Abstract
Cardiomyocyte T-tubules are important for regulating ionic flux. Bridging Integrator 1 (BIN1) is a T-tubule protein associated with calcium channel trafficking that is down-regulated in failing hearts. Here we find that cardiac T-tubules normally contain dense protective inner membrane folds that are formed by a cardiac spliced isoform of BIN1. In mice with cardiac Bin1 deletion, T-tubule folding is decreased which does not change overall cardiomyocyte morphology, but frees diffusion of local extracellular calcium and potassium ions, prolonging action potential duration, and increasing susceptibility to ventricular arrhythmias. We also find that T-tubule inner folds are rescued only by the BIN1 isoform BIN1+13+17, which promotes N-WASP dependent actin polymerization to stabilize T-tubule membrane at cardiac Z-discs. In conclusion, BIN1+13+17 recruits actin to fold T-tubule membrane, creating a fuzzy space that protectively restricts ionic flux. When BIN1+13+17 is decreased, as occurs in acquired cardiomyopathy, T-tubule morphology is altered and arrhythmias can result.
Cardiac T-tubules are highly-branched invaginations of cardiomyocyte sarcolemma. T-tubules are primarily transverse to the cardiomyocyte long axis and wrap around sarcomeric Z-discs1. As an organelle involved in the initiation of calcium transients2, the T-tubule system helps determine the strength of each heartbeat by concentrating L-type calcium channels (LTCCs) and positioning them in close proximity with ryanodine receptors at the sarcoplasmic reticulum (SR)2–4. The lumina of T-tubules are continuous with the extracellular milieu which is calcium-rich. During each heartbeat, an action potential triggers extracellular calcium entry into the cell through LTCCs, increasing local intracellular calcium, activating nearby ryanodine receptors, and inducing large calcium release from intracellular SR stores, resulting in cellular contraction. Thus, T-tubules help regulate efficient beat-to-beat calcium flux.
There is growing appreciation that diffusion between the T-tubule lumen and bulk extracellular space is restricted5–8. Even though T-tubule lumina have an overall wide diameter of 20–450 nm1, they may only be accessible to ions and small nano-particles (≤11 nm)9. T-tubule diffusion coefficients for extracellular ions are ~95 µm2/s for calcium ions7, and ~85 µm2/s for potassium ions, which are five to ten times slower than in bulk extracellular space8. At fast heart rates, rapid transmembrane flux and limited diffusion can result in depleted T-tubule lumen calcium5,10 and elevation of T-tubule lumen potassium8, affecting the driving force for trans-membrane ion flux and decreasing action potential duration11.
The current understanding of T-tubule structures includes recognition of large branch points within the T-tubule lumen1, but does not explain highly-restricted diffusion. Furthermore, in failing hearts, T-tubule remodeling is notable for even larger yet fewer T-tubules12–14. Also, in failing hearts, action potentials are prolonged15 and intracellular calcium overload occurs16, resulting in dangerous arrhythmias16. Action potential duration and calcium handling are strongly influenced by T-tubule-associated currents, but without a better understanding of T-tubule anatomy, it remains difficult to clarify the impact of T-tubules on cardiac electrophysiology or determine the impact of altered T-tubules in disease.
Recent studies suggest that the membrane scaffolding protein Bridging Integrator 1 (BIN1) can be a regulator of T-tubule structure and function. BIN1, a member of the BAR domain containing protein superfamily, can induce LTCC-enriched membrane folds in cell lines and immature muscle cells17,18. In adult cardiomyocytes, BIN1 localizes to cardiac T-tubules and facilitates cytoskeleton-based calcium channel trafficking to T-tubule membrane18. The expression of BIN1 is transcriptionally decreased in acquired human and animal heart failure, which is also associated with both intracellular accumulation of LTCCs and abnormal T-tubule morphology12,13,19,20. A case of ventricular arrhythmias associated with BIN1 mutation has been reported21.
In the present study, the anatomy and function of cardiac T-tubules were studied in young adult mice with or without cardiac deletion of Bin1. With complementary in vivo and ex vivo studies, imaging, electrophysiology, biochemistry, and mathematical modeling, we find that an alternatively-spliced cardiac isoform of BIN1, BIN1+13+17, exists in mouse heart, promotes N-WASP-dependent actin polymerization and is responsible for generating actin-organized and densely-packed T-tubule membrane folds. The folds create a physical diffusion barrier to extracellular ions and protect against arrhythmias. Our finding elucidates how cardiac T-tubule ionic concentrations can differ from bulk extracellular ionic composition, and why the T-tubule diffusion barrier disappears in heart failure, increasing the likelihood of ventricular arrhythmias.
RESULTS
Cardiomyocyte T-tubule membrane is densely folded by BIN1
Homozygous mice with global Bin1 deletion suffer perinatal death due to cardiomyopathy22. To explore the role of BIN1 in cardiac T-tubule organization, we generated a cardiac-specific deletion of Bin1 using α-myosin heavy chain αMHC-Cre+23 and loxP-flanked Bin1 lines to generate heterozygous (Bin1 HT, Bin1flox/+; αMHC-Cre+) and homozygous (Bin1 HO, Bin1flox/flox; αMHC-Cre+) mice. Use of the αMHC-Cre bypasses embryonic lethality24. At 8–12 weeks, the wildtype (WT), Bin1 HT (reduction in BIN1 similar to that in heart failure19) and Bin1 HO animals have similar overall body and heart phenotypes (Supplementary Figs. 1 and 2), consistent with high cardiac reserve typical for young adult hearts24. Adult cardiomyocytes were isolated from the WT and Bin1 HT animals and labeled with a plasma membrane lipid dye Di-8-ANNEPs for live-cell imaging by spinning disk confocal microscopy (Fig. 1a–c and Supplementary Fig. 3). We found that the distribution and regularity of T-tubules are preserved in cardiomyocytes from Bin1 HT hearts. However, T-tubule fluorescence is decreased in the Bin1 HT cells, indicating less membrane along T-tubule invaginations. Similar results were obtained when membrane structures of freshly-fixed cardiomyocytes were labeled with wheat germ agglutinin (WGA, Supplementary Fig. 3b). Electrophysiological measurement of cardiomyocyte T-tubule capacitance, which is 30% of total cardiomyocyte capacitance25, is decreased by 46% in Bin1 HT (Fig. 1d) cells with normal cell size (Supplementary Fig. 3c), confirming decreased T-tubule membrane in the cardiomyocytes that are deficient in BIN1. The decrease in T-tubule membrane occurs despite preserved cellular content of T-tubule proteins (Supplementary Fig. 4a).
Figure 1.
Cardiomyocyte T-tubules are densely folded by BIN1. (α–b) Representative confocal images (a, scale bars: 5 µm) and the fluorescent profiles (b) of live WT and Bin1 HT cardiomyocytes labeled with Di-8-ANNEPS. (c) Quantification of T-tubules peak intensity. (n = 40 from 4–5 cells, P < 0.0001). (d) Cell size normalized membrane capacitance in WT (n = 14) and Bin1 HT (n = 12) cardiomyocytes (P = 0.0181). WC indicates reported whole cell capacitance without T-tubules. (e) 2D transmission electron microscope (TEM) images (Left to right: gross morphology, transverse cross section, and axial cross section) and 3D electron tomography images (right) of WT and Bin1 HT heart sections. Scale bars (left to right): 1 µm, 250 nm, 100 nm, and 100 nm. (f) Electron density profiles (middle) across individual T-tubules marked by the lines in the images above, with average T-tubule electron density in the bottom (n = 75, P < 0.0001). (g) T-tubule lumen area of axial cross sections (n = 80, P < 0.0001). (h) Cardiomyocyte T-tubule contour score (1, circular shape and no folds and spatial complexity; 2, non-circular shape and no folds and spatial complexity; or 3, multiple folds with spatial complexity) distribution (n = 196, P < 0.0001). Data are presented as mean ± SEM, cardiomyocytes are from three mice per genotype, and six left ventricular sections from three hearts per genotype were used for TEM analysis. Student’s t-test and one way-ANOVA were used for statistical analysis.
To directly visualize the impact of Bin1 deletion on T-tubule membrane in situ, we used transmission electron microscopy (TEM, Fig. 1e–h). Freshly-isolated whole mouse hearts were perfused with a calcium-containing fixative, and the left ventricular free wall was then sectioned and post-fixed with potassium ferricyanide, which enhances membrane contrast by forming calcium-dependent extracellular precipitates26,27. We found that the gross morphology of T-tubule invaginations is preserved in Bin1 HT and HO cardiomyocytes (Fig. 1e, left). In the high-magnification transverse and axial cross sectional images of T-tubules (Fig. 1e, quantification in 1f), we observed that, relative to WT T-tubules, Bin1 HT T-tubules consistently have fewer calcium-dense membrane folds. As revealed by 3D tomography (Fig. 1e, right panel), these membrane folds divide WT T-tubule lumen into multiple spatial subdomains, which are lost in the Bin1 HT and Bin1 HO T-tubules. As a result, Bin1 HT and Bin1 HO T-tubules contain one singular luminal space, which is enlarged in Bin1 HO cardiomyocytes (quantification in Fig. 1g). To numerically compare the degree of contoured T-tubules between WT and non-dilated Bin1 HT T-tubules, we applied a T-tubule contour score to each axial cross section image of T-tubule from multiple samples (three sections per heart and two hearts per genotype), assigning a score of 1 (round T-tubule lumen without folds and spatial complexity), 2 (non-circular contoured T-tubule lumen without folds and spatial complexity), or 3 (both round and non-circular T-tubules with multiple folds and spatial complexity). We found that T-tubules in Bin1 HT cardiomyocytes are dramatically less contoured and folded (Fig. 1h). Consistent with these findings, T-tubule topology by scanning ion conductance microscopy reveals more distinct T-tubule openings with large diameters in Bin1 HT cardiomyocytes, indicating that BIN1-folded membrane creates a “fuzzy space” inside T-tubules (Supplementary Fig. 3d).
BIN1 determines the T-tubule ion diffusion barrier
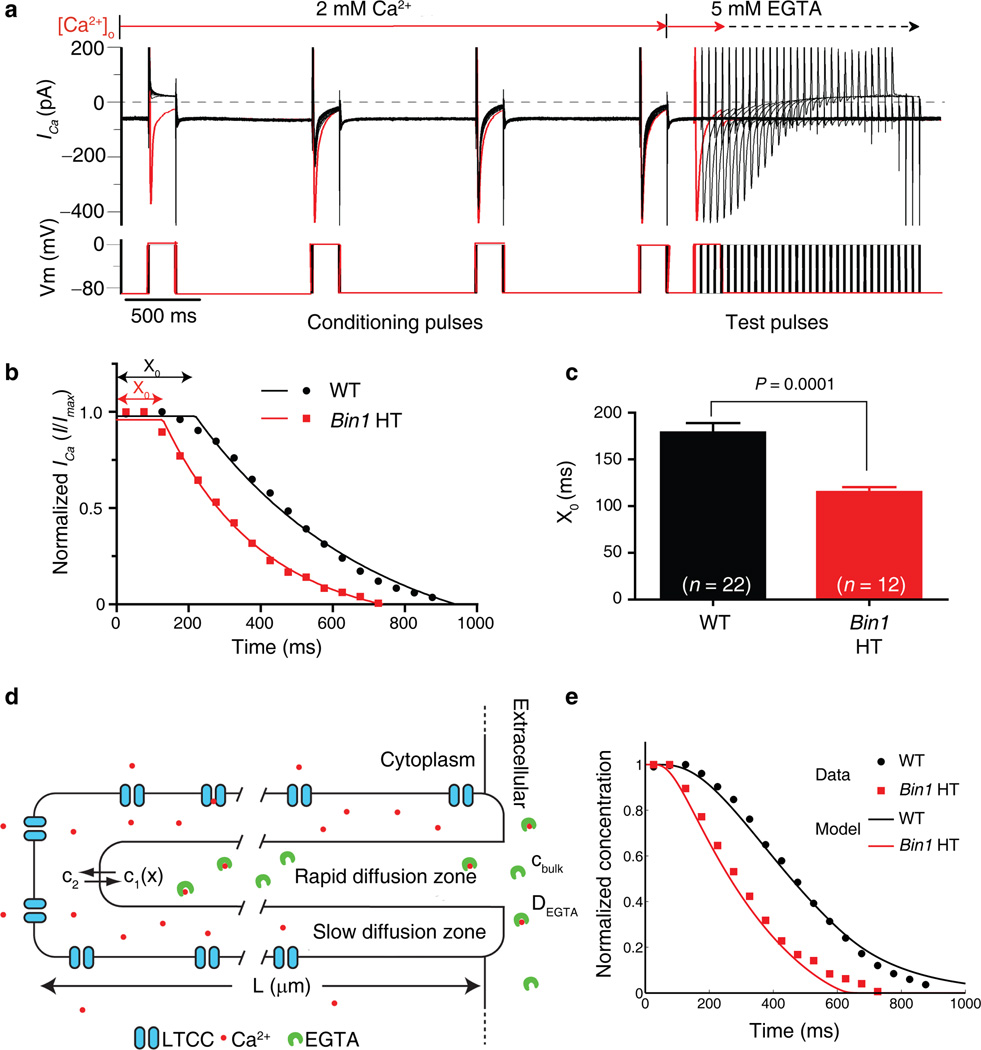
What is the function of T-tubule membrane folds? One possibility, suggested by the calcium density, is that the folds create an extracellular microenvironment distinct from the bulk extracellular space. We measured the dynamics of calcium ion diffusion into T-tubules by recording the ICa current through LTCCs while rapidly changing extracellular calcium. The baseline measurements revealed that Bin1 HT cardiomyocytes have similar total protein expression level of Cav1.2 (Supplementary Fig. 4a) and, of the membrane-inserted channels, preserved preferential localization of LTCCs to T-tubules. The Bin1 HT cardiomyocytes also have less total membrane LTCCs and less overall LTCC current (Supplementary Fig. 4b), consistent with our previous report that BIN1 helps LTCC forward trafficking18. Next, the activity of the surface LTCCs was studied by recording ICa decay kinetics following a rapid switch of extracellular perfusate from physiological calcium concentration to zero calcium (buffered with 5 mM EGTA; Fig. 2a). After an initial delay, ICa decayed exponentially until all extracellular calcium inside the T-tubules was removed or chelated (Fig. 2b). In Bin1 HT cardiomyocytes, the time length of the initial delay (X0) is shortened by 64 ms compared to that of WT cardiomyocytes (239 ± 5 ms vs. 303 ± 10 ms, Fig. 2c, P = 0.0001). To test whether shortened X0 could be due to increased ion diffusion within the T-tubules, a simple mathematical model of calcium diffusion was implemented containing a slow diffusion zone (T-tubule folds, Fig. 1eh), a rapid diffusion zone (T-tubule central lumen, tomography in Fig. 1e) and EGTA chelation (Fig. 2d). The theoretical decay curve generated from the model superimposes with the experimental data in WT cardiomyocytes. Removal of the diffusion barrier results in a shortened X0, as seen in the Bin1 HT cardiomyocytes (Fig. 2e). The autonomous mathematical algorithm that determined best-fit T-tubule diffusion and anatomical parameters yielded values that are remarkably consistent with those previously measured (Supplementary Table 1), providing computational support for a model of BIN1-induced restricted extracellular calcium diffusion.
Figure 2.
Bin1 deletion increases extracellular Ca2+ diffusion. (a) Representative patch clamp recording of the LTCC mediated ICa from a WT cardiomyocyte in response to quick change from 2 mM extracellular calcium solution to calcium free 5 mM EGTA solution. (b) Kinetics of ICa current changes using the protocol described in (a) were fitted with one plateau followed by one phase exponential decay. X0 is the initial delay before ICa decays. (c) Comparison of X0 for WT and Bin1 HT. Data are presented as mean ± SEM, P = 0.0001 by student’s t-test (cardiomyocytes are from 3 mice for each genotype). (d) A diagram describing the salient features of a mathematical model for calcium diffusion. (e) Kinetics of ICa current decay computed using the model in (d). The normalized calcium concentration in the slow diffusion zone serves as a surrogate for the calcium current since it is directly related to the inward Ca2+ driving force. The model of WT T-tubules containing a slow diffusion zone matches the experimental data (black curve – model, black circles – data). Removal of the diffusion barrier at the left side of the T-tubule in (a) results in a shorter initial delay as observed in the Bin1 HT experiments (red curve – model, red squares – data).
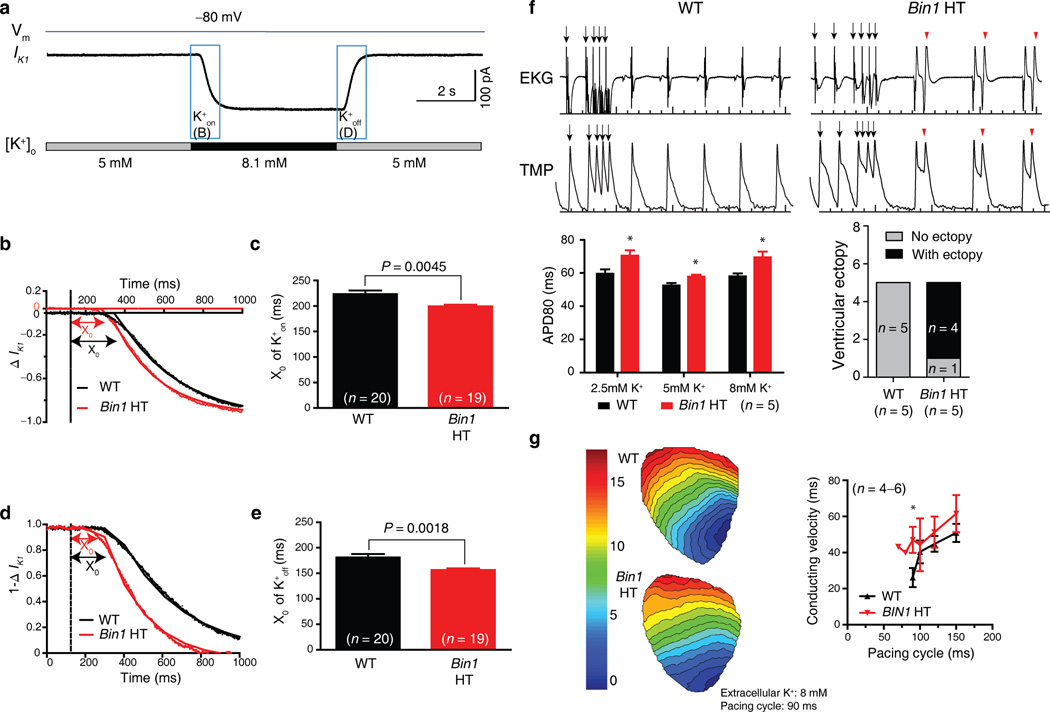
Next, we asked whether the physical barrier formed by BIN1-organized membrane folds applies to other ions. With regard to potassium ions, which can also be affected by slow diffusion8, we measured current from the inwardly rectifying potassium channel, IK1, which maintains the cardiomyocyte resting membrane potential. As indicated in Fig. 3a, steady-state IK1 and its kinetics were recorded following rapid wash-in and wash-out of K+ in T-tubules. Similar to our observations of calcium diffusion, decay of IK1 in response to an increase of extracellular potassium concentration from 5 mM to 8.1 mM (K+on) is significantly faster in Bin1 HT cardiomyocytes (Fig. 3b, c). The smaller difference of K+ diffusion (24 ms versus 64 ms for calcium) between WT and Bin1 HT likely reflects less T-tubule based enrichment of IK128,29. Similarly, IK1 response to decreases of extracellular potassium concentration from 8.1 mM back to 5 mM (K+off) is also significantly faster in Bin1 HT cardiomyocytes (Fig. 3d, e).
Figure 3.
Bin1 deletion increases extracellular K+ diffusion, prolonging action potential duration and increasing ventricular ectopy. (a) Representative patch clamp recording of IK1 current changes when quickly switching extracellular potassium concentration in a wildtype (WT) cardiomyocyte. (b) Kinetics of IK1 during K+on in WT and Bin1 HT cardiomyocytes (dotted line, dead volume time of 124 ms). (c) Comparison of the initial delay X0 of K+on for WT (n = 20) and Bin1 HT (n = 19) cardiomyocytes (P = 0.0045). (d) Kinetics of IK1 during K+off (1−∆IK1) in WT and Bin1 HT cardiomyocytes. (e) Comparison of X0 of K+off for WT (n = 20) and Bin1 HT (n = 19) cardiomyocytes (P = 0.0018). (f) Top: representative tracings of EKG (top) and TMP (transmembrane potential, bottom) from isolated and langendorff perfused WT (left) and Bin1 HT (right) hearts. Bottom: Action potential duration (APD80) is always prolonged in Bin1 HT hearts whether subjected to low (2.5 mM), normal (5 mM), and high (8 mM) potassium solution (left), and ventricular ectopy is increased in Bin1 HT hearts (right, incidence of arrhythmias during physiological buffer perfusion). (g) Ventricular activation map (left) and conduction velocity (right) of WT and Bin1 HT hearts subjected to high potassium (8 mM) perfusion (*, P < 0.05). Data are presented as mean ± SEM and cardiomyocytes are from three mice for each genotype, student’s t-test was used for statistical analysis.
A change in extracellular diffusion will affect the extracellular ion concentration, altering ion channel activity and electrophysiological parameters. We recorded electrocardiograms (EKGs) and performed whole-heart optical mapping on both WT and Bin1 HT hearts. In Bin1 HT hearts, action potentials are prolonged and ectopy from afterdepolarizations occurs both spontaneously and with induction (Fig. 3f). If limited diffusion includes an increase in extracellular potassium of the local fuzzy space, then in theory hyperkalemia will induce sodium channel block and conduction failure in WT animals before HT animals30. When the hearts were perfused with high-potassium solution, fast pacing induced conduction block more readily in WT than in HT hearts (Fig. 3g). These data support a difference in extracellular potassium accumulation in WT versus HT hearts.
Bin1 HT and HO mice have inducible ventricular arrhythmias
Given the effects of BIN1 reduction on ion homeostasis and membrane potential prolongation, we next studied in vivo arrhythmogenesis in WT, Bin1 HT and Bin1 HO mice subjected to ventricular pacing before and after intra-myocardial injection of isoproterenol. As is evident from the representative EKG recordings (Fig. 4a), sinus rhythm resumes immediately after ectopic pacing in WT mice (top panel). However, sustained monomorphic ventricular tachycardia (VT) ensues after pacing in Bin1 HT mice (middle panel), and more severe sustained polymorphic VT alternating with ventricular fibrillation (VF) occurs in Bin1 HO mice (bottom panel). The overall incidence of sustained VT (>9 consecutive wide-complex beats)31,32 or VF (Fig. 4c), as well as the frequency of pacing-induced arrhythmias (Fig. 4d), were significantly increased in Bin1 HT and HO mice. Bin1 deletion also dramatically increased isoproterenol-induced arrhythmias (Fig. 4d), which have been linked to LTCC-mediated afterdepolarizations33.
Figure 4.
Ventricular arrhythmias induced by pacing and beta adrenergic activation with isoproterenol. (a) Representative recordings of EKG following a S1–S4 stimulation protocol. Normal sinus node beats resume immediately following pacing in WT mice (top panel), sustained monomorphic ventricular tachycardia (4.5 s) was induced in Bin1 HT mice (middle panel), sustained polymorphic ventricular tachycardia (VT) alternating with ventricular fibrillation (VF) (>20s) was induced in Bin1 HO mice (bottom panel). (b) Heart rate increase (∆HR) in response to isoproterenol was analyzed and compared among the three groups (mean ± SEM, n = 3–4, P = 0.04 by one-way ANOVA). (c) Incidence of sustained VT (>9 QRS) or VF in each group (n = 3–4, P = 0.03 by chi-square). (d) The frequency of ventricular arrhythmias before and after isoproterenol treatment was quantified in each group (n = 3–4, P < 0.01 by two-way ANOVA).
BIN1+13+17 uses F-actin to maintain T-tubule folds at Z-discs
To identify the BIN1 isoform(s) responsible for T-tubule membrane folding, we explored cardiac splicing of Bin1, which is a gene encoded by 20 exons (Fig. 5a). Using PCR detection and sequencing of cloned fragments with primers flanking the alternatively spliced region between exon 10 (or exon 13) and 18 (Fig. 5b,c), we found that Bin1 message in adult mouse cardiomyocytes consists of ubiquitous Bin1 (36% of all clones) and Bin1+17 (8%), as well as the alternatively-spliced cardiac variants Bin1+13 (48%) and Bin1+13+17 (8%). Quantitative rt-PCR confirms a similar expression pattern and further identifies an increase of gene expression of all the Bin1 variants in mature adult mouse cardiomyocytes relative to neonatal cardiomyocytes (Fig. 5d). Next, we used exon 13- or 17-specific BIN1 antibodies (Fig. 5e) to localize the distribution of BIN1 isoforms in adult mouse cardiomyocytes. Both anti-BIN1–13 and anti-BIN1–17 antibodies localized to anti-α-actinin and anti-Cav1.2 identified Z-line/T-tubules regions (Fig. 5f), indicating that BIN1+13+17 resides at T-tubules.
Figure 5.
Adult mouse cardiomyocytes express four Bin1 splice variants. (a) Cartoon of Bin1 exons and the splice variants we found in adult mouse cardiomyocytes. BAR, Bin–Amphiphysin–Rvs domain; PI, phosphoinositide binding domain; CLAP, clathrin / AP2 binding region; MDB, myc-binding domain; SH3, SRC Homology 3 domain. (b) Four Bin1 splice variants with alternative inclusion of exon 13 and 17 are detected in adult mouse cardiomyocytes (A.M.C.) using PCR detection with primer sets flanking exon 10–18 or exon 13–18. (c) The percent of each Bin1 variants in adult mouse cardiomyocytes after subcloning and sequencing using PCR primer sets flanking exon 10–18. (d) Quantitative rtPCR analysis of each Bin1 variants (Bin1/HPRT1) in purified neonatal cardiomyocytes (P3, n = 2 litters with 8–10 pups each) and isolated adult mouse cardiomyocytes (n = 5 mice). (e) Western blot analysis confirms the antibody specificity of anti-exon 17 (clone 99D, Sigma) and anti-exon 13 (A#5299, Anaspec) BIN1 antibodies. All four BIN1 isoforms are detected by panBIN1 antibody (rabbit anti BIN1 SH3 domain). (f) Immunofluorescence of anti-exon 17 and anti-exon 13 labeling (red arrow, Z-line/TT region by α-actinin or Cav1.2 co-labeling) in adult mouse cardiomyocytes. (g) Representative confocal images (left, scale bars: 5 µm) and fluorescent profiles (right) of Di-8-ANNEPS membrane labeling in WT and Bin1 HT cardiomyocytes over-expressing GFP, BIN1, BIN1+13, BIN1+17, or BIN1+13+17 (n = 5 cells). Data are presented as mean +/− SEM. *, P < 0.05; **, P < 0.01, and ***, P < 0.001 by student’s t-test or two-way ANOVA.
The localization of BIN1+13+17 to T-tubules indicates a role in organizing membrane folds. Imaging of T-tubules indicates that exogenous BIN1+13+17 is the only isoform capable of fully rescuing T-tubule membrane intensity in Bin1 HT cardiomyocytes (Fig. 5g and Supplementary Fig. 5a). Conversely, in WT adult mouse cardiomyocytes, Bin1 knockdown using lentivirus-mediated shRNA targeting constitutive exon 2 or exon 13 decreases T-tubule membrane intensity, in proportion to BIN1 protein reduction (Supplementary Fig. 5b).
To understand how BIN1+13+17 organizes T-tubule membrane, we expressed GFP-tagged BIN1 isoforms in HeLa cells. Both confocal and TEM (Fig. 6a–c) imaging identified that only BIN1+13+17 induces formation of elongated, F-actin associated, membrane folds (Fig. 6d), indicating a role of actin in BIN1+13+17-mediated fold formation. Cytochalasin D induced stabilization of the barbed ends of F-actin at Z-discs is known to help preserve T-tubule structure of cultured cardiomyocytes34. We found that actin stabilization by cytochalasin D increases, whereas actin disruption by latrunculin A decreases, cardiac T-tubule membrane intensity (Supplementary Fig. 6). To further explore the differential roles of BIN1 isoforms in organizing actin, we studied their interaction with the actin polymerizing protein N-WASP, a known binding partner of the BAR domain protein Amphiphysin 135. Interestingly, although BIN1+13 does not bind to N-WASP (Fig. 6e), BIN1+13+17 does and can activate N-WASP to promote Arp2/3 nucleated actin polymerization (Fig. 6f). Biochemical GST pull-down confirms that BIN1+13+17 is associated with F-actin and α-actinin in adult mouse hearts (Fig. 6g). These data indicate that BIN1+13+17 binds to and activates N-WASP function to form elongated F-actin polymers for the development of membrane folds, and binds to Z-disc α-actinin to maintain these membrane folds, creating an ionic diffusion barrier within cardiac T-tubules (cartoon in Fig. 6h).
Figure 6.
BIN1+13+17 uses F-actin to connect to Z-disc α-actinin. (a–b) HeLa cells expressing GFP tagged BIN1, BIN1+13, BIN1+17, and BIN1+13+17 (scale bars: 10 µm) (a), with the length of folds like structure (linear streaks) quantified in (b). (Mean ± SEM; n = 20 folds from 5 cells; *** indicates P < 0.001 by one-way ANOVA). (c) TEM confirms that BIN1+13+17 but not BIN1+17 induces elongated membrane folds in HeLa cells. Scale bars: 1 µm (left) and 0.5 µm (right two panels). (d) HeLa cells expressing isoforms of GFP-BIN1 (green) and LifeAct-mCherry (red) (scale bars: 10 µm). (e) GST pulldown of GST-BIN1 isoforms and N-WASP-V5 in HeLa cells. (f) In vitro pyrene-actin polymerization assay using purified Arp2/3, N-WASP and BIN1 isoforms. Left, representative tracing of actin polymerization kinetics. Right, the Vmax data of polymerization kinetics. Data are presented as mean ± SEM (n = 5, * indicates P < 0.05 by one-way ANOVA). The negative control contains pyrene-actin alone with a GST control protein (GST-GFP, bottom black line indicated by the bottom arrow), the positive control contains pyrene-actin supplemented with Arp2/3 and VCA (active domain of N-WASP, top black line indicate by the top arrow), and the rest samples contain pyrene-actin supplemented with Arp2/3, N-WASP with GST-GFP or 1 µM GST-BIN1 isoforms. (g) Purified GST-BIN1 fusion protein pre-coated glutathione beads were added to adult heart lysates for pulldowns of α-actinin (right) or F-actin (left). (h) Schematic illustration of BIN1+13+17 forming an extracellular ionic diffusion barrier inside T-tubules.
DISCUSSION
Here we report that cardiac T-tubules are densely folded by a particular cardiac isoform of BIN1, resulting in local extracellular microenvironments that resist diffusion with bulk extracellular space with important consequences for membrane electrophysiology. The results expand the repertoire of BIN1 proteins to include a role for alternative splicing and cytoskeleton-based membrane localization to fold T-tubule membrane and regulate ionic flux. In conditions where cardiac BIN1 is transcriptionally decreased, such as heart failure19,20, the loss of a T-tubule fuzzy space in failing cardiomyocytes can increase arrhythmogenesis16,36. Our studies indicate that a loss of BIN1 increases arrhythmogenesis independent of the organ level structural changes that are associated with failing hearts.
Our results provide the first direct experimental evidence for the presence of a “slow diffusion zone”, or “fuzzy space” that has been proposed previously6, inside the T-tubule network. Future consideration of cardiomyocyte electrophysiology should take into account a separate slow diffusion zone at the inner face of T-tubules organized by membrane folds. The two zones separated by a resistant barrier have been predicted from computer models to change action potential duration11, and increase the safety of spatial propagation of cardiac action potentials37. Consistently, we found that removal of the BIN1-organized “fuzzy space” prolongs action potential duration and increases ventricular arrhythmias. Removal of the ion diffusion barrier also significantly increases cardiac sensitivity to β adrenergic receptor activation. Thus, by creating a physical barrier that impedes the access of particles9 such as catecholamine, the folded T-tubules serve as a protective mechanism to maintain cardiomyocyte homeostasis when extracellular bulk environment fluctuates. It remains unclear how the T-tubule folding affects efficient calcium-induced-calcium-release (CICR). The folds could in theory bring LTCCs closer to nearby ryanodine receptors. A recent study also suggests a direct conformational change of calcium binding activated LTCC, a mechanism in addition to local Ca2+ diffusion, may help cardiac CICR similar to the known mechanism of skeletal CICR38. Future studies of intracellular ion diffusion, and the relationship between channel density and the folds supporting the channels, in Bin1-depleted cardiomyocytes could enhance our understanding of calcium cycling.
Our study adds a cardiac-specific context to Bin1 splicing. The exon 1316 encoded clathrin binding domain targets neuronal BIN1 to clathrin-coated pits, facilitating endocytosis39 and neurotransmitter reuptake40. Exon 11 inclusion encodes a phosphoinositide binding domain that targets skeletal BIN1 to the plasma membrane for skeletal tubular biogenesis17. We found inclusion of exons 13 and 17 targets BIN1+13+17 to cardiac T-tubules for extensive membrane folding. This cardiac-specific, isoform-dependent function of BIN1 involves N-WASP-mediated actin polymerization, helping BIN1+13+17 to extend F-actin organized membrane folds. Through α-actinin binding, BIN1+13+17 stabilizes these folds at cardiac Z-discs. It may be that, in addition to skeletal tubulogenesis17, BIN1+11+17 has an analogous maintenance role in skeletal muscle41, but given the significant differences in T-tubule number, size, and location in skeletal versus cardiac muscle, careful studies are needed in each muscle subtype.
Other functions of the cardiac-expressing BIN1 isoforms, unrelated to T-tubule folding, are likely. Constitutive knockout of BIN1 causes embryonic cardiac hypertrophy and lethality22, occurring before postnatal T-tubule development. Our cardiac-specific Bin1 HO mice used the α-MHC promoter driven Cre which, in ventricular myocytes, activates in the perinatal stage. Furthermore, the most abundant cardiac isoform of BIN1 is BIN1+13, which elsewhere is associated with cell proliferation42. It is interesting that this isoform does not interact with actin but localizes to actin-free M-line regions. Given BIN1 has been implicated in roles as varied as endocytosis to transcription co-factor43,44, future studies will be helpful to define the spectrum of roles of different BIN1 isoforms in the heart.
In conclusion, we find that cardiac T-tubules are densely folded by a particular BIN1 isoform, resulting in retention of extracellular ions that are protective of heart function during stress by modulating transmembrane ion flux. We previously found that BIN1 anchors microtubules to deliver LTCCs to T-tubule membrane18. The present findings indicate that BIN1 can also create T-tubule folds to which the channels can be delivered. When BIN1 and other scaffolding proteins are reduced, such as in heart failure14,19,45, not only are potential membrane anchor points lost, the membrane fold itself is no longer present. Loss of the folds can help explain the increase in ventricular arrhythmias that occur in failing hearts46.
ONLINE METHODS
Plasmids, antibodies, and cell culture
Rabbit polyclonal antibody (A#5299, Anaspec) was raised against BIN1 peptide LRKGPPVPPPPKHTPSKEMKQEQI (epitope encoded by exon 13). We also used antibodies directed against the exon 17 encoded MYC-binding domain of BIN1 (99D, Sigma), pan-BIN1 rabbit antibody against SH3 domain (Abcam), mouse and rabbit anti α-actinin (Abcam), rabbit anti Cav1.2 (Alomone), mouse and rabbit anti V5 (Sigma), rabbit anti actin (Sigma), rabbit anti GST (Santa Cruz Inc.).
LifeAct-mCherry was described previously47. Bovine N-WASP was obtained from Addgene (Plasmid 33019: Pcs2-Bnwasp). HeLa and FT293 cells were cultured in DMEM under standard mammalian cell conditions. Lipofectamine 2000 (Invitrogen) was used for cDNA transfections in HeLa cells.
Cloning
Total RNA was extracted from isolated mouse cardiomyocytes and reverse transcribed into cDNA using iScript™ cDNA synthesis kit (BioRad). The resulting cDNA was used for PCR amplification of the middle section including alternatively spliced region using the following two primer sets: for exon 10–18 TTCACAGTCAAGGCCCAACC (Fwd) and ACCACCACAGCCGGAAGAGA (Rev); and for exon 13–18: TCCCCAAGTCCCCATCTCA (Fwd) and ACCACAGCCGGAAGAGAGCT (Rev). To further confirm the sequence, the first set of primers (exon 10–18) flanked with attB1/attB2 sites were used to PCR out the same regions for subsequent Gateway BP cloning into pDONR/Zeo (Invitrogen) to generate entry clones. A total of one hundred entry clones of the cardiac Bin1 middle section (exon 10–18, n = 100) were sequenced to identify a total of 4 distinct isoforms expressed in cardiomyocytes. The percent of clones of each variant was also analyzed and presented in Fig. 5c.
Meanwhile, to obtain Bin1 expression clones, full length Bin1 genes were amplified using primers flanked with attB1/attB2 sites. The PCR products were BP cloned into pDONR/Zeo construct to generate entry clones. The entry clones were sequenced to obtain all four cardiac BIN1 isoforms, which were subsequently inserted into pDest-eGFP-N1, pDest-mCherry-N1 (converted vectors originally from Clontech), pcDNA3.2-V5-Dest, pDest27-N-GST, and pAdeno-V5-Dest by Gateway LR cloning to generate final tagged expression clones and adenovirus.
qRT-PCR analysis of cardiac Bin1 variants in isolated adult and neonatal mouse cardiomyocytes
Adult mouse cardiomyocytes were isolated from 8 week old C57/B6 mouse hearts according to a previously established method48. The stepwise calcium reintroduction and slow centrifugation (40 g) followed by plating on laminin pre-coated dishes were used to generate a population of adult mouse cardiomyocytes with 90% as viable rod shape cells48. Following neonatal (P3) cardiomyocyte isolation18, we adopted an efficient non-genetic mitochondrial labeling approach using flow cytometry49 (BD Biosciences LSRII and FACSAriaII) to separate cardiomyocytes with the highest mitochondrial dye intensity, which were further confirmed with cardiac marker (Tnni3 and MF20) expression. Total RNA was then isolated from purified adult and neonatal cardiomyocytes using Picopure RNA extraction kit (Arcturus), and cDNA was synthesized using the BioRad iScript™ kit. To determine mRNA expression level of cardiac Bin1 variants, we designed exon junction-specific primers to quantify splice variant levels by qRT-PCR at both P3 neonatal and adult mouse cardiomyocytes. Custom Taqman exon-specific primers were generated to span exon junctions of interest (12 and 13, 17 and 18, 12 and 17, 12 and 18) using the Custom TaqMan® Assay Design Tool to ensure specificity without repeats or SNPs. Assay A uses exon12 and 13 junction to detect Bin1+13 and Bin1+13+17; Assay B uses exon 17 and 18 junction to detect both Bin1+17 and Bin1+13+17; Assay C uses exon 12 and 17 junction to detect only Bin1+17; and Assay D uses exon 12 and 18 junction to detect only the smallest Bin1 variant. The expression level of Bin1+13+17 is derived by Assay B-C, and the expression level of Bin1+13 is estimated by A-(B-C). The custom probe sequences are:
Bin1_12_13 TGACAATGCCCCTGAGAAAGGGAACAAGAGCCCGTCACCTCCTCCAGATGGCTCCCCTGCTGCTACCCCTGAGATCAGAGTGAACCATGAGCCAGAGCCGGCCAGTGGGGCCTCACCCGGGGCTACCATCCCCAAGTCCCCATCTCAGCTCCGGAAAGGCCCACCTGTCCCTCCGCCTCCCAAACACACCCCATCCAAGGAGATGAAGCAGGAGCAGATTCTCAGCCTTTTTGATGACGCATTTGTCCCTGAGATCAGCGTGACCACCCCCTCCCAG 94=ex11, 204=ex12
Bin1_12_17 TGACAATGCCCCTGAGAAAGGGAACAAGAGCCCGTCACCTCCTCCAGATGGCTCCCCTGCTGCTACCCCTGAGATCAGAGTGAACCATGAGCCAGAGCCGGCCAGTGGGGCCTCACCCGGGGCTACCATCCCCAAGTCCCCATCTCAGCCAGCAGAGGCCTCCGAGGTGGTGGGTGGAGCCCAGGAGCCAGGGGAGACAGCAGCCAGTGAAGCAACCTCC 94=ex11,190=ex16
Bin1_17_18 CCAGCAGAGGCCTCCGAGGTGGTGGGTGGAGCCCAGGAGCCAGGGGAGACAGCAGCCAGTGAAGCAACCTCCAGCTCTCTTCCGGCTGTGGTGGTGGAGACCTTCTCCGCAACTGTGAATGGGGCGGTGGAGGGCAGCGCTGGGACTGGACGCTTGGACCTGCCCCCGGGATTCATGTTCAAG 31=ex16, 141=ex17
Bin1_12_18 TGACAATGCCCCTGAGAAAGGGAACAAGAGCCCGTCACCTCCTCCAGATGGCTCCCCTGCTGCTACCCCTGAGATCAGAGTGAACCATGAGCCAGAGCCGGCCAGTGGGGCCTCACCCGGGGCTACCATCCCCAAGTCCCCATCTCAGAGCTCTCTTCCGGCTGTGGTGGTGGAGACCTTCTCCGCAACTGTGAATGGGGCGGTGGAGGGCAGCGCTGGGACTGGACGCTTGGACCTGCCCCCGGGATTCATGTTCAAG 94=ex11,203=ex17
Generation of the cardiac-specific Bin1 heterozygote mouse and cardiomyocytes isolation
All mouse procedures were reviewed and approved by the University of California and Cedars-Sinai Medical Center IACUC (Institutional Animal Care and Use Committee). Heterozygote Bin1flox (loxP sites around exon 3 of the Bin1 gene) mice were interbred with αMHC-Cre mice23 to generate cardiomyocyte specific Bin1 heterozygotes (Bin1 HT, Bin1flox/+; αMHC- Cre+) and homozygotes (Bin1 HO, Bin1flox/flox; αMHC-Cre+) (See detailed breeding strategies in Supplementary Fig.1). There are no differences in the other genotypes in the same litters (Bin1+/+; Cre−, Bin1+/+; Cre+, Bin1flox/+; Cre−, and Bin1flox/flox; Cre−) and hence data are pooled as wildtype (WT). Genotypes were confirmed by PCR to differentiate Bin1+, Bin1flox, and Cre+ alleles. To confirm the efficiency of Cre-mediated deletion of Bin1, quantitative real-time PCR using TaqMan primer/probe sets (5'FAM/3'BHQ; Applied Biosystems) spanning constitutively splice exons 2 and 3 (Mm01158690_mH) was used to measure transcription of total Bin1 gene, the house-keeping gene Hrpt1, and a cardiac specific gene TnI3 in cardiomyocyte cDNA generated from both WT and Bin1 HT mice. Quantitative real-time PCR reactions (qRT-PCR, 10 µl) were performed in a 384-well format using mix (Applied Biosystems) on an ABI 7900HT (Applied Biosystems).
Ventricular myocytes were isolated from 8–12 week old WT and Bin1 HT littermates after dissociation with collagenase II (2 mg ml−1, Worthington) using a previously described method48. Each cellular experiment represents cardiomyocytes isolated from three male and female (sex matched) mice per genotype.
Adenoviral and lentiviral infection of adult mouse cardiomyocytes
For rescue experiments, after 2 h plating, Bin1 HT cardiomyocytes were infected with GFP or BIN1 isoform overexpressing adenovirus (MOI 1000) overnight for live-cell membrane imaging with Di-8-ANNEPS or fixed in 4% PFA and labeled with Alexa647 conjugated WGA for fixed cell membrane imaging. For Bin1 knockdown experiments, cardiomyocytes were infected with (MOI 5) control, constitutive exon 2 BIN1 shRNA (5’–CCGGAGACGAAGGACGAGCAGTTTGCTCGAGCAAACTGCTCGTCCTTCGTCTTTTTTG - 3’), or exon 13 targeting BIN1 shRNA (5’ –CCGGTGACGCATTTGTCCCTGAGATCTCGAGATCTGCAGGGACAAATGCGTCATTTTTG - 3’) expressing lentivirus (Sigma) overnight. Post-viral infection, cardiomyocytes were cultured for 3 d before fixation for WGA labeled membrane intensity study. For actin experiments, fresh isolated cardiomyocytes were plated for 2 h, treated with 1 µM latrunculin A or 10 µM cytochalasin D overnight, and followed by PFA fixation and T-tubule labeling with Alexa647-conjugated WGA50.
Membrane fluorescence labeling, immunofluorescence and Imaging
Freshly isolated ventricular cardiomyocytes from both WT and Bin1 HT plasma membrane were incubated with 10 µM Di-8-ANNEPS for 20 min at room temperature. The cells were then washed with HBSS to remove the remaining dye and imaged with spinning disc confocal microscope. Di-8-ANEPPS labeled cells were excited with 488 nm laser and emitted fluorescence was collected through a 580 nm emission filter. For conjugated Wheat Germ Agglutinin (WGA) labeled membrane fluorescence, fresh isolated cardiomyocytes were fixed with 4% PFA at room temperature for 30 min and labeled with Alexa-647 conjugated WGA at 4°C overnight followed by mounting in ProLong gold and imaged using spinning disc confocal microscope.
For all immunofluorescence, cells were fixed in methanol at −20 °C for 5 min. After fixation, cardiomyocytes were permeablized and blocked with 0.5% Triton X-100 and 5% NGS in PBS for 1hr at room temperature. The cells were incubated with mouse anti BIN1 exon 17 (99D, 1:50, Sigma) and rabbit anti Cav1.2 (1:50, Alomone); or rabbit anti BIN1 exon 13 (1:50, Anaspec) and mouse anti α-actinin (1:500, Abcam) overnight at 4°C. Exogenous protein expression after adenoviral infection was confirmed by anti-V5 (mouse anti-V5, Sigma, 1:500) labeling of the tagged protein introduced by adenovirus. After several washes with PBS post primary antibody incubation, cells were then incubated with goat anti-mouse IgG conjugated to AlexaFluor 488 and goat anti-rabbit IgG conjugated to AlexaFluor 555. Cells were then fixed and mounted with DAPI containing ProLong gold.
All imaging was performed on a Nikon Eclipse Ti microscope with a 100 × 1.49 NA or 60 × 1.1 NA TIRF objectives and NIS Elements software. For BIN1 localization, isolated mouse cardiomyocytes were imaged at Z-depth increments of 0.5 µm. Deconvolution of images was performed using Autoquant software (Media Cybernetics). High-resolution cardiomyocyte images were obtained by using a spinning disc confocal unit (Yokogawa CSU10) with DPSS lasers (486, 561) generated from laser merge module 5 (Spectral applied research, CA) and captured by a high-resolution Cool SNAP HQ2 camera (Photometrics).
For live-cell imaging of BIN1-GFP and LifeAct-mCherry, HeLa cells were transfected with BIN1+17-GFP or BIN1+13+17-GFP, along with co-transfection of LifeAct-mCherry. Co-transfected cells were imaged in regular HBSS (Gibco) using a 100 × 1.49 NA TIRF objective with spinning disc confocal microscopy. Live images were captured by using a high-resolution Cool SNAP HQ2 camera (Photometrics).
Topography scans were obtained from freshly isolated adult cardiomyocytes using scanning ion conductance microscopy as described previously51.
Electron Microscopy
For transmission electron microscopy study of in situ ventricular cardiomyocyte membrane ultrastructure, whole mouse hearts were perfused with fixative (2% paraformaldehyde / 2.5% Glutaraldehyde in 0.1 M Sodium cacodylate buffer, and 50 mM CaCl2, pH 7.4) at 3 ml min−1 at room temperature for 15 min before being stored at 4 °C in fixative. T-tubules were labeled according to a previously established method27. Briefly, ultrathin heart tissue sections were cut by a Leica Ultracut S microtome and post-fixed in OsO4 (2% OsO4 + 0.8% K3Fe(CN)6 in 0.1 M cacodylate buffer (pH 7.4) (Sigma) and followed by uranyl acetate (EM Science) incubation before being dehydrated in ethanol and embedded in epon resin. Finally, tissue sections were imaged at 3,000×, 8,000× and 150,000× magnifications under a transmission electron microscope JEOL TEM1230 from the Electron Microscopy Core (EMC) of the Cell Sciences Imaging Facility (CSIF) at the Stanford University Medical Center. IMOD was used for three-dimensional reconstruction of TEM serial sections, and a two-axis tomography of T-tubules was generated according to a previously established method52,53.
Cardiomyocyte electrophysiology
Membrane capacitance and the macroscopic calcium (ICa) and the inward rectifying potassium channel mediated IK1 current were recorded by whole-cell voltage clamp of acutely isolated adult mouse cardiomyocytes. Patches were formed with borosilicate pipettes of 3–5 MΩ resistance. Data were acquired using an Axopatch 200-B patch-clamp amplifier and pClamp10 software (Molecular Devices). For membrane capacitance and ICa measurement, the internal solution contained (in mM) 130 CsCl, 10 NaCl, 10 tetraethylammonium chloride (TEA-Cl), 5 EGTA, 3 magnesium ATP and 10 HEPES, pH 7.2. For membrane capacitance measurement, the external solution contained (in mM) 140 NaCl, 5 CsCl, 2 CaCl2, 1 MgCl2, 10 glucose and 10 mM HEPES, pH 7.2. To estimate cell size, bright field images were taken before patch clamping through an Andor Clara DR-328G CCD camera in a Nikon TE2000U microscope. ImageJ software was used to measure the cell size. For ICa recording, the external solution contained (in mM) 140 TEA-Cl, 5 CsCl, 2 CaCl2, 1 MgCl2, 10 glucose and 10 HEPES, pH 7.2. To obtain the current-voltage relationship of ICa, test pulses were elicited from −90 mV to +50 mV with 10 mV increments. Both holding and repolarizing potentials were −90 mV.
To measure the diffusion barrier inside the cardiomyocyte T-tubules, ICa or IK1 was monitored while switching extracellular solutions with different calcium or potassium concentrations using a fast perfusion system (VC3-8xP pressured perfusion system, ALA Scientific Instruments). The protocol shown in Fig. 2a was used to measure calcium diffusion. If brief, the ICa was elicited by four conditioning pulses followed by a test pulse. Both conditioning and test pulses were from −80 to 0 mV for 200 ms. The delay between the conditioning pulses and the test pulse was gradually prolonged (with a step-wise 50 ms increment) during which the extracellular solution with 2 mM calcium was quickly switched to a solution with 5 mM EGTA. The change of ICa was normalized to the current of fourth conditioning pulse (IMax) and fitted with one plateau followed by one phase exponential decay. A dead time of 124 ms due to dead volume was subtracted before fitting.
The intracellular solution for measuring the inward rectifying potassium channel mediated IK1 current contains (in mM): 100 potassium gluconate, 30 KCl, 1 MgCl2, 1 EGTA, 3 sodium ATP, 10 Glucose, 10 HEPES, pH 7.3. The extracellular solution contains (in mM): 5 or 8.1 KCl, 140 NaCl, 1.8 CaCl2, 1 MgCl2, 0.2 CdCl2, 10 Glucose, 10 HEPES, pH 7.3. Kinetics of IK1 changes following a quick switch of extracellular potassium concentration from 5 mM to 8.1 mM was constantly recorded by holding the cardiomyocytes at −80 mV. The kinetics was fitted with one plateau followed by one phase exponential decay. The dead time of the perfusion system between solution switches was 124 ms (measured by monitoring changes of junction potential, data not shown), and was subtracted from the initial delay time shown in Fig. 3.
All the experiments were performed at room temperature (22–24°C). Series resistance was compensated at 60–75% and monitored during the experiment. Recordings were filtered at 2–5 KHz and digitized at 10 KHz (Digidata 1440A, Molecular Devices). Clampfit 10 (Molecular Devices), Origin 7.5 (OriginLab) and Prism (Graphpad software) were used for data analysis.
Mathematical modeling of calcium diffusion in T-tubules
A set of 1 dimensional partial differential equations (PDEs) was developed to model calcium dynamics in the T-tubule. There are three time-dependent variables: the calcium concentration in the rapid diffusion zone, c1(x,t); number of calcium ions in the slow diffusion zone, N2(t); and the concentration of EGTA in the fast diffusion zone, cE(x,t) (see Fig. 2d). In our model, the slow diffusion zone can only exchange with the rapid diffusion zone at the distal end of the T-tubule farthest from the extracellular space. The equations are:
where k is the exchange rate between calcium in the slow and rapid diffusion zones, c2 is the concentration of calcium in the slow diffusion zone (c2 = N2/V2), σ is a rate constant for Ca2+ chelation by EGTA, DCa54 and DE55 are the free diffusion constants of Ca2+ and EGTA, respectively, and τCa and τE are tortuosity factors for Ca2+ and EGTA, respectively, that account for reduced diffusion in the T-tubule. The delta function, δ(x), indicates that compartment exchange only influences c1 at the x = 0 end of the T-tubule.
The equations were solved in Matlab™ (2012a, The Mathworks, Natick, MA) using customized code. At each time step, the spatial diffusion terms for c1 and cE were first solved with a standard Crank-Nicholson tridiagonal scheme56. Next, N2(t), c1(0,t), and the chemical terms involving chelation, σcicE, were updated using a simple Euler method. The movement of ions across the boundary between slow and rapid zones involves geometric factors to properly convert the change in ion numbers in the slow diffusion space, ΔN2, to a change in concentration in the rapid diffusion zone, Δc1. Division of ΔN2 by V1, the volume of the first spatially, discretized element in the rapid diffusion zone, properly converts moles to concentration. V1 is the cross-sectional area of the rapid diffusion zone of the T-tubule, A, times the spatial discretization along the length of the tubule, dx, such that V1 = Adx. Initially, the calcium concentration was assumed to be uniform at 2 mM in both compartments, but the value was set to zero at the far right boundary, x = L (the T-tubule length), which corresponds to the bulk extracellular space. The right boundary was therefore treated as a Dirichlet boundary condition. The calcium concentration at the inner boundary, x = 0, was treated with a Neuman-type reflecting boundary condition. The EGTA concentration was initially set to zero in the rapid diffusion zone except where it was fixed to 2 mM at x = L. cE was also treated with reflecting boundary conditions at x = 0 in the same manner as c1. The convergence properties of the solution were investigated, and the discretization values shown in Supplementary Table S1 for dt and dx provide well converged solutions.
The mathematical model was fit to the wildtype data in Fig. 2e using a Nelder-Mead Simplex method56 provided in the Matlab™ package NMSMAX.m57. Six parameters indicated in Supplementary Table 1 were systematically varied, and the closeness of the modeled curve to the experimental data was calculated by using the following error function:
where i runs over all experimental data points, Yi is the experimental value of the normalized current for point i, and Y*i is normalized calcium concentration in the slow diffusion zone corresponding to the time of point i. The search algorithm was run in 10–20 iterative rounds using the best fit from one round as the initial conditions for further runs. Each round was run for 200–1000 iterations. After fitting the model to the wildtype data, the diffusion barrier between compartments was removed by increasing the value of k by a factor of 100, and the normalized concentration in the slow diffusion zone was replotted without further fitting of the data.
All code will be provided upon request to M.G. (michael.grabe@ucsf.edu).
Optical mapping
Optical mapping was performed on mouse hearts, which were rapidly harvested after anesthesia and arrested in cold cardioplegia solution. The aorta was cannulated and retrogradely perfused with 37°C modified Tyrode solution (mmol L−1: NaCl 120, NaHCO3 18, KH2PO4 1.2, MgSO4 1.2, glucose 11, CaCl2 1.8, sodium pyruvate 5, and Manitol 1, KCl 3.8), bubbled with 95% O2/5% CO2. Hearts were then placed in a temperature-controlled optical recording chamber (maintained at 37 °C) while electrocardiograms and perfusion pressures were measured continuously. Before optical recording, Tyrode solution containing voltage-sensitive dye di-4-ANEPPS (10 µl of 2.5 mM stock) was perfused through the hearts. Contractility was blocked using 5 µM blebbistatin. For studies of low and high extracellular K+ perfusion, KCl concentration in Tyrode solution was adjusted to achieve a total K+ concentration of 2.5 (low) or 8 (high) mM, as compared to normal 5 mM K+ in the normal Tyrode solution.
The optical mapping system used has been previously described58. Briefly, action potentials were simultaneously recorded at 1,000 Hz with a 100 × 100 complementary metal oxide semiconductor (CMOS) camera with a 10 × 10 mm mapping field on the epicardium of the LV anterior wall. Fluorescence was excited using a 1,000-W tungsten-halogen light source with a filter of 530 nm and transmitted with an emission long-pass filter of 630 nm. Pacing was performed from two sites sutured with silver electrodes: one is on the apex of left ventricle with pacing cycle lengths (PCL) from 150 to 70 ms, and then with S1-S2 using a basic PCL of 150 ms and S2 decremented by 2 ms until the ventricular effective refractory period was reached. After this, a train of programmed stimulation with up to three extra stimuli following a basic PCL of 150 ms and a burst pacing from 60 ms to 30 ms (decrement by 2 ms) were performed to induce ventricular arrhythmias. The other site is on the left atrium paced with simulated heart rates of 300, 400, 600, and 800 per min in order to induce ectopic ventricular beats.
Data analysis was performed using custom Matlab (Mathworks, Natick, MA) programs designed for analysis of optically recorded action potentials. Quantitative data were obtained from optically derived action potentials for each of the 10,000 pixels of the CMOS camera. Local activation was determined as the time point of maximum change in fluorescence over time (dF dt−1) for each fluorescent signal in the array. Isochronal activation maps, conduction velocity were analyzed as described previously59. The action potential duration (APD) at 80% repolarization (APD80) was determined as the time difference between the take-off potential and the point during repolarization at 20% of the maximal action potential amplitude.
In vivo cardiac electrophysiology studies in adult mice
Invasive intracardiac electrophysiology was studied in young adult mice (8 week old) following a previously established protocol with some modification32. Mice were anesthetized with 1% isofluorane in 100% oxygen, body temperature was maintained at 37°C by a heating pad, and respiration was maintained using a respirator connected to an endotracheal tube placed into mouse trachea. Electrocardiograms (EKG) were recorded using PowerLab (ADInstruments) during the whole experimental procedure. Upon lateral thoracotomy, the heart was visualized, and two platinum electrodes were placed on the apex of the left ventricle for pacing. A constant current stimulus isolator (FE180, ADInstruments, Colorado Springs, CO) was used to deliver pacing currents, which was interfaced with a software-driven programmed electrical stimulator (LabChart Pro, ADInstruments, Colorado Springs, CO). Pacing thresholds were then determined and stimulation was delivered at 0.30.4 ms pulse width, at twice of the capture threshold. Standard triple extrastimulation were performed. A drive cycle length of 80ms (S1) followed by S2 decremented by 2 ms until the ventricular effective refractory period was reached. After this, a train of programmed stimulation with three extra stimuli following S1 (S1–S4) was performed and repeated every 10 s for 10 min to determine the incidence and frequency of induced VT. After baseline experiment, isoproterenol (0.1 mg kg−1) was injected into left ventricle and 10 min were allowed to elapse for stabilization of heart rate before the same S1–S4 pacing protocol was repeated. Non-sustained VT was defined as 4–9 consecutive beats and sustained VT was defined as >9 QRS complexes not preceded by superventricular activity31. LabChart was used for data analysis.
In vitro actin complex formation using heart lysates
Adult mouse hearts were perfused through aorta to remove blood and lysed in actin polymerization buffer (in mM, 150 KCl, 20 HEPES, 2 MgCl2, 2 K2HPO4, 1% NP40, pH7.4 with halt protease inhibitor). Following a10 minute incubation on ice, the lysate was ultra-centrifuged for 1 h at 170,000g at 4°C in a bench top Beckman type Ti-50 fixed angle rotor to remove endogenous F-actin polymers. The F-actin free supernatant was collected and divided equally into four aliquots. Each of the aliquots was then supplemented with various GST-BIN1 isoform coated glutathione beads (BIN1, BIN1+13, BIN1+17, and BIN1+13+17), which were prepared fresh by incubating glutathione beads with lysates of FT293 cells overexpressing different GST-BIN1 isoform (2 h incubation at 4 °C). The F-actin free heart lysates supplemented with BIN1 beads were then rotated head-to-toe overnight (16 h) at 4 °C in the present of 25 µM phallodin and 100 µM ATP, allowing in vitro actin polymerization. Then the BIN1 coated beads were spun down and washed three times in actin polymerization buffer at 4 °C. Finally, the BIN1 and its associated protein complex were eluted from the beads in SDS-PAGE sample buffer (70 °C for 10 min) and resolved by gel electrophoresis.
Pull down between GST-BIN1 isoforms and α-actinin or N-WASP
For GST-BIN1 mediated pull down of α-actinin, adult hearts were lysed in 0.5% NP40 buffer (in mM, 150 KCl, 20 HEPES, 2 MgCl2, 2 K2HPO4, 1 DTT, 1 NaF, 0.1 Na3VO4, 0.5% NP40, pH7.4 with halt protease inhibitor). The lysates were then divided equally into five portions and incubated with purified GST-GFP, GST-BIN1, GST-BIN1+13, GST-BIN1+17, or GST-BIN1+13+17 coated glutathione beads prepared as stated in the last section. The lysates were rotated head-to-toe at 4 °C for 2 h to allow protein complex formation. BIN1 protein complex attached beads were then washed and eluted in 4× sample buffer (70°C for 10 min). The eluted samples were resolved by Western blot for the detection of α-actinin and GST-BIN1.
For BIN1 pulldown of N-WASP, HeLa cells overexpressing N-WASP-V5 and GST-BIN1 protein isoforms were lysed and incubated with glutathione beads at 4 °C for 30 min to pull down GST fusion protein complex.
In vitro actin polymerization assay
Actin Polymerization Biochem Kit, purified Arp2/3 protein complex, and purified VCA domain of N-WASP were obtained from Cytoskeleton. GST fused N-WASP and BIN1 protein isoforms were purified by glutathione beads pulldown followed by elution with 5mM glutathione. The protein was then concentrated and washed with Amicon concentrator to reach a final concentration of 10 µM in 100 mM Tris (pH 7.4) buffer to be compatible with actin assay. The manufactual actin polymerization protocol was optimized using 0.2 mg ml−1 100% pyrene labeled actin supplemented with 60 nM Arp2/3 protein complex and N-WASP (200 nM) in the presence of control GST-GFP protein or 1 µM of BIN1 isoforms. Pyrene-actin alone (supplemented with GST-GFP for a similar total protein concentration) was used as the negative control. For positive control, N-WASP was replaced with 200 nM purified VCA domain of N-WASP. Baseline pyrene fluorescence (excitation 360 nm and emission 410 nm) and 90 min polymerization kinetics were recorded with a FlexStation 3 Microplate Reader (Molecular Devices). SoftMax Pro 5.4.5 (Molecular Device) was used for data analysis including Vmax calculation.
Signal Processing and Statistical Analysis
Fluorescence intensity profiles were generated by ImageJ. Prism 5 (GraphPad) software was used for statistical analysis. For comparison between two groups, paired or unpaired two-tail student’s t-test was performed. For comparison among three and more treatment groups, one-way ANOVA followed by Dunnet post-test was performed. For ICa and IK1 decay kinetics, the curves were fit with one-plateau followed by one-phase exponential decay in Prism. For comparison of VT incidence among three genetic groups, chi-sqaure test was used, and for comparison of VT frequency before and after isoproterenol among three genetic groups, two-way ANOVA was used.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grants R01 HL094414 (R.M.S.), R37 MH065334 (L.Y.J.), R21 GM100224 (M.G.), T32 HL116273 (S.Z.) and K99/R00 HL109075-01A1 (T.T.H.), and American Heart Association (S.Z., R.M.S.). L.Y.J. is a Howard Hughes Medical Institute investigator. We thank E. Cingolani for helpful advice on designing the in vivo pacing protocol, D. Laury-Kleintop and G.C. Prendergast from Lankenau Institute for Medical Research for Bin1-loxP mice, J. Mulholland and J. J. Perrino for TEM imaging at the Electron Microscopy Core (EMC, directed by J. Mulholland) of the Cell Sciences Imaging Facility (CSIF) at the Stanford University Medical Center, J. Smyth and R. Wirka for helpful discussions, and T.S. Fong and T. Hitzeman for technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS
All authors contributed to study design, analysis of the data, and writing of the paper. T.T.H. was involved in the design and performance of all key experiments, H.Y. did the patch clamp experiments, S.Z. prepared mouse crossing, adenovirus and Bin1 cloning experiments, H.C.C. and B.S. did the in vivo arrhythmia experiments, M.K. performed the actin polymerization assay, A.B. did the SICM imaging and analysis of T-tubule topology, H.Z. did the optical mapping studies, M.G. did the mathematical modeling.
The authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
REFERENCES
- 1.Soeller C, Cannell MB. Examination of the transverse tubular system in living cardiac rat myocytes by 2-photon microscopy and digital image-processing techniques. Circ Res. 1999;84:266–275. doi: 10.1161/01.res.84.3.266. [DOI] [PubMed] [Google Scholar]
- 2.Brette F, Orchard C. T-tubule function in mammalian cardiac myocytes. Circ Res. 2003;92:1182–1192. doi: 10.1161/01.RES.0000074908.17214.FD. [DOI] [PubMed] [Google Scholar]
- 3.Cheng H, Lederer WJ, Cannell MB. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262:740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 4.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 5.Pasek M, Simurda J, Christe G. The functional role of cardiac T-tubules explored in a model of rat ventricular myocytes. Philos Transact A Math Phys Eng Sci. 2006;364:1187–1206. doi: 10.1098/rsta.2006.1764. [DOI] [PubMed] [Google Scholar]
- 6.Lederer WJ, Niggli E, Hadley RW. Sodium-calcium exchange in excitable cells: fuzzy space. Science. 1990;248:283. doi: 10.1126/science.2326638. [DOI] [PubMed] [Google Scholar]
- 7.Shepherd N, McDonough HB. Ionic diffusion in transverse tubules of cardiac ventricular myocytes. Am J Physiol. 1998;275:H852–H860. doi: 10.1152/ajpheart.1998.275.3.H852. [DOI] [PubMed] [Google Scholar]
- 8.Swift F, Stromme TA, Amundsen B, Sejersted OM, Sjaastad I. Slow diffusion of K+ in the T tubules of rat cardiomyocytes. J Appl Physiol. 2006;101:1170–1176. doi: 10.1152/japplphysiol.00297.2006. [DOI] [PubMed] [Google Scholar]
- 9.Parfenov AS, Salnikov V, Lederer WJ, Lukyanenko V. Aqueous diffusion pathways as a part of the ventricular cell ultrastructure. Biophys J. 2006;90:1107–1119. doi: 10.1529/biophysj.105.071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasek M, Simurda J, Orchard CH. Role of t-tubules in the control of trans-sarcolemmal ion flux and intracellular Ca2+ in a model of the rat cardiac ventricular myocyte. Eur Biophys J. 2012;41:491–503. doi: 10.1007/s00249-012-0804-x. [DOI] [PubMed] [Google Scholar]
- 11.Pasek M, Simurda J, Orchard CH, Christe G. A model of the guinea-pig ventricular cardiac myocyte incorporating a transverse-axial tubular system. Prog Biophys Mol Biol. 2008;96:258–280. doi: 10.1016/j.pbiomolbio.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Lyon AR, et al. Loss of T-tubules and other changes to surface topography in ventricular myocytes from failing human and rat heart. Proc Natl Acad Sci U S A. 2009;106:6854–6859. doi: 10.1073/pnas.0809777106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei S, et al. T-tubule remodeling during transition from hypertrophy to heart failure. Circ Res. 2010;107:520–531. doi: 10.1161/CIRCRESAHA.109.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner E, et al. Stimulated emission depletion live-cell super-resolution imaging shows proliferative remodeling of T-tubule membrane structures after myocardial infarction. Circ Res. 2012;111:402–414. doi: 10.1161/CIRCRESAHA.112.274530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aiba T, Tomaselli GF. Electrical remodeling in the failing heart. Curr Opin Cardiol. 2010;25:29–36. doi: 10.1097/HCO.0b013e328333d3d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sipido KR. Calcium overload, spontaneous calcium release, and ventricular arrhythmias. Heart Rhythm. 2006;3:977–979. doi: 10.1016/j.hrthm.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Lee E, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 18.Hong TT, et al. BIN1 Localizes the L-Type Calcium Channel to Cardiac T-Tubules. PLoS Biol. 2010;8:e1000312. doi: 10.1371/journal.pbio.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong TT, et al. BIN1 is reduced and Cav1.2 trafficking is impaired in human failing cardiomyocytes. Heart Rhythm. 2012;9:812–820. doi: 10.1016/j.hrthm.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyon AR, et al. Plasticity of surface structures and beta(2)-adrenergic receptor localization in failing ventricular cardiomyocytes during recovery from heart failure. Circ Heart Fail. 2012;5:357–365. doi: 10.1161/CIRCHEARTFAILURE.111.964692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohm J, et al. Case report of intrafamilial variability in autosomal recessive centronuclear myopathy associated to a novel BIN1 stop mutation. Orphanet J Rare Dis. 2010;5:35. doi: 10.1186/1750-1172-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller AJ, et al. Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol Cell Biol. 2003;23:4295–4306. doi: 10.1128/MCB.23.12.4295-4306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goonasekera SA, et al. Decreased cardiac L-type Ca(2)(+) channel activity induces hypertrophy and heart failure in mice. J Clin Invest. 2012;122:280–290. doi: 10.1172/JCI58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai M, Hussain M, Orchard CH. Excitation-contraction coupling in rat ventricular myocytes after formamide-induced detubulation. Am J Physiol. 1999;277:H603–H609. doi: 10.1152/ajpheart.1999.277.2.H603. [DOI] [PubMed] [Google Scholar]
- 26.Hepler PK. Membranes in the mitotic apparatus of barley cells. J Cell Biol. 1980;86:490–499. doi: 10.1083/jcb.86.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franzini-Armstrong C. Simultaneous maturation of transverse tubules and sarcoplasmic reticulum during muscle differentiation in the mouse. Dev Biol. 1991;146:353–363. doi: 10.1016/0012-1606(91)90237-w. [DOI] [PubMed] [Google Scholar]
- 28.Komukai K, Brette F, Yamanushi TT, Orchard CH. K(+) current distribution in rat sub-epicardial ventricular myocytes. Pflugers Arch. 2002;444:532–538. doi: 10.1007/s00424-002-0851-8. [DOI] [PubMed] [Google Scholar]
- 29.Pasek M, et al. Quantification of t-tubule area and protein distribution in rat cardiac ventricular myocytes. Prog Biophys Mol Biol. 2008;96:244–257. doi: 10.1016/j.pbiomolbio.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Shaw RM, Rudy Y. Electrophysiologic effects of acute myocardial ischemia. A mechanistic investigation of action potential conduction and conduction failure. Circ Res. 1997;80:124–138. doi: 10.1161/01.res.80.1.124. [DOI] [PubMed] [Google Scholar]
- 31.Mathur N, et al. Sudden infant death syndrome in mice with an inherited mutation in RyR2. Circ Arrhythm Electrophysiol. 2009;2:677–685. doi: 10.1161/CIRCEP.109.894683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannankeril PJ, et al. Mice with the R176Q cardiac ryanodine receptor mutation exhibit catecholamine-induced ventricular tachycardia and cardiomyopathy. Proc Natl Acad Sci U S A. 2006;103:12179–12184. doi: 10.1073/pnas.0600268103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marban E, Robinson SW, Wier WG. Mechanisms of arrhythmogenic delayed and early afterdepolarizations in ferret ventricular muscle. J Clin Invest. 1986;78:1185–1192. doi: 10.1172/JCI112701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Q, et al. Functional and morphological preservation of adult ventricular myocytes in culture by sub-micromolar cytochalasin D supplement. J Mol Cell Cardiol. 2012;52:113–124. doi: 10.1016/j.yjmcc.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Yamada H, et al. Dynamic interaction of amphiphysin with N-WASP regulates actin assembly. J Biol Chem. 2009;284:34244–34256. doi: 10.1074/jbc.M109.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaur N, Rudy Y, Hool L. Contributions of ion channel currents to ventricular action potential changes and induction of early afterdepolarizations during acute hypoxia. Circ Res. 2009;105:1196–1203. doi: 10.1161/CIRCRESAHA.109.202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JM, Bursac N, Henriquez CS. A computer model of engineered cardiac monolayers. Biophys J. 2010;98:1762–1771. doi: 10.1016/j.bpj.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gez LS, Hagalili Y, Shainberg A, Atlas D. Voltage-driven Ca(2+) binding at the L-type Ca(2+) channel triggers cardiac excitation-contraction coupling prior to Ca(2+) influx. Biochemistry. 2012;51:9658–9666. doi: 10.1021/bi301124a. [DOI] [PubMed] [Google Scholar]
- 39.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler MH, et al. Amphiphysin II (SH3P9-BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohm J, et al. Altered splicing of the BIN1 muscle-specific exon in humans and dogs with highly progressive centronuclear myopathy. PLoS Genet. 2013;9:e1003430. doi: 10.1371/journal.pgen.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge K, et al. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci U S A. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- 44.Elliott K, et al. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene. 1999;18:3564–3573. doi: 10.1038/sj.onc.1202670. [DOI] [PubMed] [Google Scholar]
- 45.Kashef F, et al. Ankyrin-B protein in heart failure: identification of a new component of metazoan cardioprotection. J Biol Chem. 2012;287:30268–30281. doi: 10.1074/jbc.M112.368415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beuckelmann DJ, Nabauer M, Erdmann E. Alterations of K+ currents in isolated human ventricular myocytes from patients with terminal heart failure. Circ Res. 1993;73:379–385. doi: 10.1161/01.res.73.2.379. [DOI] [PubMed] [Google Scholar]
- 47.Smyth JW, et al. Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma membrane. Circ Res. 2012;110:978–989. doi: 10.1161/CIRCRESAHA.111.257964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connell TD, Rodrigo MC, Simpson PC. Isolation and culture of adult mouse cardiac myocytes. Methods Mol Biol. 2007;357:271–296. doi: 10.1385/1-59745-214-9:271. [DOI] [PubMed] [Google Scholar]
- 49.Hattori F, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 50.Leach RN, Desai JC, Orchard CH. Effect of cytoskeleton disruptors on L-type Ca channel distribution in rat ventricular myocytes. Cell Calcium. 2005;38:515–526. doi: 10.1016/j.ceca.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Novak P, et al. Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nat Methods. 2009;6:279–281. doi: 10.1038/nmeth.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 53.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 54.Shorten PR, Soboleva TK. Anomalous ion diffusion within skeletal muscle transverse tubule networks. Theor Biol Med Model. 2007;4:18. doi: 10.1186/1742-4682-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kits KS, de Vlieger TA, Kooi BW, Mansvelder HD. Diffusion barriers limit the effect of mobile calcium buffers on exocytosis of large dense cored vesicles. Biophys J. 1999;76:1693–1705. doi: 10.1016/S0006-3495(99)77328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Press W. Numerical Recipes: The Art of Scientific Computing. Cambridge University Press; 2007. [Google Scholar]
- 57.Higham NJ. Optimization by direct search in matrix computations. SIAM J. Matrix Anal. Appl. 1993;14:317–333. [Google Scholar]
- 58.Nguyen DT, Ding C, Wilson E, Marcus GM, Olgin JE. Pirfenidone mitigates left ventricular fibrosis and dysfunction after myocardial infarction and reduces arrhythmias. Heart Rhythm. 2010;7:1438–1445. doi: 10.1016/j.hrthm.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 59.Salama G, Kanai A, Efimov IR. Subthreshold stimulation of Purkinje fibers interrupts ventricular tachycardia in intact hearts. Experimental study with voltage-sensitive dyes and imaging techniques. Circ Res. 1994;74:604–619. doi: 10.1161/01.res.74.4.604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.