Abstract
Purpose
To confirm that aneuploidy candidate genes are detectable in the first polar body (PB1) of MII oocytes and to investigate the age-dependent molecular changes in PB1.
Methods
Aged (12-to 15-mo-old) and young (2-mo-old) mice were administered pregnant mare’s serum gonadotropin (PMSG) and human chorionic gonadotrophin (hCG). MII oocytes were obtained and the first PB was removed. mRNA from each PB and its sibling oocyte was reverse transcribed. Real-time PCR was performed to quantify the expression of six genes (BUB1, CDC20, Filia, MCAK, SGOL1, SMC1A) in single PB.
Results
We first demonstrated that detection and quantification of transcripts associated with aneuploidy in single mouse oocyte and sibling PB1 is possible and the relative abundance of mRNA transcripts in a single PB faithfully reflects the relative abundance of that transcript in its sibling oocyte. We further found that transcript levels were significantly lower in aged PBs compared with young PBs (P<0.05).
Conclusions
Our results suggest that the detection and analysis of polar body mRNA may provide insight in age-related aneuploidy in oocyte. This analysis is a novel concept to investigate the genesis of chromosome abnormality and could potentially assist in the characterization of mechanisms underlying key molecular origin of female meiotic aneuploidy, which would be of great scientific and clinical value.
Keywords: Infertility, Meiosis, Genes expression, Aneuploidy, Age
Chromosome aneuploidy is the major cause of infertility, fetal loss, or birth defects and increases exponentially with maternal age in the decade preceding the menopause [1]. More recently, investigations of gametes and preimplantation embryos conceived using assisted reproductive technology (ART) have identified aneuploidy as the leading impediment to successful pregnancies in this setting [2]. Currently, the molecular mechanisms involved in the onset of aneuploidy in mammalian oocytes are not fully understood. However, evidence from studies in humans and mice suggests that the genesis of aneuploidy is not due to a single causal factor but a multi-step process caused by errors at several distinct stages of oogenesis, which involves a complex constellation of effects that begins in utero, continues throughout the reproductive lifespan of the woman, is exacerbated by age and is facilitated by the unique features of cell cycle control in the oocyte [1, 3].
Susceptibility of aneuploidy is likely to occur during oocyte development and maturation. During this time, Oocyte accumulates large members of mRNA transcripts and proteins. These raw materials implement basic biosynthetic processes in the early embryo, direct the first mitotic divisions, and support early embryonic development [4]. It is reasonable to assume that some oocytes may acquire different amounts of transcripts and proteins that, in turn, affect chromosome stability [5]. Dupont et al. reported that oocytes with differential aneuploidy rates display different gene expression patterns that may be associated with their susceptibility to chromosomal errors [6]. Fragouli et al. compared the quantities of mRNA transcripts in normal and aneuploidy oocytes and indicated that aneuploidy is associated with altered transcript levels affecting a subset of genes [7]. So the analysis of transcript levels of a number of genes important for chromosome stability in individual oocytes may provide valuable insights into the etiology of aneuploidy during early development.
One obvious drawback to quantifying gene expression profile is the risk of damaging the oocyte during sampling. The polar body (PB) is a cell created by asymmetric division of the oocyte at the time of meiosis and produces a readily accessible test source of genomic material that can be sampled as a proxy for the “sibling” oocyte for diagnostic purposes. Reich et al. demonstrated that the transcriptome of a human PB accurately reflects its sibling oocyte [8, 9]. The ability to quantify mRNA in individual cells—as small as a single PB—opens up the possibility that we can detect and compare individual differences in gene expression in the PB without harming the oocyte.
It is now well established that aneuploidy dramatically increases as women age. As in humans, advanced maternal age is associated with an increase in aneuploidy incidence in naturally aged mice [10, 11]. So the combination of genetically altered animals and natural aging model helps us understand the age-dependent increase of in aneuploidy and the alternation of gene expression patterns.
Using a natural reproductive aging mouse model, we have set out to examine the expression of six genes in individual PBs and their sibling MII oocytes from both young and aged mice. The genes tested have significance in a range of important processes that can lead to aneuploidy, including maintenance of accurate chromosomal segregation and cell cycle control (Table 1). The number of transcripts derived from each gene was assessed using real-time RT– PCR, a highly accurate method for quantification of nucleic acids. In this way, we first compared transcript abundance between PBs and their sibling oocytes, and then compared the expression of candidate genes in PB from young and aged mice determined gene expression patterns associated with their susceptibility to aneuploidy.
Table 1.
Genes selected for testing in individual PBs and sibling oocytes
| Gene | Function | Accession No. |
|---|---|---|
| BUB1 | SAC proteins. Required for recruiting other SAC proteins to the kinetochore. Required for SAC in meiosis I and meiosis II. | NM_009772.2 |
| CDC20 | Binds to APC/C; interaction with the MCC inhibits APC/C activity. | NM_023223.2 |
| Filia | Help maintain chromosome stability and euploidy in early-cleavage mouse embryogenesis. The depletion of Filia impairs preimplantation embryo development with a high incidence of aneuploidy | NM_025890.3 |
| MCAK | Depolymerises microtubules in response to a lack of tension. Also required for poleward chromosome movement. | NM_134471.4 |
| SGOL1 | Shugoshin-like 1. Proteins that protects centromeric cohesion from phosphorylation during the prophase pathway and from separate during meiosis I. | NM_028232.2 |
| SMC1A | Cohesin complex subunits protein during mitosis | NM_019710.2 |
Materials and methods
Animals
We obtained Institutional Review Board (IRB) permission to perform the animal experiments in this study. CD1 mice were housed and bred in a controlled barrier facility within Northwestern University’s Center for Comparative Medicine (Chicago, IL) in a temperature- and light-controlled environment (12L:12D) and were provided with food and water ad libitum. All mice were maintained in accordance with the policies of Northwestern University’s Animal Care and Use Committee and National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Collection and culture of MII oocytes
MII oocytes were collected from young (2-month-old) and aged (12- to 15-month-old) female mice after superovulation as previously described [12]. Oocyte-cumulus cell complexes were recovered from ampullae into Leibovitz L15 medium containing 1 % fetal bovine serum (FBS) 14 h after human chorionic gonadotropin (hCG) administration. Oocytes were dissociated from the surrounding cumulus cells using 0.3 mg/ml hyaluronidase (Sigma, St. Louis, MO) in order to identify MII oocytes. After the removal of cumulus-corona cells, only those MII oocytes that had extruded a PB were collected.
PB and sibling oocyte separation
MII oocytes that had extruded an intact PB of normal size were collected. Each MII oocyte was briefly treated with acidic Tyrode’s solution (Sigma) to remove its zona pellucida and then subjected to gentle pipetting to separate the PB from its coupled “sibling” oocyte. Single PBs in 0.5 μl PBS were transferred to the bottom of 0.5-ml thin-wall Eppendorf tubes containing 1.5 μl lysis buffer (0.8 % Igepal, 1 U RNAsin/μl, 5nM DTT) [13]. The Sibling oocytes were then randomly subjected to either fix for immunocytochemistry and chromosome counting or transfer to an identical lysis solution for RT-PCR. The lysed specimens were stored at −20 °C.
Immunocytochemistry and chromosome counting
To determine the chromosome count, eggs were fixed individually in freshly prepared 2 % paraformaldehyde in PBS for 20 min, permeabilized in PBS containing 0.3 % bovine serum albumin (BSA) and 0.1 % Triton X-100 for 15 min, and blocked in PBS containing 0.3 % BSA and 0.01 % Tween-20 (blocking solution). Cells were incubated in a 1: 50 dilution of human CREST autoimmune serum (Immunovision, Springdale, AZ) overnight at 4 °C followed by washes in blocking solution. The primary antibody was detected using an Alexa-Fluor 488 conjugated goat anti-human secondary antibody (1:200; Invitrogen Carlsbad, CA). Images were obtained at 0.5-μm intervals to span the entire region of the MII spindle using a Leica TCS SP5X laser scanning confocal microscope (Leica, Manheim, Germany) under a 63X oil immersion objective and processed using ImageJ software [14].
Reverse transcription (RT)
Total mRNA within lysed single PBs and their sibling oocytes was reverse transcribed using the AccuScript High Fidelity 1st Strand cDNA Synthesis Kit (Stratagene, La Jolla, CA) without prior RNA purification from the lysate. To normalize for variations in mRNA content among individual PB and oocyte lysates, 106 copies of a plasmid-derived RNA transcript (pw109, Perkin-Elmer GeneAmp RNA PCR kit) were added to each sample prior to RT as an exogenous control for RNA recovery and efficiency of reverse transcription [15]. Samples with less pw109 RNA than controls were excluded from further study. RT was performed by adding the following to each sample: 7.7 μl nuclease-free water, 2 μl 10× RT buffer, 0.8 μl dNTP (2.5 mM of each dNTP), and 3 μl random primers (0.1 μg/μl). Reactions were incubated at 65 °C for 5 min then cooled to room temperature (approximately 5 min). Next, 2 μl of 100 mM dithiothreitol, 0.5 μl RNAse inhibitor (40 U/μl), and 1 μl reverse transcriptase (50 U/μl) per sample were added; the reactions were incubated at 42 °C for 60 min and then terminated by heating to 70 °C for 15 min.
Real-time quantitative PCR (qPCR)
Gene expression levels in PB1 and sibling oocytes were determined for six candidate genes (Table 1). Real-time qPCR was performed using the ABI PRISM 7900 sequence detection system (Applied Biosystems, Forest City, CA). For each reaction, 0.5 μl cDNA, 0.5 μl primers, 5 μl TaqMan Universal PCR Master Mix (Applied Biosystems), and 4 μl nuclease-free water were added to a final volume of 10 μl. PCR cycling conditions were 95 °C for 10 min, followed by 50 amplification cycles of 95 °C for 15 s and 60 °C for 1 min. To maximize accuracy, each sample was run three times with a negative control of reaction mixture with no cDNA added.
Statistical analysis
Statistical comparisons for qRT-PCR results were analyzed using t-test and one-way ANOVA. Chi square analysis was used to analyze categorical data. Linear regression analysis was performed to test if mean oocyte Ct value could predict detection of mRNA in sibling polar bodies. The calculation of gene expression was as fold change (2−ΔΔCt) and relative mRNA level [ΔCt = Ct(Target) − Ct(18s). The lower ΔCt means the higher mRNA expression lever in this study]. Data were reported as mean ± SD. P < 0.05 was considered statistically significant. All statistical calculations were performed using the software GraphPad Prism version 4.0.
Results
Reduced egg numbers, increased aneuploidy from superovulated older mice
Mice used in our studies were 12 to 15-month-old, corresponding to woman ages 35–40 based on a linear extrapolation estimate. We first confirmed that increasing age is associated with a decrease in ovulation. The mean number of eggs ± SEM recovered from aged mice only 4.1 ± 0.7 eggs, compared with female aged 2-month-old, with 23.7 ± 4.1 eggs. Reduced numbers in all aged mice suggest that the ovarian follicular reserve had become nearly exhausted.
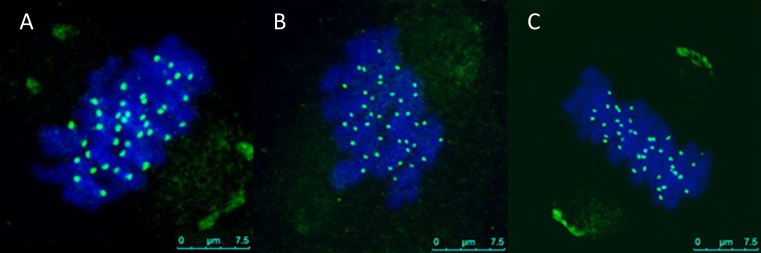
We next ascertained whether an increase in maternal age is associated with an increase incidence in aneuploidy. A z-plane series of confocal sections through each meiotic spindle were taken in order to count chromosomes and kinetochores. Euploid mouse eggs would contain a total of 20 pairs of sister chromatids with 40 kinetochores, with two sister kinetochores on each sister chromatid pair, but aneuploidy eggs would deviate from these numbers. Chromosome count revealed the aneuploidy rate in young mice was 5 % (1/20), and significantly increases to 45.8 % in eggs obtained from old mice (11/24). In the 11 aneuploidy eggs from old mice, we observed no particular bias for hypoploidy (<40 sister chromatids, n = 6) vs. hyperploidy (>40 sister chromatids, n = 5) (Fig. 1).
Fig. 1.
Determining chromosome number in mouse metaphase II oocytes (a–c). To count chromosomes following live imaging, eggs were fixed and stained for kinetochores using human CREST autoimmune antiserum (green) and for chromosomes using Hoechst (blue). Representative projected images from old eggs are shown a: euploid (n = 40); b: hypoploid (n = 39); c: hyperploid (n = 41). n = CREST-positive foci (40 CREST-positive foci are counted in a euploid oocyte). Bar = 7.5 μm
Analysis of relative mRNA expression levels in oocyte and PB1
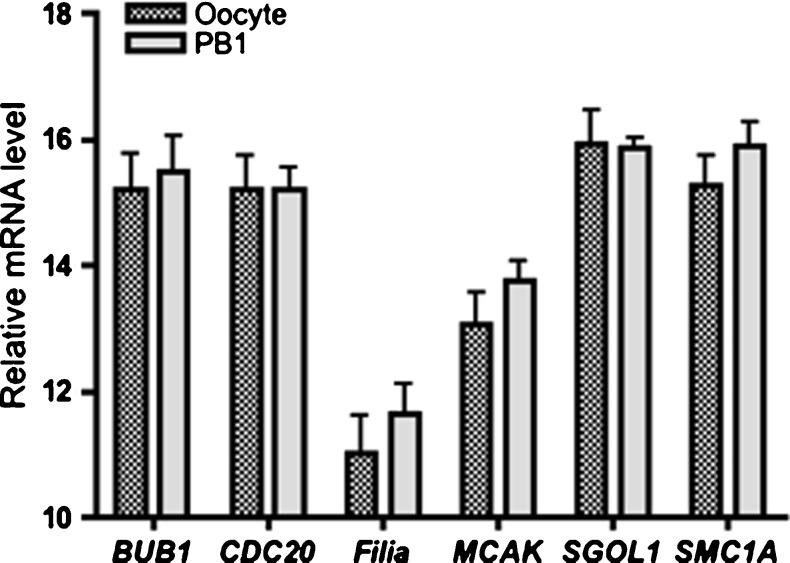
We first investigated the presence of mRNA to test the feasibility of our quantification methods. The expression of transcripts was analyzed in individual PB1 and sibling oocyte (n = 10 in each group) using real-time qPCR. mRNA was detected and quantified for all of the six candidate genes in both oocyte and single PB1. None of the qPCR reactions amplified product from the negative controls. About 98.7 % of replicates from single oocyte samples yielded an exponential amplification curve for its candidate gene. In single PB1 samples, transcripts of all the six selected candidate genes were detected by real-time qPCR. However, the amplification rate was lower in the PB1 compared with its sibling oocyte (84.7 vs. 98.7 %). A strong inverse correlation was noted between the mean oocyte Ct value and the probability of detection that transcript in a sibling polar body. Genes that were more reliably detected in polar bodies had consistently more abundant transcripts (lower mean oocyte and polar body Ct values) (Pearson’s correlation coefficient: −0.93, P < 0.05). The difference of relative levels of all six candidate mRNA transcripts was not significant between oocyte and sibling PB1 (Fig. 2).
Fig. 2.
The relative abundance of each of the six candidate genes in PB1 compared with their sibling oocyte (n = 10). Δ Ct = Ct(Target) − Ct(18s). Error bars represent SD
Relative mRNA expression levels in PBs from young vs. aged oocytes
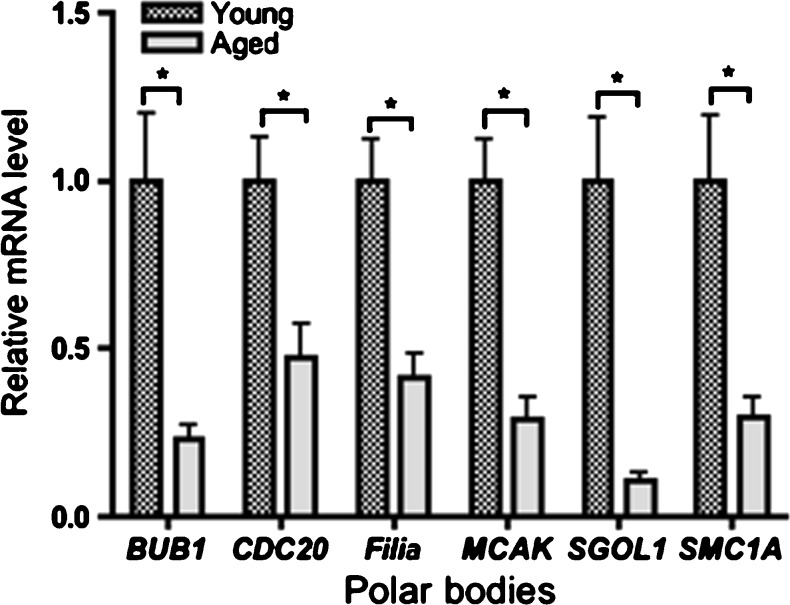
We use young and age PB1 (n = 20 in each group) to identify the changes in candidate gene expression that may contribute to the age-associated increase in aneuploidy. Transcripts of the six selected candidate genes were detected by real-time qPCR in individual PBs from both young and aged mice. PBs from aged mice had significantly lower levels of all six transcripts compared with PBs from young mice (P < 0.05). We set the normalized level of each transcript in the PBs from young eggs as 1, and then determined the fold change in transcript abundance in PBs from aged eggs. The change in transcript level was between 0.23-fold (BUB1) and 0.47-fold (CDC20) in aged group (Fig. 3).
Fig. 3.
Candidate genes expression in normal PB from young and aged mice (n = 20). Results are normalized to each transcript in PB from young mice as 1 (the calibrator). PBs from aged mice had significantly lower levels of all six transcripts compared with PBs from young mice. Fold change: 2− ΔΔCt; ΔΔCt = [Ct(Target) − Ct(18s)]aged − [Ct(Target) − Ct(18s)]young. Error bars represent SD. * significance relative to young mice (P < 0.05)
Discussion
Maternal age-related aneuploidy is well known. Most studies suggest that the expected oocyte aneuploidy rate for women under the age of 25 years is ~5 %, increasing to 10 ~ 25 % by the early 30 s and exceeding 50 % in the oocytes of women over 40 years [2]. In the present study, we demonstrated that the oocytes from aged mice exhibited an increased rate of aneuploidy than oocytes from young mice (45.8 % vs. 5 %). Merriman et al. measured the aneuploidy in mice eggs and revealed a low rate, ~3–4 % in mice aged 1 and 3 months, rising to 12.5 % by 9 month old, and to 37.5 % at 12 month. 15-month-old mice had the highest rate of aneuploidy, peaking at 60 % [11]. Our data and others demonstrated that mouse is a suitable model to study the molecular basis for the age-related effects on aneuploidy [10].
In female meiosis, the two meiotic divisions are unequal resulting in the extrusion from the oocyte of two small polar bodies (PB). Others have demonstrated that the chromosomes within the PB are identical to those in the oocyte [16]. A limited number of genes associated with aneuploidy have been assessed in human oocyte [17, 18]. This is the first report, to our knowledge, documenting the quantification of transcripts associated with aneuploidy in mouse PB1, and providing an insight into the relationship between gene expression in PB1 and age-related aneuploidy in mouse oocyte.
Candidate genes were chosen because they are known to have key roles in maintaining the fidelity of chromosome segregation [19–21]. The cohesin complex, a ring-like protein structure, is composed of four subunits: SMC1, SMC3, kleisin and STAGE. Recent data strongly suggest that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocyte [19, 22]. Sister chromatids are held together by cohesin on their centromeres and pericentromeric regions from the moment of the establishment of the cohesion during prenatal development until the resumption of meiosis in sexually mature females. This chromosome behavior, particular for meiosis, would not be possible without the preservation of a sufficient amount of cohesion. Cohesin present on sister chromatid centromeres has to be protected against separase cleavage in anaphase I, to prevent precocious segregation of sister chromatids. The shugoshins (Sgo1 and Sgo2) are the proteins which, among their other functions, protect cohesin against proteolysis [23]. A reduced level of cohesion and insufficient centromere protection, could contribute to the rise of aneuploidy with age [24]. SAC is the key regulatory mechanism in dividing cells, which controls anaphase entry by monitoring chromosome attachment to spindle microtubules. The SAC ensure faithful chromosome segregation. SAC can detect unattached microtubules or loss of tension, and cause a metaphase ‘arrest’ until all of the microtubules from the spindle poles capture the kinetochores and the chromosomes are congressed to the equatorial plate. The SAC is silenced and the anaphase promoting complex/cyclosome (APC/C) is activated, resulting in the onset of chromosome segregation and entry of the cell into anaphase [25]. Budding uninhibited by benomyl (BUB1) is involved in the regulation of the spindle assembly checkpoint, which is responsible for delaying anaphase initiation if defects in the alignment of chromosomes at the metaphase plate are detected [17, 25]. APC-CDC20 is required for the onset of anaphase [26]. MCAK is involved in spindle regulation, chromosome congression and cell-cycle control, and that reduction in mRNA and protein in a context of permissive SAC predispose to aneuploidy [27]. Disruptions of either complex predispose oocyte to an increased incidence of aneuploidy [10, 28].
In our study, we first demonstrated that detection and quantification of transcripts associated with aneuploidy in single mouse oocyte and sibling PB1 is possible and the relative abundance of mRNA transcripts in a single PB faithfully reflects the relative abundance of that transcript in its sibling oocyte. Reich et al. reported similar results for human oocytes and their sibling PBs. They analyzed over 12,700 unique mRNAs and miRNAs from oocyte samples and compared them with 5,431 mRNAs recovered from the sibling PBs. Their results demonstrated that detection and quantification of mRNA in human PBs is possible and that the human PB mRNA transcriptome reflects that of its sibling MII oocyte [9]. This work and our data demonstrated that PB1 mRNA is being considered as a proxy for the oocyte [29].
In this study, we also found that the abundance of transcripts was lower but detectable in aged PBs versus young PBs. Despite the long-established association between advanced maternal age and the incidence of aneuploidy, the molecular link between female age and germ line genomic instability has remained elusive. The influence of maternal age on global gene expression has been offered as an explanation for the increase in aneuploidy with age [30]. Hamatani et al. found a number of genes involved in cell cycles, DNA stability and chromosome had lower expression with aging [31]. Pan et al. compared the expression profiling of young and old oocytes in mice reveals that transcripts encoding several key components of the SAC, including BUB1 and CDC20, were misexpressed in old oocytes, suggesting that the SAC may be perturbed in old oocytes. They also reveal the similar changes in regulating kinetochore/spindle microtubule interactions, such as MCAK [10]. The patterns of expression in the MII oocytes were mirrored in their PB. So it is not surprise to find that the abundance of transcripts of BUB1, CDC20 and MCAK in PB is lower in aged animals compared with young animals in our study. Filia play a role in maintaining chromosome stability and euploidy in early-cleavage mouse embryogenesis. We found Filia transcripts were significantly decreased in aged PB1. The depletion of maternal stores of Filia causes a high incidence of aneuploidy that results from abnormal spindle assembly, chromosome misalignment, and SAC inactivation [32]. Our prior work demonstrated that there is a significant decrease in the transcript levels of oocyte-specific genes in aged vs. young PB [12]. Taken together a decline in a large number of different processes, including reduction in chromosome cohesion, alteration in cytoskeletal function and permissive checkpoint control, leading to decline in general fitness of older oocytes, may be contribute to age-related aneuploidy and decline in egg quality [33]. Future studies are needed to further evaluate which factors determine oocyte aneuploidy and how these molecules interact with each other during meiosis. The recent advent of improved live-cell imaging, combined with the ability to address the role of specific proteins using gene manipulation and specific inhibition, will accelerate our understanding of aneuploidy.
The PB gene expression approach has major potential benefits. Gene orchestrates virtually every aspect of cellular life. Knowledge of those genes active at a given moment of time in PB may reveal information concerning the biological processes occurring within oocyte. There is growing interest in how information on gene expression could be clinically applied in order to improve the success rates of ART. Indeed it is conceivable that particular patterns of gene expression may be indicative of embryo viability per se. Current evaluations of embryo viability are based on morphology. Analysis transcripts in PB may supplement information on chromosomal status or development competence of a given oocyte and its resulting embryo, which would be of great benefit in IVF treatment, allowing the embryos with the highest implantation potential to be prioritized for transfer to the uterus [18, 34].
In conclusion, this is first study to report, to our knowledge, that provides an insight into the relationship of mRNAs in the PB1 and aneuploidy using a natural reproductive aging mouse model. This analysis is a novel concept to investigate the genesis of chromosome abnormality and could potentially assist in the characterization of mechanisms underlying key molecular origin of female meiotic aneuploidy, which would be of great scientific and clinical value.
Acknowledgments
Financial support
This work was supported by National Institutes of Health through the Eunice Kennedy Shriver National Institute of Child Health and Human Development: RL1HD058295 and U54HD041857.
Disclosure statement
Z.-X. J has nothing to disclose. M. X. has nothing to disclose. T. K.W. has nothing to disclose.
References
- 1.Nagaoka SI, Hassold TJ, et al. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13(7):493–504. [DOI] [PMC free article] [PubMed]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91. [DOI] [PubMed]
- 3.Handyside AH. Molecular origin of female meiotic aneuploidies. Biochim Biophys Acta. 2012;1822(12):1913–20. [DOI] [PubMed]
- 4.Obradors A, Rius M, et al. Errors at mitotic segregation early in oogenesis and at firstmeiotic division in oocytes fromdonor females: comparative genomic hybridization analyses in metaphase II oocytes and their first polar body. Fertil Steril. 2010;93(2):675–9. [DOI] [PubMed]
- 5.Jones KT, Lane SI. Chromosomal, metabolic, environmental, and hormonal origins of aneuploidy in mammalian oocytes. Exp Cell Res. 2012;318(12):1394–9. [DOI] [PubMed]
- 6.Dupont C, Harvey AJ, et al. Expression profiles of cohesins, shugoshins and spindle assembly checkpoint genes in rhesus macaque oocytes predict their susceptibility for aneuploidy during embryonic development. Cell Cycle. 2012;11(4):740–8. [DOI] [PMC free article] [PubMed]
- 7.Fragouli E, Bianchi V, et al. Transcriptomic profiling of human oocytes: association of meiotic aneuploidy and altered oocyte gene expression. Mol Hum Reprod. 2010;16(8):570–82. [DOI] [PubMed]
- 8.Klatsky PC,Wessel GM, et al. Detection and quantification of mRNA in single human polar bodies: a minimally invasive test of gene expression during oogenesis.Mol Hum Reprod. 2010;16(12):938–43. [DOI] [PubMed]
- 9.Reich A, Klatsky P, et al. The transcriptome of a human polar body accurately reflects its sibling oocyte. J Biol Chem. 2011;286(47): 40743–9. [DOI] [PMC free article] [PubMed]
- 10.Pan H,Ma P, et al. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol. 2008;316(2):397–407. [DOI] [PMC free article] [PubMed]
- 11.Merriman JA, Jennings PC, et al. Effect of aging on superovulation efficiency, aneuploidy rates, and sister chromatid cohesion in mice aged up to 15 months. Biol Reprod. 2012;86(2):49. [DOI] [PubMed]
- 12.Jiao ZX, Xu M, et al. Age-associated alteration of oocyte-specific gene expression in polar bodies: potential markers of oocyte competence. Fertil Steril. 2012;98(2):480–6. [DOI] [PMC free article] [PubMed]
- 13.ZuccottiM, BoianiM, et al.Mouse Xist expression begins at zygotic genome activation and is timed by a zygotic clock.Mol Reprod Dev. 2002;61(1):14–20. [DOI] [PubMed]
- 14.Duncan FE, Chiang T, et al. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod. 2009;81(4):768–76. [DOI] [PMC free article] [PubMed]
- 15.Steuerwald N, Cohen J, et al. Quantification of mRNA in single oocytes and embryos by real-time rapid cycle fluorescencemonitored RT-PCR. Mol Hum Reprod. 2000;6(5):448–53. [DOI] [PubMed]
- 16.Verlinsky Y, Cieslak J, et al. Preimplantation diagnosis of common aneuploidies by the first- and second-polar body FISH analysis. J Assist Reprod Genet. 1998;15(5):285–9. [DOI] [PMC free article] [PubMed]
- 17.Steuerwald N, Cohen J, et al. Association between spindle assembly checkpoint expression and maternal age in human oocytes.Mol Hum Reprod. 2001;7(1):49–55. [DOI] [PubMed]
- 18.Wells D, Bermudez MG, et al. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum Reprod. 2005;20(5):1339–48. [DOI] [PubMed]
- 19.Jessberger R. Age-related aneuploidy through cohesion exhaustion. EMBO Rep. 2012;13(6):539–46. [DOI] [PMC free article] [PubMed]
- 20.Howe K, FitzHarris G. Recent insights into spindle function in mammalian oocytes and early embryos. Biol Reprod. 2013;89(3):71doi:10.1095/biolreprod.113.112151. [DOI] [PubMed]
- 21.Pellestor F, Andreo B, et al. Maternal aging and chromosomal abnormalities: new data drawn from in vitro unfertilized human oocytes. Hum Genet. 2003;112(2):195–203. [DOI] [PubMed]
- 22.Chiang T, Duncan FE, et al. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol. 2010;20(17):1522–8. [DOI] [PMC free article] [PubMed]
- 23.Lister LM, Kouznetsova A, et al. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol. 2010;20(17):1511–21. [DOI] [PubMed]
- 24.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21(7):405–12. [DOI] [PubMed]
- 25.Sun SC, Kim NH. Spindle assembly checkpoint and its regulators in meiosis. Hum Reprod Update. 2012;18(1):60–72. [DOI] [PubMed]
- 26.Jin F, Hamada M, et al. Cdc20 is critical for meiosis I and fertility of female mice. PLoS Genet. 2010;6(9):e1001147. [DOI] [PMC free article] [PubMed]
- 27.Eichenlaub-Ritter U, Staubach N, et al. Chromosomal and cytoplasmic context determines predisposition to maternal age-related aneuploidy: brief overview and update onMCAK in mammalian oocytes. Biochem Soc Trans. 2010;38(6):1681–6. [DOI] [PubMed]
- 28.Mailhes JB. Faulty spindle checkpoint and cohesion protein activities predispose oocytes to premature chromosome separation and aneuploidy. Environ Mol Mutagen. 2008;49(8):642–58. [DOI] [PubMed]
- 29.Jiao ZX, Woodruff TK. Follicle microenvironment-associated alterations in gene expression in the mouse oocyte and its polar body. Fertil Steril. 2013;99(5):1453–9. [DOI] [PMC free article] [PubMed]
- 30.Jones KT. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update. 2008;14(2):143–58. [DOI] [PubMed]
- 31.Hamatani T, Falco G, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13(19):2263–78. [DOI] [PubMed]
- 32.Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A. 2009;106(18):7473–8. [DOI] [PMC free article] [PubMed]
- 33.Vogt E, Kirsch-Volders M, et al. Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error. Mutat Res. 2008;651(1–2):14–29. [DOI] [PubMed]
- 34.Jiao ZX, Xu M, et al. Detection and quantification of maternal-effect gene transcripts in mouse second polar bodies: potential markers of embryo development competence. Fertil Steril. 2013;99(7):2055–61. [DOI] [PMC free article] [PubMed]