamura T, Doyle A, Ishikawa M, de Vega S, Fukumoto S, Yamada Y. Pannexin 3 regulates intracellular ATP/cAMP levels and promotes chondrocyte differentiation. J Biol Chem. 2010;285:18948–58. doi: 10.1074/jbc.M110.127027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa M, Iwamoto T, Nakamura T, Doyle A, Fukumoto S, Yamada Y. Pannexin 3 functions as an ER Ca(2+) channel, hemichannel, and gap junction to promote osteoblast differentiation. J Cell Biol. 2011;193:1257–74. doi: 10.1083/jcb.201101050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanden Abeele F, Bidaux G, Gordienko D, Beck B, Panchin YV, Baranova AV, Ivanov DV, Skryma R, Prevarskaya N. Functional implications of calcium permeability of the channel formed by pannexin 1. J Cell Biol. 2006;174:535–46. doi: 10.1083/jcb.200601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penuela S, Gehi R, Laird DW. The biochemistry and function of pannexin channels. Biochim Biophys Acta. 2013;1828:15–22. doi: 10.1016/j.bbamem.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Billaud M, Lohman AW, Straub AC, Looft-Wilson R, Johnstone SR, Araj CA, Best AK, Chekeni FB, Ravichandran KS, Penuela S, et al. Pannexin1 regulates α1-adrenergic receptor- mediated vasoconstriction. Circ Res. 2011;109:80–5. doi: 10.1161/CIRCRESAHA.110.237594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson RJ, Zhou N, MacVicar BA. Ischemia opens neuronal gap junction hemichannels. Science. 2006;312:924–7. doi: 10.1126/science.1126241. [DOI] [PubMed] [Google Scholar]
- 14.Santiago MF, Veliskova J, Patel NK, Lutz SE, Caille D, Charollais A, Meda P, Scemes E. Targeting pannexin1 improves seizure outcome. PLoS One. 2011;6:e25178. doi: 10.1371/journal.pone.0025178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulbransen BD, Bashashati M, Hirota SA, Gui X, Roberts JA, MacDonald JA, Muruve DA, McKay DM, Beck PL, Mawe GM, et al. Activation of neuronal P2X7 receptor-pannexin-1 mediates death of enteric neurons during colitis. Nat Med. 2012;18:600–4. doi: 10.1038/nm.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, Armstrong AJ, Penuela S, Laird DW, Salvesen GS, et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467:863–7. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandilos JK, Chiu YH, Chekeni FB, Armstrong AJ, Walk SF, Ravichandran KS, Bayliss DA. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J Biol Chem. 2012;287:11303–11. doi: 10.1074/jbc.M111.323378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boassa D, Ambrosi C, Qiu F, Dahl G, Gaietta G, Sosinsky G. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J Biol Chem. 2007;282:31733–43. doi: 10.1074/jbc.M702422200. [DOI] [PubMed] [Google Scholar]
- 19.Boassa D, Qiu F, Dahl G, Sosinsky G. Trafficking dynamics of glycosylated pannexin 1 proteins. Cell Commun Adhes. 2008;15:119–32. doi: 10.1080/15419060802013885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Straub AC, Billaud M, Johnstone SR, Best AK, Yemen S, Dwyer ST, Looft-Wilson R, Lysiak JJ, Gaston B, Palmer L, et al. Compartmentalized connexin 43 s-nitrosylation/denitrosylation regulates heterocellular communication in the vessel wall. Arterioscler Thromb Vasc Biol. 2011;31:399–407. doi: 10.1161/ATVBAHA.110.215939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nogueira L, Figueiredo-Freitas C, Casimiro-Lopes G, Magdesian MH, Assreuy J, Sorenson MM. Myosin is reversibly inhibited by S-nitrosylation. Biochem J. 2009;424:221–31. doi: 10.1042/BJ20091144. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida T, Inoue R, Morii T, Takahashi N, Yamamoto S, Hara Y, Tominaga M, Shimizu S, Sato Y, Mori Y. Nitric oxide activates TRP channels by cysteine S-nitrosylation. Nat Chem Biol. 2006;2:596–607. doi: 10.1038/nchembio821. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci U S A. 2006;103:1295–300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O’Rourke B, et al. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–50. doi: 10.1016/S0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohman AW, Weaver JL, Billaud M, Sandilos JK, Griffiths R, Straub AC, Penuela S, Leitinger N, Laird DW, Bayliss DA, et al. S-nitrosylation inhibits pannexin 1 channel function. J Biol Chem. 2012;287:39602–12. doi: 10.1074/jbc.M112.397976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson WV, Heath EC. Evidence for posttranslational O-glycosylation of fetuin. Biochemistry. 1986;25:5518–25. doi: 10.1021/bi00367a026. [DOI] [PubMed] [Google Scholar]
- 27.Johnstone SR, Billaud M, Lohman AW, Taddeo EP, Isakson BE. Posttranslational modifications in connexins and pannexins. J Membr Biol. 2012;245:319–32. doi: 10.1007/s00232-012-9453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swayne LA, Sorbara CD, Bennett SA. Pannexin 2 is expressed by postnatal hippocampal neural progenitors and modulates neuronal commitment. J Biol Chem. 2010;285:24977–86. doi: 10.1074/jbc.M110.130054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Penuela S, Bhalla R, Gong XQ, Cowan KN, Celetti SJ, Cowan BJ, Bai D, Shao Q, Laird DW. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci. 2007;120:3772–83. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
- 31.Hård K, Damm JB, Spruijt MP, Bergwerff AA, Kamerling JP, Van Dedem GW, Vliegenthart JF. The carbohydrate chains of the beta subunit of human chorionic gonadotropin produced by the choriocarcinoma cell line BeWo. Novel O-linked and novel bisecting-GlcNAc-containing N-linked carbohydrates. Eur J Biochem. 1992;205:785–98. doi: 10.1111/j.1432-1033.1992.tb16843.x. [DOI] [PubMed] [Google Scholar]
- 32.Wilkins PP, McEver RP, Cummings RD. Structures of the O-glycans on P-selectin glycoprotein ligand-1 from HL-60 cells. J Biol Chem. 1996;271:18732–42. doi: 10.1074/jbc.271.31.18732. [DOI] [PubMed] [Google Scholar]
- 33.Bargiotas P, Krenz A, Hormuzdi SG, Ridder DA, Herb A, Barakat W, Penuela S, von Engelhardt J, Monyer H, Schwaninger M. Pannexins in ischemia-induced neurodegeneration. Proc Natl Acad Sci U S A. 2011;108:20772–7. doi: 10.1073/pnas.1018262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le DA, Wu Y, Huang Z, Matsushita K, Plesnila N, Augustinack JC, Hyman BT, Yuan J, Kuida K, Flavell RA, et al. Caspase activation and neuroprotection in caspase-3- deficient mice after in vivo cerebral ischemia and in vitro oxygen glucose deprivation. Proc Natl Acad Sci U S A. 2002;99:15188–93. doi: 10.1073/pnas.232473399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bargiotas P, Krenz A, Monyer H, Schwaninger M. Functional outcome of pannexin-deficient mice after cerebral ischemia. Channels (Austin) 2012;6:453–6. doi: 10.4161/chan.22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bond SR, Lau A, Penuela S, Sampaio AV, Underhill TM, Laird DW, Naus CC. Pannexin 3 is a novel target for Runx2, expressed by osteoblasts and mature growth plate chondrocytes. J Bone Miner Res. 2011;26:2911–22. doi: 10.1002/jbmr.509. [DOI] [PubMed] [Google Scholar]
- 37.Yan Q, Feng Q, Beier F. Reduced chondrocyte proliferation, earlier cell cycle exit and increased apoptosis in neuronal nitric oxide synthase-deficient mice. Osteoarthritis Cartilage. 2012;20:144–51. doi: 10.1016/j.joca.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Yan Q, Feng Q, Beier F. Endothelial nitric oxide synthase deficiency in mice results in reduced chondrocyte proliferation and endochondral bone growth. Arthritis Rheum. 2010;62:2013–22. doi: 10.1002/art.27486. [DOI] [PubMed] [Google Scholar]
- 39.Krischel V, Bruch-Gerharz D, Suschek C, Kröncke KD, Ruzicka T, Kolb-Bachofen V. Biphasic effect of exogenous nitric oxide on proliferation and differentiation in skin derived keratinocytes but not fibroblasts. J Invest Dermatol. 1998;111:286–91. doi: 10.1046/j.1523-1747.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- 40.Frank S, Kämpfer H, Podda M, Kaufmann R, Pfeilschifter J. Identification of copper/zinc superoxide dismutase as a nitric oxide-regulated gene in human (HaCaT) keratinocytes: implications for keratinocyte proliferation. Biochem J. 2000;346:719–28. doi: 10.1042/0264-6021:3460719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowan KN, Langlois S, Penuela S, Cowan BJ, Laird DW. Pannexin1 and Pannexin3 exhibit distinct localization patterns in human skin appendages and are regulated during keratinocyte differentiation and carcinogenesis. Cell Commun Adhes. 2012;19:45–53. doi: 10.3109/15419061.2012.712575. [DOI] [PubMed] [Google Scholar]
- 42.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–23. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 43.Stennicke HR, Salvesen GS. Caspase assays. Methods Enzymol. 2000;322:91–100. doi: 10.1016/S0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- 44.Duncan JS, Turowec JP, Duncan KE, Vilk G, Wu C, Lüscher B, Li SS, Gloor GB, Litchfield DW. A peptide-based target screen implicates the protein kinase CK2 in the global regulation of caspase signaling. Sci Signal. 2011;4:ra30. doi: 10.1126/scisignal.2001682. [DOI] [PubMed] [Google Scholar]
- 45.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–7. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]