Abstract
The stroma is a key component of the lymph node structure and function. However, little is known about its origin, exact cellular composition and the mechanisms governing its formation. Lymph nodes are always encapsulated in adipose tissue and we recently demonstrated the importance of this relation for the formation of lymph node stroma. Adipocyte precursor cells migrate into the lymph node during its development and upon engagement of the Lymphotoxin-b receptor switch off adipogenesis and differentiate into lymphoid stromal cells (Bénézech et al.14). Based on the lymphoid stroma potential of adipose tissue, we present a method using a lymph node/fat pad chimera that allows the lineage tracing of lymph node stromal cell precursors. We show how to isolate newborn lymph nodes and EYFP+ embryonic adipose tissue and make a LN/ EYFP+ fat pad chimera. After transfer under the kidney capsule of a host mouse, the lymph node incorporates local adipose tissue precursor cells and finishes its formation. Progeny analysis of EYFP+ fat pad cells in the resulting lymph nodes can be performed by flow-cytometric analysis of enzymatically digested lymph nodes or by immunofluorescence analysis of lymph nodes cryosections. By using fat pads from different knockout mouse models, this method will provide an efficient way of analyzing the origin of the different lymph node stromal cell populations.
Keywords: Immunology, Issue 82, Adipose Tissue, Mesenchymal Stromal Cells, Immune System, Lymphoid Tissue, Lymph Nodes, Lymph node development, lymph node stromal cells, lymph node transplantation, immune responses, adipose tissue, adipose tissue stromal cells, stem cells
Introduction
Lymph nodes (LNs) are key organs of the immune system situated at strategic sites in the body, along the lymphatic vasculature network. They enable filtration of antigens and pathogens and provide a site for antigen presentation to lymphocytes and induction of adaptive immune responses. The stroma, which forms the basic structure of the LN and orchestrates the movement of the different hematopoietic participants of the adaptive immune response, is central to the function of these organs. Different populations of stromal cells supply essential and specific cues for the movements, localization, survival, proliferation and maturation of the hematopoietic component of the immune system1-3. Adult LN stromal cells fall in three categories: the blood endothelial cells, the lymphatic endothelial cells and fibroblasts. These three categories encompass heterogeneous populations. The fibroblastic populations contain fibroblastic reticular cells (FRC), follicular dendritic cells (FDC), marginal reticular cells (MRC), while the fibroblasts forming the capsule, the medulla and other cells which are not yet identified1-5. The origins and the mechanisms governing the maturation of the different LN stromal cell populations are unclear and the absence of specific markers allowing the fate mapping of specific LN stromal cell populations rends their study particularly difficult. However, a full understanding of the ontogeny of LN stromal cells is necessary to the comprehension of adaptive immune responses, the mechanisms contributing to tolerance and is at the basis of the development of artificial LNs.
So far, the study of LN stromal cell origin and development has been mostly limited to the direct assessment of LN development in embryos and newborns in wild type mice and different knockout mouse strains6-9. These approaches are limited by the embryonic and perinatal lethality of some of the mouse strains carrying deletions in genes important for LN development. Moreover, some of the genes essential for lymphoid tissue development are also involved in a wide range of biological process as it is the case for RANK10-11 or NF-κB212. To address these issues, isolation and transplantation of embryonic LNs under the kidney capsule of a host mouse have been performed12-13. This technique allows, for example, the transfer of genetically modified embryonic LNs in a wild type environment to assess organ development and the recruitment and organization of host cells. However, the growth of embryonic LN grafted under the kidney capsule of an adult host is impaired, thus limiting the use of this technique.
LNs and fat deposits are anatomically closely associated and they develop simultaneously during embryogenesis. We recently demonstrated that the association LN/adipose tissue plays a crucial role in the provision of stromal cell progenitors for the LNs. In particular, signaling through the LTβR controls the fate of adipocyte precursor cells by blocking adipogenesis and instead promoting maturation towards a LN stromal cell phenotype14. Here we describe a method based on generation of LN-fat pad chimeras allowing the tracing of adipose tissue derived cells in the developing LN. This method will be useful to determine the contribution of adipose tissues to different LN stromal cell populations and combined with the use of tissues from genetically modified mouse strains, will allow a better understanding of the mechanisms controlling the differentiation of the different LN stromal cell subsets.
Protocol
Mice were bred and maintained under SPF conditions in the Biomedical Service Unit at the University of Birmingham according to UK Home Office and local ethics committee regulations. All procedures described in this protocol are covered under a Project License approved by both local ethics committee and the Home Office.
1. Isolation of Newborn LNs
Sacrifice the newborn mice by cervical dislocation.
Section the head, and open the body with scissors from the top of the thoracic region to the bottom of the abdominal region and remove carefully all the viscera (heart, lungs, liver, intestine, kidney, bladder) from the abdominal cavity.
Transfer the body into a 90 mm Petri dish containing RF10 media (RPMI1640 media supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM L-glutamine). From this step onwards, the tissues will be kept under sterile conditions and handled in a tissue culture hood.
Transfer the body into a new 90 mm Petri dish containing RF10.
Under the dissecting microscope, detach carefully the peritoneum from the skin in the inguinal region. The inguinal LN is situated at the intersection of three blood vessels in the fat pad.
Remove carefully the LN. Make sure that all adipose tissue is removed. Transfer the LN into a 50 mm Petri dish containing RF10 media. Keep the tissues on ice while continuing with the procedure.
2. Isolation of E18.5 Fat Pads
- To generate mouse embryos at day 18.5 of gestation (E18.5), set up timed pregnancies by placing a female and a male mouse per cage overnight. Separate the mice in the morning and check for vaginal plugs (gestational day E0.5).
- Euthanize the plugged females at day 18.5 of pregnancy by cervical dislocation.
- Remove the embryos and placed them in a 90 mm Petri Dish containing RF10.
From this step onwards, tissues should be handled using sterile technique in the tissue culture hood. Under the dissecting microscope, section the head, open the embryos with scissors from the thoracic region to the bottom of the abdominal region and remove carefully all the viscera (heart, lungs, liver, intestine, kidney, bladder) from the body cavity.
Clean the body from all remaining visceral tissues and wash it in PBS to eliminate all traces of blood that could make the dissection more difficult.
Transfer the clean bodies into a fresh 90 mm Petri dish containing RF10.
Carefully detach the peritoneum from the skin in the inguinal region. The inguinal LN is situated at the intersection of three blood vessels in the fat pad.
Carefully remove the LN and discard it. It is very important to completely remove the LN to ensure that the fat pad used for the chimeras is not contaminated by any lymphoid stromal cells.
Remove the inguinal fat pad and transfer it into a 50 mm Petri dish containing RF10 media. Keep the tissues on ice while continuing with the procedure.
3. Generation of the LN-fat Pad Chimera
- Prepare the in vitro organ culture system15.
- Sponge and filters preparation: This can be done in advance and sponges and filters stored sterile in a Petri dish in the culture hood.
- Cut the Vulkan Underwrap in 1-1.5 cm2 pieces
- Boil the sponges for 2 hr in distilled water.
- Let the sponges dry for a couple of hours in the cell culture hood.
- Boil the filters for 20 min.
- Let the filters dry for a couple of hours in the cell culture hood.
- Prepare DMEM media complemented with 10% fetal bovine serum, 10 mM HEPES, 1x MEM Nonessential Amino Acid Solution, 50 mM 2-Mercaptoethanol, 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM L-Glutamine.
- Add 2 ml of media in a 50 mm Petri dish.
- Arrange one sponge at the bottom of the Petri dish. Wet both sides of the sponge by submerging them in the media.
- Place one filter on top of the sponge. The filter is positioned at the liquid-air interface.
Under the dissecting microscope, carefully reassociate one newborn LN with one embryonic fat pad.
Place the LN-fat pad chimera on top of the filter.
Transfer the Petri dish in a rectangular plastic box with a couple of holes on the lid and containing water at the bottom to ensure humidity.
Seal the lid to the box with tape. Leave the two holes in the lid open.
Transfer the box to a cell culture incubator at 37 °C with 5% CO2. Allow the box to equilibrate for 2 hr, before sealing the holes in the lid with tape.
Incubate the tissues for at least 2 days to allow the LN to attach to the fat pad and be transferable under the kidney capsule.
4. Transfer of the LN-fat Pad Chimera Under the Kidney Capsule of Host Mice
Collect the LN-fat pad chimeras from the filter and transfer them under the kidney capsule of host mice. The kidney capsule transfer method has already been described 16.
5. Isolation of the Transferred LNs
After 3-4 weeks under the kidney capsule, the LN-fat pads chimeras are ready to be harvested. Euthanize the host mice using approved guidance.
Remove the kidney from the host mice.
Under the dissecting microscope, isolate the LN-fat pad chimera and dissect out the LN.
6. Enzymatic Digestion of the Transferred LNs for Flow-Cytometric Analysis
Make one incision in the LN with small scissors to allow the enzymes to penetrate the organ and facilitate the digestion.
Place one LN in 1.5 ml Eppendorf containing 600 μl of digestion buffer (RPMI 1% FCS, 2.5 mg/ml Collagenase D, 100 μg/ml DNase I).
Incubate for 30 min at 37 °C on an agitating thermal block. Pipette up and down to help dissociation of tissues every 10 min.
Add 6 μl EDTA 0.5 M to the tube and pipette up and down to finish the dissociation of the tissues.
Centrifuge for 5 min at 490 x g.
Resuspend the cell pellet in the staining solution (PBS, 0.1% BSA, 5% rat serum and 5% mouse serum) containing the combination of primary antibodies.
Incubate for 30 min on ice.
Wash twice in PBS
Incubate with secondary antibodies for 20 min if required.
Wash 2x in PBS.
Analyze on flow-cytometer.
7. Analysis of the Transferred LNs by Immunofluorescence
Fixation for EYFP conservation during the freezing and cryosections process: Place the LN in a solution of PBS, 4% PFA and 10% sucrose and leave for 3-4 hr.
Put a single small drop of cold O.C.T. compound on a small piece of aluminum foil. Avoid air bubbles. Carefully place the LN without any liquid in the middle of the drop.
Freeze the organ by placing the aluminum foil on dry ice.
Cut 8-10 μm sections onto extra adhesives glass slides using a cryostat. Let the slides dry for 2 hr before storing them at -20 °C.
Staining procedures can vary according to primary antibodies used and can be optimized from the following protocol. Stain the sections with staining solution (PBS, 1% BSA) containing the combination of primary antibodies.
Incubate for 1 hr at room temperature in a humidified chamber.
Wash twice in PBS for 5 min
Incubate with secondary antibodies for 1 hr.
Wash twice in PBS for 5 min.
Mount slides in mounting media containing DAPI.
Capture images using a fluorescence or confocal microscope.
Representative Results
Three weeks after transplantation, the LN/fat pad chimera is recovered from the kidney. The chimera is now very similar to a normal LN in its own fat pad, and the LN is visible in the center of the adipose tissue (Figure 1). If the LN can't be identified, it is possible that it was lost during the transplantation on the kidney. When assessing the role of genes potentially important in LN development, the LNs recovered may remain very small and more difficult to find. This is the case when adipose tissue from LTβR-/- mice is reassociated with newborn LNs14.
Careful isolation of the LN allows further analysis of the progeny of EYFP+ adipose derived cells. Cryosections and immunofluorescence analysis of the LN reveal that EYFP+ adipose derived cells migrate into the LN where they contribute to the Gp38+ERTR-7+ LN stromal cell network (Figure 2A). Flow-cytometric analysis confirms that an important fraction of LN stromal cells derives from local EYFP+ adipose tissue progenitor cells, contributing to 30% of the CD45-Gp38+CD31- fibroblastic fraction and 10% of the CD45-Gp38-CD31+ blood endothelial cell fraction (Figure 2B). Up to 80% of the CD45-Gp38+CD31- fibroblastic fraction can derived from adipose tissue precursor cells demonstrating the crucial role played by adipose tissue in sustaining the growth of the LN stroma.
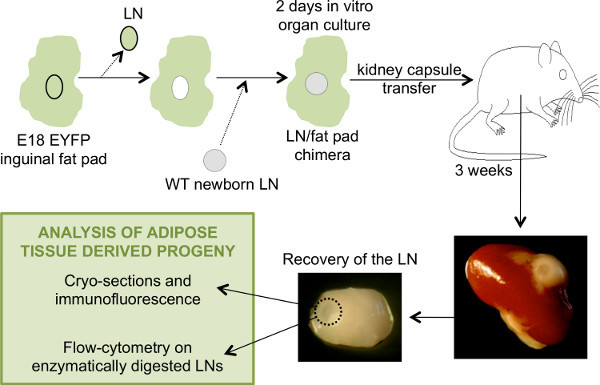 Figure 1. General schematic of the steps involved in the generation of LN/fat pads chimeras and the analysis of the LNs recovered after transplant. To generate a chimeric eYFP- LNs-eYFP+ fat pads, the LN from an E18.5 eYFP+ fat pad is removed and replaced by a WT eYFP- newborn LN. The chimeric LN-fat pad is placed in culture for two days before transplantation under the kidney capsule of an adult WT host mouse. After 3 weeks, the chimera is recovered and the LN isolated for analysis. Click here to view larger image.
Figure 1. General schematic of the steps involved in the generation of LN/fat pads chimeras and the analysis of the LNs recovered after transplant. To generate a chimeric eYFP- LNs-eYFP+ fat pads, the LN from an E18.5 eYFP+ fat pad is removed and replaced by a WT eYFP- newborn LN. The chimeric LN-fat pad is placed in culture for two days before transplantation under the kidney capsule of an adult WT host mouse. After 3 weeks, the chimera is recovered and the LN isolated for analysis. Click here to view larger image.
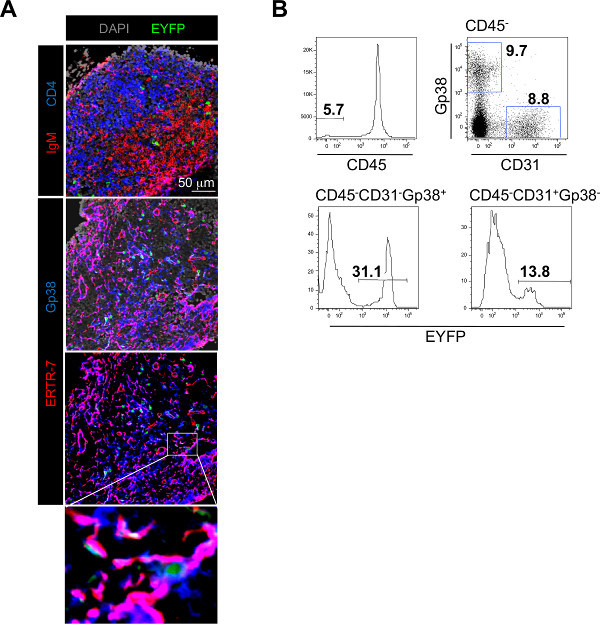 Figure 2. Typical experimental results obtained using the LN/fat pad chimera method. A) Immunofluorescence staining of frozen sections of a LN recovered from a LN-EYFP+ fat pad chimera showing the colonization of the LN by host derived IgM+ B cells in red and CD4+ T cells in blue and the presence of EYFP+ cells in green (top panel). The fibroblastic stromal cell network is stained with Gp38/Podoplanin in blue and ERTR-7 in red. EYFP+ cells derived from the fat pads in green are integrated to the stromal cell network (middle, bottom panels and enlargement). B) Flow-cytometric analysis of single cell suspension of enzymatically digested LNs recovered from LN-EYFP+ fat pad chimeras stained with CD45, CD31 and Gp38/Podoplanin. Percentage of CD45- stromal cells is shown in the histogram on the top row. Percentages of CD31-Gp38+ fibroblastic stromal cells and CD31+Gp38- blood endothelial cells in the CD45- fraction are shown in the dot plot on the top row. Percentages of fat pad derived-EYFP+ cells in the CD31-Gp38+ fibroblastic stromal cell fraction (bottom row, left histogram) and in the CD31+Gp38- blood endothelial cell fraction (bottom row, right histogram) are shown. Click here to view larger figure. Click here to view larger image.
Figure 2. Typical experimental results obtained using the LN/fat pad chimera method. A) Immunofluorescence staining of frozen sections of a LN recovered from a LN-EYFP+ fat pad chimera showing the colonization of the LN by host derived IgM+ B cells in red and CD4+ T cells in blue and the presence of EYFP+ cells in green (top panel). The fibroblastic stromal cell network is stained with Gp38/Podoplanin in blue and ERTR-7 in red. EYFP+ cells derived from the fat pads in green are integrated to the stromal cell network (middle, bottom panels and enlargement). B) Flow-cytometric analysis of single cell suspension of enzymatically digested LNs recovered from LN-EYFP+ fat pad chimeras stained with CD45, CD31 and Gp38/Podoplanin. Percentage of CD45- stromal cells is shown in the histogram on the top row. Percentages of CD31-Gp38+ fibroblastic stromal cells and CD31+Gp38- blood endothelial cells in the CD45- fraction are shown in the dot plot on the top row. Percentages of fat pad derived-EYFP+ cells in the CD31-Gp38+ fibroblastic stromal cell fraction (bottom row, left histogram) and in the CD31+Gp38- blood endothelial cell fraction (bottom row, right histogram) are shown. Click here to view larger figure. Click here to view larger image.
Discussion
In this article we presented a method to assay and quantify the contribution of adipose tissue progenitor cells to the developing LNs and two techniques that allow the analysis of their progeny. Dissection of embryonic fat pads and newborns LNs are delicate and require manual skills gained by a lot of practice prior to the generation of the actual LN-fat pad chimera. To control the quality of the dissections, flow-cytometric analysis can be performed on the fat pads and LNs. Embryonic fat pad preparation should be devoid of lymph nodes and should not contain any CD45+CD4+IL-7Ra+ lymphoid tissue inducer cells. Newborns LN preparations should be completely free of adipose tissue and contain a high proportion of CD45-CD31-ICAM-1highVCAM-1high lymphoid tissue organizer cells6,13-14. Standard sterile techniques for tissue culture and surgery should be followed to reduce the risk of contamination.
Engagement of the LTβR on immature LN stromal cells induces their differentiation into fully mature LN stromal cells. Using this in vivo system, we previously demonstrated that fat pads derived from a mouse deficient for LTβR are not able to sustain the growth of the LN. Therefore, this technique can be used with fat pads derived from different knock-out mouse models to assess the need for adipose tissue derived cells in the development of different LN stromal cell subsets. It is not known yet whether adipose tissue can give rise to all LN stromal cell populations. Combinations of LNs and fat pad of different ages could be used to determine the presence of progenitor cells at varying time points. This approach can also be combined with the use of recipient mice expressing another cell marker, such as RFP, to compare the requirement in local and distant progenitor cells for the LN growth.
During an immune response, the LN undergoes a dramatic expansion, including a marked remodeling and expansion of the stromal cell network. However, very little is known about the mechanisms regulating these processes. This in vivo system, could be used with adult adipose tissue and adults LNs in combination with a systemic immunization17 to assess the role of adipose tissue derived cells in the remodeling of the LN stroma during immune responses.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We are grateful to the personnel of the Biomedical Services Unit of the University of Birmingham for taking care of our animal colonies. This work was supported by the EU FP7 integrated project INFLACARE to JC.
References
- Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat. Rev. Immunol. 2009;9:618–629. doi: 10.1038/nri2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal R, Mebius RE. Stromal cell-immune cell interactions. Annu. Rev. Immunol. 2011;29:23–43. doi: 10.1146/annurev-immunol-031210-101357. [DOI] [PubMed] [Google Scholar]
- Turley SJ, Fletcher AL, Elpek KG. The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nat. Rev. Immunol. 2010;10:813–825. doi: 10.1038/nri2886. [DOI] [PubMed] [Google Scholar]
- Katakai T, et al. Organizer-like reticular stromal cell layer common to adult secondary lymphoid organs. J. Immunol. 2008;181:6189–6200. doi: 10.4049/jimmunol.181.9.6189. [DOI] [PubMed] [Google Scholar]
- Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 2007;8:1255–1265. doi: 10.1038/ni1513. [DOI] [PubMed] [Google Scholar]
- Benezech C, et al. Ontogeny of stromal organizer cells during lymph node development. J. Immunol. 2010;184:4521–4530. doi: 10.4049/jimmunol.0903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T, et al. Presumptive lymph node organizers are differentially represented in developing mesenteric and peripheral nodes. J. Immunol. 2004;173:2968–2975. doi: 10.4049/jimmunol.173.5.2968. [DOI] [PubMed] [Google Scholar]
- Avan de Pavert S, Mebius RE. New insights into the development of lymphoid tissues. Nat. Rev. Immunol. 2010;10:664–674. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- Vondenhoff MF, et al. LTbetaR signaling induces cytokine expression and up-regulates lymphangiogenic factors in lymph node anlagen. J. Immunol. 2009;182:5439–5445. doi: 10.4049/jimmunol.0801165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougall WC, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. Regulation of peripheral lymph node genesis by the tumor necrosis factor family member TRANCE. J. Exp. Med. 2000;192:1467–1478. doi: 10.1084/jem.192.10.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher D, et al. A stroma-derived defect in NF-kappaB2-/- mice causes impaired lymph node development and lymphocyte recruitment. J. Immunol. 2004;173:2271–2279. doi: 10.4049/jimmunol.173.4.2271. [DOI] [PubMed] [Google Scholar]
- White A, et al. Lymphotoxin a-dependent and -independent signals regulate stromal organizer cell homeostasis during lymph node organogenesis. Blood. 2007;110:1950–1959. doi: 10.1182/blood-2007-01-070003. [DOI] [PubMed] [Google Scholar]
- Benezech C, et al. Lymphotoxin-beta receptor signaling through NF-kappaB2-RelB pathway reprograms adipocyte precursors as lymph node stromal cells. Immunity. 2012;37:721–734. doi: 10.1016/j.immuni.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G, Jenkinson EJ, Moore NC, Owen JJ. MHC class II-positive epithelium and mesenchyme cells are both required for T-cell development in the thymus. Nature. 1993;362:70–73. doi: 10.1038/362070a0. [DOI] [PubMed] [Google Scholar]
- Szot GL, Koudria P, Bluestone JA. Transplantation of pancreatic islets into the kidney capsule of diabetic mice. J. Vis. Exp. 2007. p. e404. [DOI] [PMC free article] [PubMed]
- Krautler NJ, et al. Follicular dendritic cells emerge from ubiquitous perivascular precursors. Cell. 2012;150:194–206. doi: 10.1016/j.cell.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]


