Abstract
Cancer immune evasion is an emerging hallmark of disease progression. Functional studies to understand the role of myeloid-derived suppressor cells (MDSC) in the tumor microenvironment however, are limited by the lack of available specific cell surface markers. We adapted a competitive peptide phage display platform to identify candidate peptides binding MDSC specifically and generated peptide-Fc fusion proteins (peptibody). In multiple tumor models peptibody injection iv completely depleted blood, splenic, and intratumoral MDSC in tumor-bearing mice, without affecting proinflammatory immune cell types, such as dendritic cells. While control Gr-1 antibody depleted primarily granulocytic MDSC, peptibodies depleted both granulocytic and monocytic subsets. Remarkably, peptibody treatment was associated with inhibition of tumor growth in vivo, which was superior to Gr-1. Immunoprecipitation of MDSC membrane proteins identified S100 family proteins as candidate targets. Our strategy may be useful to identify novel diagnostic and therapeutic surface targets on rare cell subtypes, including human MDSC.
Activating the immune system has emerged as promising cancer treatments. Recent positive Phase III clinical trials of therapeutic cancer vaccines include FDA-approved Sipuleucel-T prostate cancer vaccine, melanoma peptide vaccines, and personalized lymphoma vaccines 1-3. However, tumor-induced immune suppression remains an obstacle limiting their potency. MDSC are heterogeneous cells that co-express Gr-1 and CD11b myeloid lineage differentiation markers 4-6. Functional studies showed that MDSC are potent inhibitors of T-cells in mice 7-10, but more specific surface markers that would allow isolation of viable cells would facilitate additional studies to precisely understand tumor-MDSC interactions in the microenvironment. New specific markers are also needed for targeting MDSC in vivo to test the hypothesis that MDSC inhibition enhances antitumor immunity 4,10.
RESULTS
Identification of mouse MDSC-binding peptides
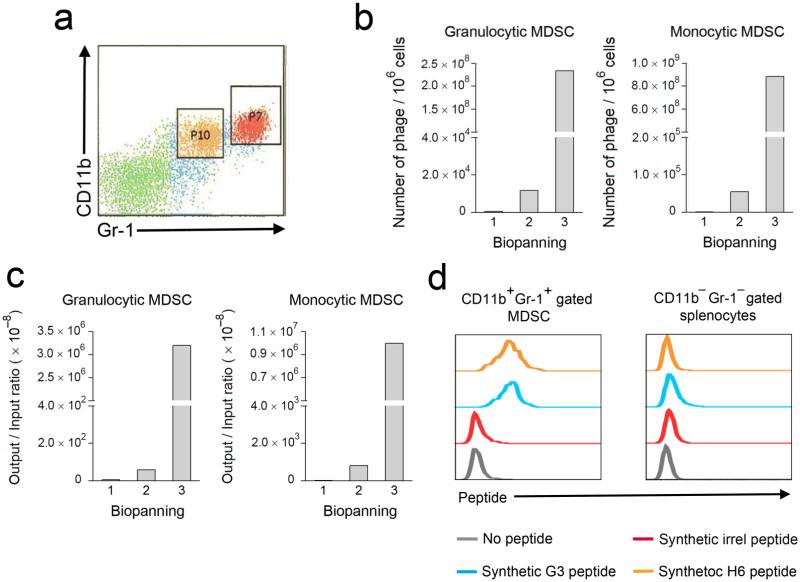
MDSC frequency is low in naïve C57BL/6 mice; however, after transplantation of syngeneic EL4 thymomas MDSC are increased accounting for approximately 10% of total splenocytes5. Splenic MDSC from EL4-bearing mice consist of two distinct subpopulations characterized by Gr-1highCD11b+ granulocytic (P7) and Gr-1intCD11b+ monocytic (P10) staining (Fig. 1a). With the goal of selecting specifically binding peptide ligands by phage display, we separated Gr-1 and CD11b labeled MDSC from non-MDSC by cell sorting after incubation with a Ph.D.-12 peptide phage library. Phage eluted from granulocytic or monocytic MDSC were then expanded by 3 rounds of competitive biopanning. We analyzed enrichment by the number of phage eluted from 106 MDSC (Fig. 1b) and by phage output, normalized to the initial input of 2 × 1010 phage (Fig. 1c).
Figure 1. Identification and characterization of MDSC-binding peptides.
(a) Identification of Gr-1+CD11b+ MDSC in spleens of C57BL/6 mice (n = 5) challenged subcutaneously with EL4 mouse lymphoma cells for 3 weeks. Double positive cells contain 2 distinct populations including Gr-1highCD11b+ granulocytic (P7) and Gr-1intCD11b+ monocytic (P10) MDSC subsets. (b–c) Biopanning with Ph.D.-12 peptide phage display library on Gr-1 and CD11b labeled splenocytes showed enriched phage eluted from sorted MDSC subsets. Biopanning enrichment was expressed in either “Number of plaques / 106 cells” or phage “Output / Input ratio” (× 10–8). (d) Binding of synthetic FITC-conjugated G3 and H6 peptides on Gr-1+CD11b+ gated MDSC from EL4-bearing C57BL/6 mice (n = 4), compared with Gr-1−CD11b− gated non-MDSC splenocytes. A non-specific peptide (irrel peptide) was used as a negative control to exclude non-specific binding. The data are representative of 3 identical experiments.
Sequencing of enriched phage revealed over-represented peptide sequences, and two predominant peptides (H6 and G3) were selected for further study (Table 1). Each clone that was expanded and tested for binding to MDSC revealed the specificity of both H6 and G3 phage for MDSC, without significant binding to non-MDSC (not shown). Additional candidate MDSC-binding phage were isolated, but were not given further consideration, because their binding was not specific. Corresponding synthetic FITC-conjugated H6 and G3 peptides bound specifically to Gr-1+ CD11b+ gated MDSC, but not Gr-1− CD11b− non-MDSC splenocytes from EL4-bearing mice (Fig. 1d).
Table 1.
Amino acid sequences of MDSC-binding peptides identified by phage display
| Granulocytic MDSC (66 coloniess) | Monocytic MDSC (48 colonies) | |||
|---|---|---|---|---|
| Peptide | Positive colonies | Percentage | Positive colonies | Percentage |
| MEWSLEKGYTIK (H6) | 24 | 36.36% | 25 | 52.08% |
| WGWSLSHGYQVK (G3) | 5 | 7.58% | 10 | 20.83% |
Generation of MDSC-specific peptibodies
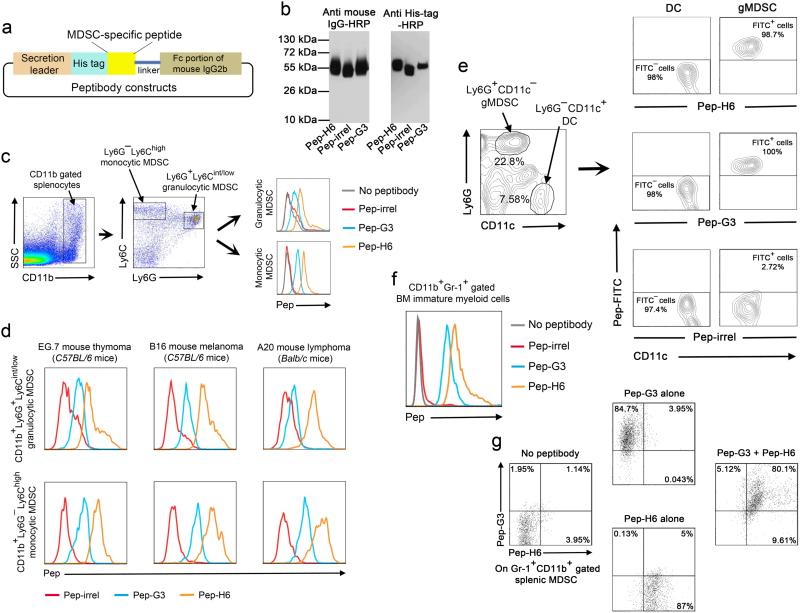
We genetically fused sequences encoding H6 and G3 peptides with the Fc portion of mouse IgG2b to generate peptibodies (Pep-H6 and -G3, respectively) (Fig. 2a). Control peptibodies (Pep-irrel) were also made using non-specific sequences. Then we produced recombinant peptibodies from 293T mammalian cells, followed by purification of the peptibodies by Protein A chromatography and characterization by Western blot using anti-mouse IgG and anti-His tag antibodies (Fig. 2b). These peptibodies were conjugated with fluorescein isothiocyanate (FITC) and analyzed for binding specificity on CD11b+Ly6G−Ly6Chigh monocytic MDSC and CD11b+Ly6G+Ly6Cint/low granulocytic MDSC from EL4-bearing mice. Gating on these well-defined subpopulations, we conclusively showed that Pep-H6 and Pep-G3 bind both monocytic and granulocytic MDSC (Fig. 2c), (Supplementary Fig.1a). However, the expression of the Pep-G3 target seems to be lower in granulocytic MDSC than in monocytic MDSC.
Figure 2. Generation and characterization of MDSC-specific peptibodies.
(a) Schematic representation of peptibody construction. (b) Characterization of recombinant peptibodies (Pep-H6, Pep-G3 and a control Pep-irrel) that were purified using Protein A chromatography. The identity of peptibodies was verified by Western blot using HRP-conjugated anti-mouse IgG (left) or anti-His tag antibodies (right). (c) Binding of FITC-conjugated Pep-H6 or Pep-G3 on CD11b+Ly6G+Ly6Cint/low gated granulocytic MDSC and CD11b+Ly6G−Ly6Chigh gated monocytic MDSC in splenocytes from EL4-bearing C57BL/6 mice (n = 5). (d) Binding of the peptibodies with granulocytic (upper panel) and monocytic (lower panel) MDSC subsets in splenocytes from different species of mice (C57BL/6 and Balb/c) challenged with various tumors (n = 3 for each tumor type). (e) Characterization of binding specificity of the peptibodies on Ly6G + CD11c − gated granulocytic MDSC (gMDSC) versus CD11c+Ly6G− gated DC in splenocytes pooled from EL4-bearing C57BL/6 mice (n = 5). (f) Identification of peptibody binding on CD11b+Gr-1+ immature myeloid cells in the bone marrow from EL4-bearing C57BL/6 mice (n = 5). (g) Co-staining of APC-labeled Pep-G3 and FITC-labeled Pep-H6 with CD11b+Gr-1+ MDSC in splenocytes pooled from EL4-bearing C57BL/6 mice (n = 5). The data represent 3 independent experiments.
In addition to EL4, peptibodies stained splenic MDSC from mice bearing EG.7 thymoma, B16 melanoma and A20 lymphoma (Fig. 2d), (Supplementary Fig. 1b). Interestingly, neither peptibody stained Ly6G−CD11c+ dendritic cells (DC), also of myeloid origin (Fig. 2e). Furthermore, peptibodies did not bind lymphocytes including B, T and NK cells (Pep-G3 bound a small proportion of T and NK cells) (Supplementary Fig. 2). However, we observed specific staining of Gr-1+ CD11b+ immature myeloid cells in bone marrow with peptibodies in EL4-bearing mice (Fig. 2f). When co-staining MDSC with both peptibodies, there was nearly complete overlap between Pep-H6 and -G3 binding populations (Fig. 2g).
Peptibodies deplete MDSC in vivo and retard tumor growth
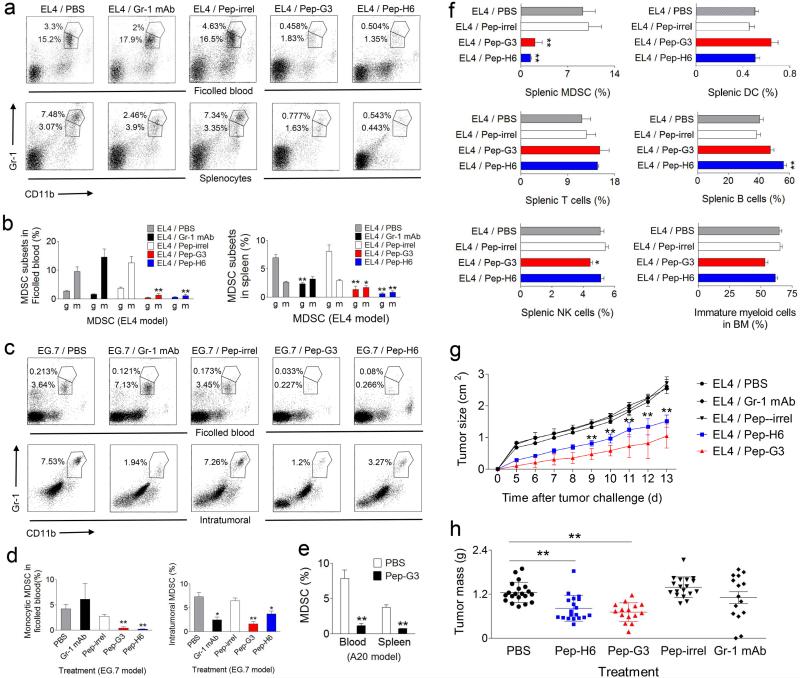
We next determined the effect of peptibody treatment on MDSC in vivo. Intravenous injection of 50 μg Pep-H6 and Pep-G3 completely depleted MDSC in blood and spleens of EL4 tumor-bearing mice, compared with control peptibody (Pep-irrel) or untreated mice (Fig. 3a,b). We found that whereas our peptibodies depleted both monocytic and granulocytic MDSC subsets, Gr-1 mAb depleted granulocytic, but not monocytic, MDSC in both EL4 and EG.7 models. This distinguishing effect of peptibody treatment on monocytic MDSC was especially apparent when blood was first subjected to Ficoll sedimentation to remove granulocytes and granulocytic MDSC (Fig. 3a-d). Importantly, we observed that peptibody treatment depleted intratumoral MDSC in both EG.7 (Fig. 3c,d) and EL4 models (Supplementary Fig. 3). Peptibodies also depleted blood and splenic MDSC in mice bearing A20 lymphomas (Fig. 3e).
Figure 3. Peptibodies specifically depleted tumor-induced MDSC in multiple tumor models and inhibited tumor growth in vivo.
(a–b) Depletion of Gr-1+CD11b+ MDSC in the blood and spleens in EL4-challenged C57BL/6 mice (n = 5 per group) after treatment with 50 μg peptibody i.v. for 3 consecutive days. Control mice received Gr-1 depleting mAb (positive control), irrelevant control peptibody (Pep-irrel) or PBS. Plots are shown for individual representative mice (a) and composite results (b) in a representative experiment out of five, with percentages of MDSC subsets indicated (g = granulocytic, m = monocytic) (mean ± s.e.m). (c–d) Depletion of MDSC in the blood and subcutaneous tumors of EG.7-challenged C57BL/6 mice (n = 5 per group) by peptibody treatment. Percentage of Gr-1+CD11b+ MDSC from ficolled blood and single cell suspensions prepared from harvested tumors is shown for individual representative mice (c) and composite results (mean ± s.e.m) (d) in a representative experiment out of two. (e) Peptibody treatment depleted MDSC in vivo from the blood and spleens of A20 lymphoma-challenged Balb/c mice (data pooled from 2 experiments). (f) Frequencies of Gr-1+CD11b+ MDSC, Ly6G−CD11c+ DC, CD3+ T cells, CD19+ B cells and CD3− CD49b+ NK cells in spleens and Gr-1+CD11b+ immature myeloid cells in the bone marrow from peptibody-treated, EL4-bearing C57BL/6 mice. Data are shown as the mean ± s.e.m of 5 mice per group. (g–h) Inhibition of EL4 tumor growth in C57BL/6 mice following every other day peptibody treatment. Tumor size is shown as the mean ± s.d of 5 mice per group in a representative experiment out of four (g). Tumor mass data are pooled results from 4 independent experiments (h). * P < 0.05, ** P < 0.01 compared with tumor-challenged mice without peptibody treatment (PBS) by two-tailed Student’s t-test.
Peptibodies specifically depleted MDSC in tumor-bearing mice without affecting other proinflammatory cells including DC and lymphocytes (T, B and NK cells), consistent with their lack of binding, although Pep-G3 treatment was associated with a minor reduction in NK cells (Fig. 3f). Interestingly, although staining of Gr-1+ CD11b+ immature myeloid cells from bone marrow was observed with peptibodies, peptibody treatment of tumor-bearing mice did not deplete myeloid precursor cells in this compartment either (Fig. 3f). Analysis of other blood cells subsets revealed associated depletion of elevated numbers of neutrophils in tumor-bearing, compared with naïve mice, but no effect on red blood cells or platelets (Supplementary Table 1).
Finally, to test the hypothesis that MDSC depletion can augment antitumor immunity, we administered peptibodies to EL4-bearing mice every other day to achieve a continuous depletion of systemic MDSC. Using this strategy, treatment with Pep-H6 or Pep-G3 alone significantly delayed tumor development, measured by tumor size (Fig. 3g) or tumor mass (Fig. 3h), compared with untreated or Pep-irrel-treated control mice. Importantly, while both peptibodies clearly inhibited tumor growth in vivo, Gr-1 mAb treatment was associated with less consistent inhibition of tumor growth. This effect of peptibody treatment correlated with MDSC depletion at the end of the two-week treatment (Supplementary Fig. 4).
Cell surface-bound alarmin is a candidate target
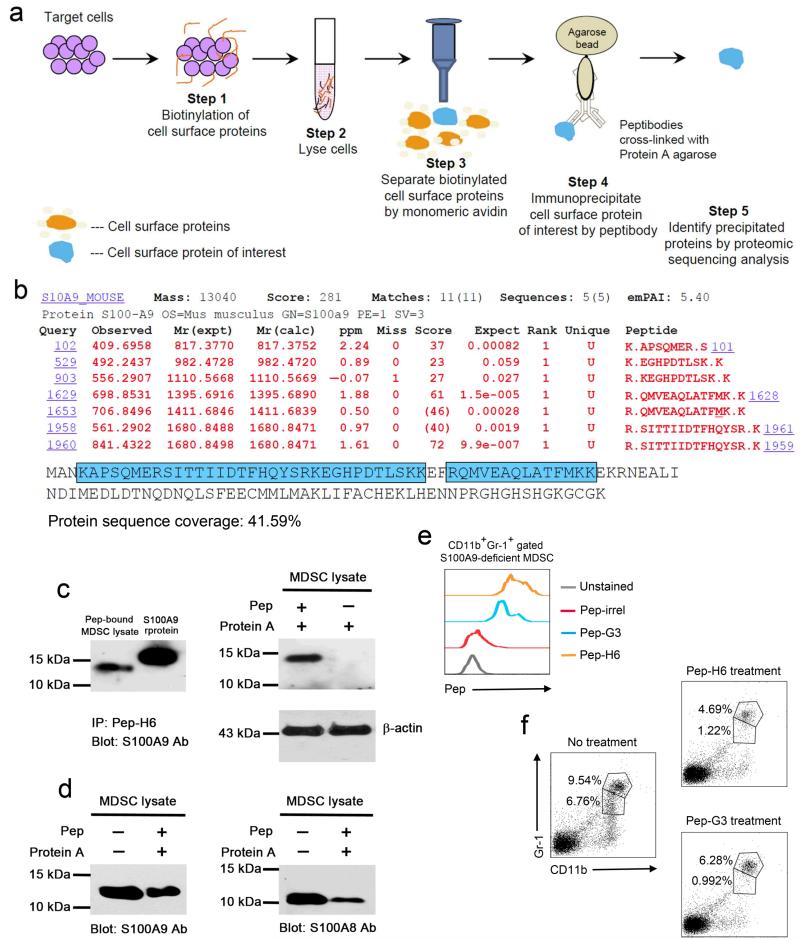
To explore the target of our peptibody, we developed a strategy to identify peptibody-bound proteins on the surface of MDSC (Fig. 4a). Specifically, surface proteins on sorted Gr-1+CD11b+ MDSC were biotinylated and subsequently captured by monomeric avidin after cell lysis. Then, we used Pep-H6 that was immobilized on Protein A to immunoprecipitate the surface protein of interest. Proteomic analysis of eluted proteins suggested that S100A9 was the source protein, with sequence coverage above 40% (Fig. 4b). Consistent with these results, immunoprecipitation studies (without prior biotinylation) showed that eluted proteins from Pep-H6-bound, sorted MDSC co-migrated with recombinant S100A9 (6 × His tagged) and was recognized by S100A9 antibodies (Fig. 4c), but no signal was detected when using lysates from MDSC without peptibody, excluding contamination by intracellular S100A9 (Fig. 4c). Furthermore, such direct immunoprecipitation of MDSC cell surface proteins with Pep-H6 revealed a protein band that more closely co-migrated with native S100A9 identified by immunoblotting of MDSC total cell lysates (Fig. 4d). Immunoprecipitation experiments suggested that the peptibody also recognizes S100A8, consistent with the formation of S100A9 and S100A8 dimers (Fig. 4d). Immunoprecipitation with Pep-G3 also suggested binding to S100A9 and S100A8 proteins (Supplementary Fig. 5). These results suggested either cross-reactivity with S100A8 or perhaps recognition of a combinatorial determinant on the S100A9 and S100A8 complex.
Figure 4. Peptibodies recognize extracellular S100 family proteins on the surface of MDSC.
(a) Schematic representation of a strategy for peptibody-based isolation of candidate cell type-specific surface markers. (b) Proteomic analysis from sorted Gr-1 + CD11b + splenic MDSC from EL4-bearing C57BL/6 mice revealing predominant peptides with homology to S100A9. The data are representative of 2 independent experiments. (c) Identification of S100A9 protein in Protein A eluates of Pep-H6-bound, sorted MDSC lysate (without biotinylation) by Western blot. Recombinant mouse S100A9 protein served as a positive control (left panel), and lysates from unbound MDSC were negative controls (right panel). Input lysates were blotted with actin as an internal control. (d) Detection of both S100A9 and S100A8 proteins in Protein A eluates of Pep-H6-bound, sorted MDSC lysate by Western blot. All Western blot data shown are representative of 3 individual experiments. (e) Binding of Pep-H6 and Pep-G3 peptibodies with CD11b+Gr-1+ gated splenic MDSC form EL4-bearing, S100A9-deficient C57BL/6 mice (n = 3). The data are representative of 2 independent experiments. (f) Frequencies of CD11b+Gr-1+ splenic MDSC from EL4-bearing, S100A9-deficient C57BL/6 mice (n = 3) after peptibody treatment as in (3a).
We then tested our peptibodies in S100A9-deficient mice and observed that peptibodies could bind MDSC (Fig. 4e) and also partially deplete MDSC in vivo (Fig. 4f). Taking these results together, the most likely explanation is that peptibodies are cross-reactive for S100A9 and S100A8. This explanation is consistent with the fact that S100A9-deficient mice express S100A811. Unfortunately, S100A8-deficient mice show an early embryonic lethal phenotype precluding further analysis for our study. Thus, further characterization of the specificity of our peptibodies for S100 family protein(s) must await the availability of S100A9 and conditional S100A8 double knockout mice.
DISCUSSION
We adapted a competitive peptide phage display platform to generate novel diagnostic and therapeutic MDSC-specific peptide-Fc fusion (peptibody) reagents. Peptibodies successfully depleted blood and splenic MDSC in multiple syngeneic tumor models, as well as intratumoral MDSC. Superiority of the peptibody therapeutic effects over an available Gr-1 mAb was suggested by their ability to deplete both granulocytic and monocytic MDSC subsets (Gr-1 depleted primarily granulocytic MDSC) and more consistent tumor inhibition in vivo.
To evaluate potential off-target activity of our specific peptibodies, we also assessed potential depletion of normal cell types, including those of myeloid origin. With the exception of transient reduction of blood neutrophils, and possibly monocytes, no depletion of DC, T, B, NK cells, or Gr1+CD11b+ immature myeloid precursor cells in the bone marrow was observed in tumor-bearing mice. We also observed that in naïve mice, peptibody treatment reduced circulating Gr-1+CD11b+ splenocytes (Supplementary Fig. 6a), but did not affect immature myeloid cells in the bone marrow (Supplementary Fig. 6b), suggesting that peptibody treatment may not have an effect on myeloid-lineage cells other than systemic (or intratumoral) MDSC. Consistent with this possibility, systemic MDSC numbers recovered 3 days after single dose treatment (not shown). The precise explanation for lack of depletion of myeloid cells in the bone marrow, despite binding by peptibody, is unclear. However, peptibodies were engineered to express mouse IgG2b Fc portion with the goal of enhancing antibody-dependent cell-mediated cytotoxicity (ADCC). One possible explanation is that the effector cells (mature NK and macrophages) required for ADCC are present in insufficient numbers in the bone marrow. Finally, peptibody-treated mice suffered no visible adverse effects or constitutional signs of cachexia or ruffled appearance, compared with control naïve or tumor-bearing mice, and had otherwise normal blood counts. Altogether, the data suggest limited off-target effects of peptibody treatment.
The identification of surface S100A9 and A8 as potential peptibody targets validates our protocol as a strategy for cell type-specific surface marker discovery. The S100 family of calcium binding proteins are intracellular molecules released to the extracellular milieu by myeloid cells in response to inflammation and function as proinflammatory danger signals (alarmins)12. Though there is limited evidence showing that soluble S100A9 binds with MDSC in vitro13, it is likely that MDSC secrete S100A9 in an autocrine feedback mechanism through receptors. The precise nature of the peptibody target remains to be elucidated. Although the epitope(s) recognized may be S100 protein-derived, because our protocol used viable cells to screen the peptide library, combinatorial native conformational epitopes comprised of the S100-receptor complex would have also been preserved. Given that our peptibody did not bind or deplete DC, it is tempting to speculate that the cell surface receptor for S100A9 and S100A8 on MDSC is as yet unidentified or different from those on DC, such as RAGE and TLR4. We also acknowledge that it is formally possible that other MDSC-specific targets exist that were not detected by our methods, as biotinylation might mask some epitopes.
Hypothetically, a MDSC depleting peptibody could cause tumor regression by (1) depleting MDSC by complement-dependent cytotoxicity or ADCC, (2) inducing apoptosis by blocking the binding of S100 family protein with its receptor(s) on MDSC or on tumor cells14, 15 and interfering with S100-induced survival signals, and (3) inducing direct cytotoxicity against tumor cells14,15. In order to rationally combine peptibody treatment to reverse immune suppression with other immunotherapy strategies, it will also be important to formally demonstrate that inhibition of tumor growth after MDSC depletion is immune-mediated. Future studies are needed to further investigate these possibilities. Finally, human MDSC targets are needed. Indeed, S100A9 was recently reported on human MDSC isolated from patients with colon cancer16. Additional studies are in progress, applying the technology platform described de novo to identify novel targets on human MDSC.
Online Methods
Phage display
Total splenocytes from EL4 tumor-bearing C57BL/6 mice were blocked with formaldehyde-inactivated M13 phage and 2.4G2 antibody (BD Biosciences, San Jose, CA), followed by staining with anti-CD11b-APC and anti-Gr-1-FITC (BD Biosciences). Gr-1 and CD11b labeled splenocytes were then incubated with 2×1010 Ph.D.-12 peptide phage display library (New England BioLabs Inc., Ipswich, MA) for 1 h at 4 °C. Gr-1highCD11b+ granulocytic and Gr-1intCD11b+ monocytic MDSC subsets were sorted separately and their bound phage was eluted, titered and amplified for the next round of biopanning. After the 3rd round of biopanning, predominant MDSC-binding peptides were identified by PCR analysis of 66 and 48 individual phage eluted from granulocytic and monocytic MDSC, respectively. The PCR product of each individual phage, generated using specific primers spanning encoded peptides, was sequenced.
MDSC specificity of synthetic peptide
FITC-conjugated H6 or G3 synthetic peptides were made by Pi Proteomics, LLC (Huntsville, AL). After blocking with 2.4G2 antibody, splenocytes from EL4-bearing C57BL/6 mice were co-stained with anti-CD11b-APC, anti-Gr-1-PE and FITC-conjugated peptides. H6 and G3 peptides were analyzed for their binding to Gr-1+CD11b+ gated MDSC, compared with Gr-1−CD11b− gated non-MDSC splenocytes. A non-specific peptide (irrel peptide) was used as a negative control to exclude non-specific binding. FACS analysis was performed using LSRFortessa cell analyzer (BD Biosciences) and results were analyzed using FlowJo software.
Generation and production of MDSC-specific peptibody
Synthetic, complementary double-stranded oligo nucleotides encoding H6 or G3 peptide were fused with a human IL-2 signal peptide and 6 × His tag and then cloned into EcoRI and BglII sites of pINFUSE-mIgG2b-Fc2 vector (InvivoGen, San Diego, CA). For initial characterization, we produced recombinant peptibodies by transfecting 293T human embryonic kidney cells with the plasmid constructs using Lipofectamine 2000 kit (Life Technologies, Grand Island, NY). Peptibodies excreted into growth media were purified using Protein A chromatography (GE Healthcare Life Sciences, Pittsburgh, PA). The identity of peptibodies was verified by Western blot using anti-His-HRP (BD Biosciences) or anti-mouse IgG-HRP antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) (Supplementary Table 1). Recombinant peptibodies used in all in vivo studies were produced by Aldevron (Madison, WI).
Animals used for in vivo studies
Six-week-old C57BL/6 and Balb/c female mice were purchased from US National Cancer Institute. S100A9-deficient female mice were a generous gift from Dr. Donna Kusewitt (M.D. Anderson Cancer Center, Smithville, TX) by permission of Dr. Johannes Roth (University of Muenster, Germany) who originally generated the model. Mice were maintained in a pathogen-free mouse facility according to institutional guidelines. All animal studies were approved by the Institutional Animal Care and Use Committee at MD Anderson Cancer Center.
FACS analysis of binding specificity of peptibodies
Recombinant peptibodies were conjugated with FITC or APC using FluoReporter Protein Labeling kits (Life Technologies). All other fluorophore-labeled monoclonal antibodies used for immune staining of cell type-specific markers including CD11b, Gr-1, Ly6C. Ly6G, CD3, CD19, CD49b and CD11c were purchased from BD Biosciences and BioLegend (San Diego, CA) (Supplementary Table 1). To determine the binding pattern of Pep-H6 and -G3, splenocytes from EL4-bearing C57BL/6 mice, EG.7-bearing C57BL/6 mice, B16-bearing C57BL/6 mice and A20-bearing Balb/c mice were co-stained with anti-CD11b-APC, anti-Ly6G-PE, anti-Ly6C-PerCP and FITC-conjugated peptibodies. Peptibody binding was analyzed on CD11b+Ly6G+Ly6Cint/low gated granulocytic MDSC and CD11b+Ly6G−Ly6Chigh gated monocytic MDSC, respectively. To further characterize the binding specificity of peptibodies, splenocytes from EL4-bearing C57BL/6 mice were co-stained with anti-CD11c-PerCP, anti-Ly6G-PE and peptibody-FITC. CD11c+Ly6G− DC were analyzed for peptibody binding. In a separate experiment, splenocytes from EL4-bearing C57BL/6 mice were co-stained for CD19-APC, CD3-PerCP, CD49b-PE and peptibody-FITC. CD3+ T cells, CD19+ B cells and CD3−CD49b+ NK cells were gated and analyzed for peptibody binding. Likewise, bone marrow cells from EL4-bearing C57BL/6 mice were isolated and co-stained with anti-CD11b-APC, anti-Gr-1-PE and FITC-conjugated peptibodies. CD11b+Gr-1+ immature myeloid cells were analyzed for their binding to peptibodies. Finally, to determine whether Pep-H6-bound population would be overlapped with Pep-G3-bound populations, splenocytes from EL4-bearing C57BL/6 mice were co-stained with anti-Gr-1-PE, anti-CD11b-PerCP, and either APC-labeled Pep-G3 or FITC-labeled Pep-H6, or both. CD11b+Gr-1+ gated MDSC were analyzed for peptibody binding.
In vivo MDSC depletion
Groups of 5 C57BL/6 mice were challenged s.c. with 106 EL4 or EG.7 mouse thymoma tumor cells on day 0. Tumor-bearing mice were treated i.v. with 50 μg of peptibodies per day for 3 consecutive days (days 17, 18 and 19), and then sacrificed on day 20 to harvest blood and spleens. Control mice received Gr-1 monoclonal antibodies (clone 1A8, BioXcell, West Lebanon, NH), irrelevant control peptibody (Pep-irrel) or PBS. Ficolled blood and splenocytes were stained for Gr-1-PE and CD11b-APC to identify MDSC. To prepare single cell suspensions from harvested tumors, EL4 or EG.7 subcutaneous tumors were cut into small pieces of 2–4 mm and digested with the enzyme mix (40 min at 37 °C) provided in “Tumor Dissociation Kit” (Miltenyi Biotec Inc., Auburn, CA). Tumors were dissociated into single cell suspension with a gentleMACS Dissociator (Milteny Biotec Inc.). The tumor cells were then stained for Gr-1-PE and CD11b-APC to identify intratumoral MDSC. To determine whether the in vivo depletion effect of peptibodies is MDSC-specific, splenocytes from peptibody-treated, EL4-bearing C57BL/6 mice were co-stained for CD11b-APC, CD11c-PerCp and Ly6G-PE and myeloid cells were gated based on forward and side scatter profile. Ly6G−CD11c+ DC were enumerated and their representative frequencies were calculated by multiplying the myeloid cell frequency in total splenocytes. Splenocytes were also stained for CD19-APC, CD3-PerCP and CD49b-PE, and frequencies of CD3+ T cells, CD19+ B cells and CD3−CD49b+ NK cells in total splenocytes were analyzed. Bone marrow cells were stained for CD11b-APC and Gr-1-PE. Double positive immature myeloid cells were enumerated.
In vivo therapeutic studies
Groups of 5 C57BL/6 mice were challenged with 106 EL4 tumor cells on Day 0. Twenty-four hours later, mice started to receive 50 μg of peptibodies per day, every other day for 2 weeks. Control mice received Gr-1 monoclonal antibodies (BioXcell), Pep-irrel or PBS. Tumor dimensions were measured daily with caliper to monitor growth. On day 14, mice were sacrificed and spleens and subcutaneous tumors were isolated. Tumor mass was measured on analytical balances. Splenocytes were stained for MDSC using anti-Gr-1-PE and anti-CD11b-APC antibodies.
Identification of targets for peptibodies on the surface of MDSC
Splenocytes from day 21 EL4-bearing C57BL/6 mice were immunolabeled for Gr-1-PE and CD11b-APC and double-positive MDSC were sorted. Cell surface proteins on sorted cells were biotinylated with EZ-Link Amine-PEG-Biotin kit (Thermo Scientific, Rockford, IL) and precipitated by monomeric avidin after cell lysis. A sequential 2nd immunoprecipitation was performed using Pep-H6 immobilized on Protein A-agarose. After washing away unbound proteins, the eluate was analyzed by proteomic sequencing in the Proteomic core at MD Anderson Cancer Center.
Immunoprecipitation and Western Blot
To confirm the results of proteomic analysis, total cell lysates prepared from Pep-H6-bound, sorted MDSC (without biotinylation) were loaded onto a protein A column. The eluate was separated by SDS-PAGE followed by immunoblotting with S100A9 antibodies (Abcam, Cambridge, MA) (Supplementary Table 1). Recombinant mouse S100A9 protein (rprotein) (Fitzgerald, Acton, MA) served as a positive control, and lysates from unbound MDSC were negative controls. Input lysates were blotted with actin as an internal control. Western blot was also performed on the Protein A eluates of Pep-H6-bound MDSC lysates or total cell lysates alone using anti-S100A8 antibodies (Abcam).
S100A9-deficient mouse studies
Groups of 3 S100A9-deficient C57BL/6 mice were challenged s.c. with 106 EL4 tumors on day 0. Three weeks later, splenocytes were harvested and co-stained with anti-CD11b-APC, anti-Gr-1-PE and peptibody-FITC. CD11b+Gr-1+ MDSC were analyzed for peptibody binding. In a separate experiment, EL4-bearing S100A9-deficient C57BL/6 mice were treated i.p. with 50 μg of peptibodies for 3 consecutive days (days 17, 18 and 19). Splenocytes were harvested on day 20 and stained for MDSC, as above.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a Developmental Research Program award to H. Qin from US National Cancer Institute Specialized Programs of Research Excellence in Lymphoma (P50 CA136411). We thank D. Hawke for performing proteomic sequencing and analyzing results, D. Kusewitt for generous assistance with S100A9-deficient mice and D. Gwak for the editing assistance.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Cheever MA, Higano CS. PROVENGE (Sipuleucel-T) in prostate cancer: the first FDA-approved therapeutic cancer vaccine. Clin Cancer Res. 2011;17:3520, 3526. doi: 10.1158/1078-0432.CCR-10-3126. [DOI] [PubMed] [Google Scholar]
- 2.Schwartzentruber DJ, et al. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med. 2011;364:2119–2127. doi: 10.1056/NEJMoa1012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster SJ, et al. Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma. J Clin Oncol. 2011;29:2787–2794. doi: 10.1200/JCO.2010.33.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–179. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- 5.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peranzoni E, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol. 2010;22:238–244. doi: 10.1016/j.coi.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 7.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez PC, et al. Regulation of T cell receptor CD3zeta chain expression by L-arginine. J Biol Chem. 2002;277:21123–21129. doi: 10.1074/jbc.M110675200. [DOI] [PubMed] [Google Scholar]
- 9.Ochoa AC, Zea AH, Hernandez C, Rodriguez PC. Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin Cancer Res. 2007;13:721s–726s. doi: 10.1158/1078-0432.CCR-06-2197. [DOI] [PubMed] [Google Scholar]
- 10.Ugel S, et al. Therapeutic targeting of myeloid-derived suppressor cells. Curr Opin Pharmacol. 2009;9:470–481. doi: 10.1016/j.coph.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Vogl T, et al. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood. 2004;104:4260–4268. doi: 10.1182/blood-2004-02-0446. [DOI] [PubMed] [Google Scholar]
- 12.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha P, et al. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9 doi: 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallberg E, et al. S100A9 interaction with TLR4 promotes tumor growth. PLoS One. 2012;7:e34207. doi: 10.1371/journal.pone.0034207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao F, et al. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology. 2012;136:176–183. doi: 10.1111/j.1365-2567.2012.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.