Abstract
The DEG/ENaC gene family of ion channels is characterized by a high degree of structural similarity and an equally high degree of diversity concerning the physiological function. In humans and rodents, the DEG/ENaC family comprises 2 main subgroups: the subunits of the epithelial Na+ channel (ENaC) and the subunits of the acid sensing ion channels (ASICs). The bile acid-sensitive channel (BASIC), previously known as BLINaC or INaC, represents a third subgroup within the DEG/ENaC family. Although BASIC was identified more than a decade ago, very little is known about its physiological function. Recent progress in the characterization of this neglected member of the DEG/ENaC family, which is summarized in this focused review, includes the discovery of surprising species differences, its pharmacological characterization, and the identification of bile acids as putative natural activators.
Keywords: acid-sensing ion channel, ASIC, BASIC, bile acid, BLINaC, cholangiocyte, DEG/ENaC, ENaC, epithelial Na+ channel
Introduction
Members of the degenerin/epithelial Na+ channel (DEG/ENaC) family of ion channels have been subjects of numerous studies for more than two decades and significant progress has been made in understanding the physiological role of these channels, their regulation, and their gating mechanisms.1 In addition, the understanding of structure-function relationships has been significantly advanced by the crystallization of chicken ASIC1a in the desensitized state 5 y ago.2
The function of DEG/ENaC channels ranges from Na+-absorption and homeostasis through mechanosensation to neuronal transmission. Accordingly, diverse gating mechanisms such as constitutive activity, mechanical activation and ligand activation have evolved within the channel family.3-5 The functional heterogeneity within the DEG/ENaC family is a key feature and is in contrast to the high degree of structural similarity among the members of the family: each DEG/ENaC subunit is composed of 2 transmembrane segments linked by an extracellular domain, the N- and the C-terminal domains protrude into the cytosol.1
Concerning subunits from humans and rodents and based on the number of publications, the subunits of the epithelial Na+ channel (ENaC) and the acid-sensing ion channels (ASICs), two subfamilies within the DEG/ENaC family, were the main focus of research. Only a few studies addressed another member of the family—the bile acid-sensitive ion channel (BASIC). However, over the last few years several studies were published that could intensify the interest in this channel. This short review summarizes the recent progress made on BASIC, the “ignored cousin” of the ASICs and ENaC.
BASIC's Place within the DEG/ENaC Gene Family of Ion Channels
In humans, nine genes constitute the DEG/ENaC family.1 The genes ACCN1 to ACCN4 (Amiloride-sensitive Cation Channel, Neuronal) encode at least 8 proteins: ASIC1a and ASIC1b, 2 splice variants of gene ACCN2, ASIC2a and ASIC2b, splice variants of gene ACCN1, ASIC3a, ASIC3b and ASIC3c, splice variants of gene ACCN3 and ASIC4.6-14 The genes SCNNA to SCNND (Sodium Channel, Non-Neuronal) encode the α-, β-, γ-, and δ-subunits of the epithelial Na+ channel (ENaC), respectively.3,15-17 ASIC subunits can form functional hetero- and homotrimeric channels9,12 while functional ENaCs are formed by heterotrimeric αβγ- or δβγ-subunit compositions.3,17 ASICs are activated by protons: a sudden increase in proton concentration leads to fast opening and rapid desensitization of ASICs.12 ENaC on the other hand is a constitutively active channel that is mainly regulated by proteolytic cleavage and surface expression.18-20
On a phylogenetic tree, members of the DEG/ENaC gene family from human and rodents localize to three different branches (Fig. 1). The ASIC subunits form 1 branch, the ENaC subunits form the other major branch and BASIC represents the third branch. The BASIC protein is derived from the gene ACCN5 and in public databases it is often referred to as ASIC5, reflecting the closer relationship of BASIC to ASIC subunits rather than ENaC subunits. However, BASIC is not sensitive to protons,21,22 which is the key feature of most ASICs except ASIC2b and ASIC4.8,9 The amino acid sequence of BASIC is approximately 30% identical to ASICs, but only approximately 20% identical to ENaC subunits.21 Among the BASICs from mouse, rat, and human, the degree of identity is relatively high: 97% between rat and mouse BASIC and 79% between human BASIC and rat or mouse BASIC.21,22
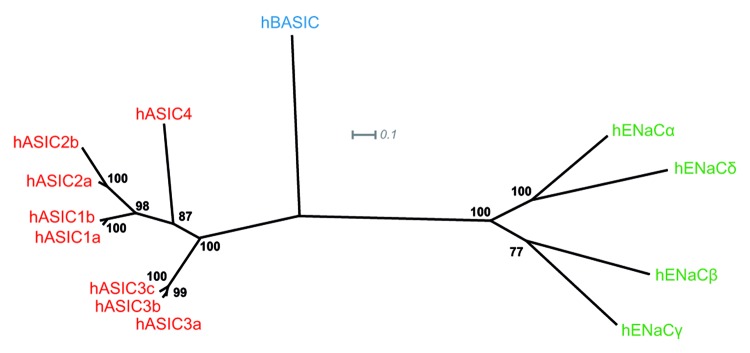
Figure 1. BASIC represents a subgroup within the human DEG/ENaC gene family of ion channels. The phylogenetic tree of human DEG/ENaC gene family members displays the genetic distances of BASIC to ASIC and ENaC subunits. Branch lengths are proportional to the evolutionary distance. Support values are indicated. Scale bar illustrates amino acid substitutions per site. The tree was created by maximum likelihood (ML) analysis with TREEPUZZLE and edited with Dendroscope3. ML analysis is based on protein alignment using ClustalX with sequences of human BASIC, ASICs, and ENaCs. Highly divergent sequences were deleted for improving alignment results. Accession numbers: hASIC1a U78180, hASIC1b HM991481, hASIC2a U50352, hASIC2b NP_899233.1, hASIC3a NP_004760, hASIC3b NP_064717; hASIC3c NP_064718, hASIC4 AJ271643, hBASIC AJ252011, αhENaC X76180, βhENaC X87159, γhENaC X87160, δhENaC U38254.
Cloning and First Electrophysiological Characterization of BASIC
The cloning of the degenerins from Caenorhabditis elegans by Driscoll and Chalfie4 and the independent cloning of the α-subunit of ENaC by Canessa et al.15 and Lingueglia et al.16 was the trigger for the identification of numerous further members of the DEG/ENaC family from different species with numbers still rising. In vertebrates the ASICs are probably the largest group within the DEG/ENaC family to date that was identified based on sequence homology to ENaC.12 While ENaC is expressed mainly in epithelia and crucial for vectorial Na+ transport, ASICs are expressed in the central and peripheral nervous system where they are involved in various neuronal processes including nociception, mechano- and chemosensation, neuromodulation and degeneration.23 BASIC was first cloned in 1999 from rat and mouse brain cDNA libraries using degenerate oligonucleotides based on conserved sequences of various known DEG/ENaC subunits.21 At the time of its initial cloning it was named brain-liver-intestine Na+ channel (BLINaC) according to its expression pattern as RT-PCR analysis from mouse tissues revealed that basic was predominantly expressed in the brain, the liver and the intestine. Furthermore basic expression was found in testis and to a weaker extent in heart, kidney, lung, and thymus. The basic expression in rat was very similar, however, it was absent from testis. basic was also present in isolated mouse and rat hepatocytes.21 Shortly after the cloning of mouse and rat BASIC, the human ortholog was described.22 It was cloned from a genomic library and the site of expression was studied by northern blot and RT-PCR. Interestingly the human basic transcript was only found along the intestinal tract, particularly in the small intestine, and to a lesser extent in testis but unlike mouse and rat, not in brain and liver. This channel was, therefore, named human intestine Na+ channel (hINaC).22 The different expression patterns of BASIC from human, rat, and mouse are summarized in Table 1.
Table 1. Comparison of the expression pattern and the pharmacological and biophysical properties of BASIC from human, rat, and mouse.
| hBASIC (hINaC) | rBASIC (rBLINaC) | mBASIC (mBLINaC) | ||
| Expression | intestine22 | brain, liver (cholangiocytes33), intestine, heart kidney, lung, thymus21 | brain, liver (cholangiocytes33), intestine, testis, heart kidney, lung, thymus21 | |
| Pharmacology | IC50 [Ca2+]e | 20 μM25 | 2 μM24,26 | 2.3 mM24 |
| IC50 AMI | > 1 mM22 | > 1 mM21,24 | 7 μM24 | |
| IC50 DIMI | 3.4 μM25 | 3.5 μM29 | 2.1 μM29 | |
| EC50 BA | ≈2 mM25 | ≈2 mM26,33 | n.a. | |
| EC50 FFA | ≈10 mM25 | ≈1.5 mM29 | n.a. | |
| Biophysical properties | constitutive activity | low22,25 | low21,24 | high24 |
| ion selectivity | • non-selective (whole oocyte)22,25 • Na+-selective in low [Ca2+]e25 • Na+-selective in outside-out patch25 |
• non-selective (whole oocyte)21,24 • Na+-selective in low [Ca2+]e21,24 |
• Na+-selective (whole oocyte)24 | |
Abbreviations: [Ca2+]e, extracellular Ca2+; AMI, amiloride; DIMI, diminazene; BA, bile acids; FFA, flufenamic acid; n.a., not applicable
The electrophysiological characterization of rat and human BASIC, respectively, proved to be difficult. On the one hand, when expressed in Xenopus oocytes only small constitutive inward currents were observed. Amiloride in millimolar concentrations only weakly blocked this current and the reversal potential was indicative of a pore, which is non-selective for Na+ over K+. On the other hand, when expressed in COS or SF9 cells no currents were observed.21,22 BASIC from mouse was not further characterized electrophysiologically by Sakai et al.21
The first electrophysiological recordings of rat BASIC in COS cells and Xenopus oocytes were enabled by the introduction of a gain-of-function mutation at the so-called deg-position (A443) shortly before the second transmembrane domain. This mutation induced large constitutive currents that were fully inhibited by micromolar concentrations of amiloride and that were highly selective for Na+ over K+, typical characteristics of DEG/ENaC channels.21 This activation by mutation showed that BASIC had the potential of forming a functional homomeric ion channel. Nevertheless the physiological role of the channel could not be determined based on these findings. A similar mutational approach with human BASIC had the same result: the channel was active and highly selective for Na+ over K+ after introducing a mutation at the deg-position.22
Channel Activity: A Question of Species
More than a decade after its cloning the first electrophysiological study focusing on mouse BASIC was published.24 Surprisingly the channel exhibited completely different electrophysiological characteristics in comparison to its human and rat orthologs: mouse BASIC showed high constitutive activity while BASIC from rat and human showed only a weak constitutive activity (Table 1). Current amplitudes of mouse BASIC recorded from Xenopus oocytes were in a similar range as currents recorded from oocytes expressing ENaC. Because mouse and rat BASIC share 97% of their amino acids this result was not expected. Using a chimeric approach with mouse and rat BASIC, one amino acid (position 387) in the extracellular domain was identified, which changed the properties of rat BASIC from a near silent, non-selective channel to a highly active, Na+-selective channel.24 In the crystal structure of the related chicken ASIC1a, the amino acid corresponding to that of position 387 — a serine in mouse BASIC and an alanine in rat BASIC—is localized to the palm domain and approximately 50 Å above the pore domain. This excludes that changes in the pore region are responsible for the different electrophysiological characteristics but suggests that the amino acid substitution between rat and mouse BASIC influences the electrophysiological features of the channel by an allosteric mechanism.24 Interestingly, the human BASIC has a serine at position 387 like mouse BASIC, yet it is silent at rest like rat BASIC.
Pharmacological Characteristics of BASIC
BASIC from rat and human is inhibited by physiological concentrations of extracellular divalent cations, rendering the channels nearly silent. Consequently, the removal of divalent cations induces robust currents.24-26 Apparent affinities of rat BASIC for Ca2+ and Mg2+ are 13 and 79 μM,24,26 respectively, and of human BASIC 18 and 51 μM,25 respectively. Under normal physiological conditions these channels would always be in an inactive, nearly silent state. Removal of divalent cations not only increased the activity of rat and human BASIC but also increased the selectivity of the channel for Na+ over K+, similar to the introduction of the mutation at the deg-position.24,25 In contrast, the constitutively active BASIC from mouse is not potently inhibited by physiological concentrations of extracellular divalent cations, since its apparent affinity for extracellular divalent cations is almost 250-fold lower compared with BASIC from rat and human. The difference in cation affinity is determined by the amino acid at position 387.24 Since divalent cations also stabilize the resting state of ASICs,27,28 the allosteric control of gating by divalent cations seems to be conserved between these channels.
The inhibition by the diuretic amiloride is a hallmark feature of all DEG/ENaC channels.1 BASIC from mouse, rat and human, however, show interesting differences. While rat and human BASIC in their resting state are only weakly inhibited by millimolar concentrations of amiloride,21,22,24 mouse BASIC is already inhibited by low micromolar concentrations of amiloride.24 This pharmacological difference is also determined by the amino acid at position 387.24
BASIC from mouse, rat and human not only show strong differences in their pharmacological profile but also similarities. They are potently inhibited by diarylamidines, a class of anti-protozoal molecules and the related substance nafamostat.29 Apparent affinity of BASIC for diarylamidines and nafamostat is in the low micromolar range and similar for mouse, rat, and human BASIC.29 Diarylamidines and nafamostat likely function as pore blockers as their block is voltage-dependent and they compete with amiloride for its binding site,29 which is in the pore region. These molecules do not inhibit the epithelial Na+ channel,30 which makes them an ideal pharmacological tool to distinguish between BASIC and ENaC currents and analyze BASIC's physiological role in native tissue. The pharmacological and biophysical properties of BASIC from human, rat and mouse are summarized in Table 1.
Another substance that was recently identified as a BASIC inhibitor is 4-aminopyridine (4-AP),31 a known blocker of voltage-dependent K+ channels (Kv channel), which is used to target symptoms of the neuroinflammatory disease multiple sclerosis (MS).32 When applied to CHO cells expressing rat BASIC containing the gain-of-function mutation at the deg-position, 4-AP inhibited BASIC currents. Apparent affinity for 4-AP is 1.7 mM, and thus, significantly lower than the affinity for diarylamidines or amiloride. Similar to the inhibition of BASIC by diarylamidines and amiloride, the block of BASIC by 4-AP is voltage-dependent.31
Ever since its cloning in 1999, it was suspected that BASIC requires the presence of an extracellular ligand in order to be activated. This suspicion was substantiated by the finding that fenamates, notably flufenamic acid (FFA), induced large currents when applied at millimolar concentrations to oocytes expressing rat or human BASIC.25,29 Interestingly, similar to the removal of divalent cations, the selectivity for Na+ over K+ was increased by FFA. For mouse BASIC, FFA had no stimulatory effect.29
Are Bile Acids Natural Ligands of BASIC?
The activation of BASIC by the introduction of a mutation at the deg-position and the properties of this mutated channel—high Na+-selectivity and high affinity inhibition by amiloride—tempted Sakai et al. to speculate that BASIC could represent another ligand-gated DEG/ENaC channel.21 The activation of BASIC by FFA supported this speculation.29 However, several molecules including cholesterol, ATP, ADP, and acetylcholine were tested for a stimulatory effect on BASIC but failed to activate the channel. Similarly, peptides such as vasoactive intestinal peptide, calcitonin gene-related peptide, angiotensin II, N-formyl-methionyl-leucyl-phenylalanine (fMLP), FMRFamide, and related peptides did not activate BASIC, and neither did thrombin treatment, hypertonic stress, acidification, or mechanical membrane stretch.21
The immunohistochemical identification of cholangiocytes as main site of BASIC expression in the liver resulted in the identification of a new natural class of rat and human BASIC activators.33 Cholangiocytes are the epithelial cells of bile ducts and at their apical side, where BASIC can be found, they are in direct contact with bile and thus with millimolar concentrations of various bile acids. After demonstrating that diluted pig bile had the potential to activate BASIC, bile acids were soon identified as the active compound in pig bile. The sensitivity for bile acids led to the renaming of the channel, previously termed BLINaC, to bile acid-sensitive ion channel - BASIC.33
A large variety of bile acids exist in nature and the composition of bile acids in bile varies between species. Hyodeoxycholic and chenodeoxycholic acid, two important bile acids in pig bile that are also present in rat bile, are the most potent activators of BASIC when applied alone. Half maximal activation of BASIC by HDCA was reached at 2.1 mM. Co-application of both bile acids has a potentiating effect. The predominant bile acid in rat bile, β-muricholic acid does not activate BASIC. Interestingly, HDCA only weakly but CDCA and deoxycholic acid (DCA) strongly activated human BASIC.25,33 Since HDCA is absent from human bile and CDCA and DCA are important human bile acids, it is tempting to speculate that the BASIC channel has adapted to the species-specific bile acid composition. The constitutively active mouse BASIC is not affected by bile acids.33
In a follow-up study, ursodeoxycholic acid (UDCA), which is used to treat various liver diseases including gallstones and primary biliary cirrhosis,34 was also shown to potently activate BASIC with a similar apparent affinity (EC50: 2.5 mM) as HDCA.26
In a recent study, human BASIC was characterized on the single channel level. Different from observations in whole oocytes, human BASIC was highly selective for Na+. Removal of divalent cations has two effects on BASIC: it increased single channel conductance and open probability. In contrast, bile acids mainly increased open probability. The increased open probability is primarily due to a reduction of the single channel closed time. Taken together these results suggest that bile acids and removal of divalent cations activate BASIC by destabilizing the closed state of the channel.25
Bile acids are natural compounds that activate BASIC in the Xenopus oocyte expression system, the question of whether they are also the natural endogenous ligand of BASIC is still awaiting an answer.
Outlook
The physiological role of BASIC
What is the possible physiological role of BASIC in cholangiocytes and other tissues that express the channel? Cholangiocytes form a predominantly secretory epithelium mainly secreting HCO3-, Cl-, and water, which guarantees a constant bile flow and inhibits the precipitation of bile acid and thus gallstone formation.35,36 A Na+ channel like BASIC located at the apical membrane, which is activated by increasing concentrations of bile (acids), would allow Na+-absorption and thus counteract the secretory efforts of the biliary epithelium. Nonetheless it is possible that under certain circumstances, for example in the presence of somatostatin, a peptide hormone which can decrease secretory processes and even induce absorptive processes in cholangiocytes,37 BASIC could be required for Na+-absorption.
In the intestine, Na+ can be reabsorbed by various mechanisms including nutrient coupled Na+ absorption, electrogenic absorption and electroneutral absorption. Similar to ENaC in the colon38 BASIC could serve as an additional pathway of electrogenic Na+-absorption, which is dependent on the presence of bile acids. Since bile acids reach the intestine together with nutrients, bile acids might function as an indicator for the passage of nutrients and thus trigger Na+-absorption upon the passage of nutrients. Like ASIC1a, BASIC is also permeable for protons,24,39 which might contribute to its physiological function.
In rat and mouse, BASIC is also found in the brain.21 Bile acids do not reach concentrations in the brain that would be sufficient to activate BASIC in either physiological or in pathophysiological conditions. However, molecules related to bile acids and cholesterol, such as the neurosteroids or intermediate molecules of the cholesterol synthesis pathway are known ion channel modulators40,41 and present in the brain. They could represent another class of BASIC activators.
Finally, the exact subcellular location of BASIC is not clear. In Xenopus oocytes, the channel reaches the plasma membrane but in other cells BASIC-specific currents could not be recorded so far. This could of course be due to silent channels located in the plasma membrane, as suggested by currents in COS cells after activation of BASIC by the gain-of-function mutation.21,22 But it is also possible that the channel is mainly located and has a function within the cell and not in the plasma membrane. Such a phenomenon was only recently described for the related DEG/ENaC channel ASIC1a which was not only found in the plasma membrane but also in the mitochondria of cortical neurons, where it is associated with adenine nucleotide translocase and possibly involved in mitochondrial permeability.42
The mechanism underlying the activation of BASIC
How do bile acids and artificial ligands activate BASIC? Is the channel ligand-gated and are bile acids its natural ligand? Are modifications of the membrane induced by bile acids responsible for the activation of BASIC and the channel, thus, a membrane-sensor? So far, no clear answers can be given to these questions. Several findings from related DEG/ENaC family members speak in favor of a direct ligand-based activation of the channel and just as many findings support the hypothesis that BASIC senses modifications of the membrane. In the DEG/ENaC family, ligand-based activation can be found for example in the ASIC subgroup with the simplest form of ligand possible: protons.12 It is also found in HyNaCs (Hydra Na+ channels) from the freshwater polyp Hydra magnipapillata and in FaNaCs (FMRFamide-gated Na+ channel) from snails, which are activated by ligands far more complex: neuropeptides.43-45
Some DEG/ENaC channels are modulated by their membranous environment. Cholesterol for example is required for full activity of αβγENaC46 and membrane-active substances like the lanthanoide gadolinium (Gd3+) and chlorpromazine have been shown to influence the activity of ENaC.47 Pharmacological approaches as well as biophysical approaches are currently being undertaken to reveal the mechanism of bile acid activation of BASIC. Sakai et al. tested whether membrane stretch applied in the outside-out patch clamp configuration influences rat BASIC activity and did not observe any activation upon stretch application.21 This could mean that the bile acid activation of BASIC—assuming that it is mediated by membrane modifications—is indirect and does not involve lateral stretch but other membranous changes, for example alterations of the charges in the hydrophilic region of the plasma membrane.
BASIC: Forever a poor cousin?
Despite the fact that we are still far away from assigning BASIC a clear physiological function, compared with the established roles for ENaC and to a certain degree also for ASICs, the recent progress shows that BASIC is an interesting target of research, suggesting that BASIC will not remain the poor cousin of the DEG/ENaC family.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Verena Schwartz for comments on the manuscript. Wiemuth D is supported by a grant from the Deutsche Forschungsgemeinschaft (WI 4176/1–1).
Glossary
Abbreviations:
- ASIC
acid-sensing ion channel
- BASIC
bile acid-sensitive ion channel
- BLINaC
brain-liver-intestine Na+ channel
- β−MCA
β-muricholic acid
- CDCA
chenodeoxycholic acid
- ENaC
epithelial Na+ channel
- FaNaC
FMRFamide-gated Na+ channel
- HDCA
hyodeoxycholic acid
- HyNaC
Hydra Na+ channel
- INaC
intestine Na+ channel
- UDCA
ursodeoxycholic acid
References
- 1.Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–67. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- 2.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–23. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 3.Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, Rossier BC. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367:463–7. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 4.Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–93. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 5.Wemmie JA, Chen J, Askwith CC, Hruska-Hageman AM, Price MP, Nolan BC, Yoder PG, Lamani E, Hoshi T, Freeman JH, Jr., et al. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron. 2002;34:463–77. doi: 10.1016/S0896-6273(02)00661-X. [DOI] [PubMed] [Google Scholar]
- 6.Waldmann R, Champigny G, Voilley N, Lauritzen I, Lazdunski M. The mammalian degenerin MDEG, an amiloride-sensitive cation channel activated by mutations causing neurodegeneration in Caenorhabditis elegans. J Biol Chem. 1996;271:10433–6. doi: 10.1074/jbc.271.18.10433. [DOI] [PubMed] [Google Scholar]
- 7.Chen CC, England S, Akopian AN, Wood JN. A sensory neuron-specific, proton-gated ion channel. Proc Natl Acad Sci U S A. 1998;95:10240–5. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gründer S, Geissler HS, Bässler EL, Ruppersberg JP. A new member of acid-sensing ion channels from pituitary gland. Neuroreport. 2000;11:1607–11. doi: 10.1097/00001756-200006050-00003. [DOI] [PubMed] [Google Scholar]
- 9.Lingueglia E, de Weille JR, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem. 1997;272:29778–83. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 10.Price MP, Snyder PM, Welsh MJ. Cloning and expression of a novel human brain Na+ channel. J Biol Chem. 1996;271:7879–82. doi: 10.1074/jbc.271.14.7879. [DOI] [PubMed] [Google Scholar]
- 11.Waldmann R, Bassilana F, de Weille J, Champigny G, Heurteaux C, Lazdunski M. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J Biol Chem. 1997;272:20975–8. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 12.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386:173–7. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 13.Delaunay A, Gasull X, Salinas M, Noël J, Friend V, Lingueglia E, Deval E. Human ASIC3 channel dynamically adapts its activity to sense the extracellular pH in both acidic and alkaline directions. Proc Natl Acad Sci U S A. 2012;109:13124–9. doi: 10.1073/pnas.1120350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deval E, Gasull X, Noël J, Salinas M, Baron A, Diochot S, Lingueglia E. Acid-sensing ion channels (ASICs): pharmacology and implication in pain. Pharmacol Ther. 2010;128:549–58. doi: 10.1016/j.pharmthera.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–70. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 16.Lingueglia E, Voilley N, Waldmann R, Lazdunski M, Barbry P. Expression cloning of an epithelial amiloride-sensitive Na+ channel. A new channel type with homologies to Caenorhabditis elegans degenerins. FEBS Lett. 1993;318:95–9. doi: 10.1016/0014-5793(93)81336-X. [DOI] [PubMed] [Google Scholar]
- 17.Waldmann R, Champigny G, Bassilana F, Voilley N, Lazdunski M. Molecular cloning and functional expression of a novel amiloride-sensitive Na+ channel. J Biol Chem. 1995;270:27411–4. doi: 10.1074/jbc.270.46.27411. [DOI] [PubMed] [Google Scholar]
- 18.Hughey RP, Bruns JB, Kinlough CL, Harkleroad KL, Tong Q, Carattino MD, Johnson JP, Stockand JD, Kleyman TR. Epithelial sodium channels are activated by furin-dependent proteolysis. J Biol Chem. 2004;279:18111–4. doi: 10.1074/jbc.C400080200. [DOI] [PubMed] [Google Scholar]
- 19.Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature. 1997;389:607–10. doi: 10.1038/39329. [DOI] [PubMed] [Google Scholar]
- 20.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–36. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai H, Lingueglia E, Champigny G, Mattei MG, Lazdunski M. Cloning and functional expression of a novel degenerin-like Na+ channel gene in mammals. J Physiol. 1999;519:323–33. doi: 10.1111/j.1469-7793.1999.0323m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer L, Sakai H, Mattei M, Lazdunski M, Lingueglia E. Molecular cloning, functional expression and chromosomal localization of an amiloride-sensitive Na(+) channel from human small intestine. FEBS Lett. 2000;471:205–10. doi: 10.1016/S0014-5793(00)01403-4. [DOI] [PubMed] [Google Scholar]
- 23.Wemmie JA, Taugher RJ, Kreple CJ. Acid-sensing ion channels in pain and disease. Nat Rev Neurosci. 2013;14:461–71. doi: 10.1038/nrn3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiemuth D, Gründer S. A single amino acid tunes Ca2+ inhibition of brain liver intestine Na+ channel (BLINaC) J Biol Chem. 2010;285:30404–10. doi: 10.1074/jbc.M110.153064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefèvre CM, Diakov A, Haerteis S, Korbmacher C, Gründer S, Wiemuth D. Pharmacological and electrophysiological characterization of the human bile acid-sensitive ion channel (hBASIC) Pflugers Arch. 2013 doi: 10.1007/s00424-013-1310-4. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 26.Wiemuth D, Sahin H, Lefèvre CM, Wasmuth HE, Gründer S. Strong activation of bile acid-sensitive ion channel (BASIC) by ursodeoxycholic acid. Channels (Austin) 2013;7:38–42. doi: 10.4161/chan.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babini E, Paukert M, Geisler HS, Gründer S. Alternative splicing and interaction with di- and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1) J Biol Chem. 2002;277:41597–603. doi: 10.1074/jbc.M205877200. [DOI] [PubMed] [Google Scholar]
- 28.Immke DC, McCleskey EW. Protons open acid-sensing ion channels by catalyzing relief of Ca2+ blockade. Neuron. 2003;37:75–84. doi: 10.1016/S0896-6273(02)01130-3. [DOI] [PubMed] [Google Scholar]
- 29.Wiemuth D, Gründer S. The pharmacological profile of brain liver intestine Na+ channel: inhibition by diarylamidines and activation by fenamates. Mol Pharmacol. 2011;80:911–9. doi: 10.1124/mol.111.073726. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Qiu L, Li M, Dürrnagel S, Orser BA, Xiong ZG, MacDonald JF. Diarylamidines: high potency inhibitors of acid-sensing ion channels. Neuropharmacology. 2010;58:1045–53. doi: 10.1016/j.neuropharm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boiko N, Kucher V, Eaton BA, Stockand JD. Inhibition of neuronal degenerin/epithelial Na+ channels by the multiple sclerosis drug 4-aminopyridine. J Biol Chem. 2013;288:9418–27. doi: 10.1074/jbc.M112.449413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judge SI, Bever CT., Jr. Potassium channel blockers in multiple sclerosis: neuronal Kv channels and effects of symptomatic treatment. Pharmacol Ther. 2006;111:224–59. doi: 10.1016/j.pharmthera.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Wiemuth D, Sahin H, Falkenburger BH, Lefèvre CM, Wasmuth HE, Gründer S. BASIC--a bile acid-sensitive ion channel highly expressed in bile ducts. FASEB J. 2012;26:4122–30. doi: 10.1096/fj.12-207043. [DOI] [PubMed] [Google Scholar]
- 34.Beuers U. Drug insight: Mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:318–28. doi: 10.1038/ncpgasthep0521. [DOI] [PubMed] [Google Scholar]
- 35.Bogert PT, LaRusso NF. Cholangiocyte biology. Curr Opin Gastroenterol. 2007;23:299–305. doi: 10.1097/MOG.0b013e3280b079fb. [DOI] [PubMed] [Google Scholar]
- 36.Marinelli RA, Larusso NF. Solute and water transport pathways in cholangiocytes. Semin Liver Dis. 1996;16:221–9. doi: 10.1055/s-2007-1007234. [DOI] [PubMed] [Google Scholar]
- 37.Gong AY, Tietz PS, Muff MA, Splinter PL, Huebert RC, Strowski MZ, Chen XM, LaRusso NF. Somatostatin stimulates ductal bile absorption and inhibits ductal bile secretion in mice via SSTR2 on cholangiocytes. Am J Physiol Cell Physiol. 2003;284:C1205–14. doi: 10.1152/ajpcell.00313.2002. [DOI] [PubMed] [Google Scholar]
- 38.Kunzelmann K, Mall M. Electrolyte transport in the mammalian colon: mechanisms and implications for disease. Physiol Rev. 2002;82:245–89. doi: 10.1152/physrev.00026.2001. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Gründer S. Permeating protons contribute to tachyphylaxis of the acid-sensing ion channel (ASIC) 1a. J Physiol. 2007;579:657–70. doi: 10.1113/jphysiol.2006.120733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belelli D, Lambert JJ. Neurosteroids: endogenous regulators of the GABA(A) receptor. Nat Rev Neurosci. 2005;6:565–75. doi: 10.1038/nrn1703. [DOI] [PubMed] [Google Scholar]
- 41.Paul SM, Doherty JJ, Robichaud AJ, Belfort GM, Chow BY, Hammond RS, Crawford DC, Linsenbardt AJ, Shu HJ, Izumi Y, et al. The major brain cholesterol metabolite 24(S)-hydroxycholesterol is a potent allosteric modulator of N-methyl-D-aspartate receptors. J Neurosci. 2013;33:17290–300. doi: 10.1523/JNEUROSCI.2619-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YZ, Zeng WZ, Xiao X, Huang Y, Song XL, Yu Z, Tang D, Dong XP, Zhu MX, Xu TL. Intracellular ASIC1a regulates mitochondrial permeability transition-dependent neuronal death. Cell Death Differ. 2013;20:1359–69. doi: 10.1038/cdd.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dürrnagel S, Kuhn A, Tsiairis CD, Williamson M, Kalbacher H, Grimmelikhuijzen CJ, Holstein TW, Gründer S. Three homologous subunits form a high affinity peptide-gated ion channel in Hydra. J Biol Chem. 2010;285:11958–65. doi: 10.1074/jbc.M109.059998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golubovic A, Kuhn A, Williamson M, Kalbacher H, Holstein TW, Grimmelikhuijzen CJ, Gründer S. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem. 2007;282:35098–103. doi: 10.1074/jbc.M706849200. [DOI] [PubMed] [Google Scholar]
- 45.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–3. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 46.Krueger B, Haerteis S, Yang L, Hartner A, Rauh R, Korbmacher C, Diakov A. Cholesterol depletion of the plasma membrane prevents activation of the epithelial sodium channel (ENaC) by SGK1. Cell Physiol Biochem. 2009;24:605–18. doi: 10.1159/000257516. [DOI] [PubMed] [Google Scholar]
- 47.Awayda MS, Shao W, Guo F, Zeidel M, Hill WG. ENaC-membrane interactions: regulation of channel activity by membrane order. J Gen Physiol. 2004;123:709–27. doi: 10.1085/jgp.200308983. [DOI] [PMC free article] [PubMed] [Google Scholar]


