Abstract
An on-chip lectin microarray based glycomic approach is employed to identify glyco markers for different gastritis and gastric cancer. Changes in protein glycosylation have impact on biological function and carcinogenesis. These altered glycosylation patterns in serum proteins and membrane proteins of tumor cells can be unique markers of cancer progression and hence have been exploited to diagnose various stages of cancer through lectin microarray technology. In the present work, we aimed to study the alteration of glycan structure itself in different stages of gastritis and gastric cancer thoroughly. In order to perform the study from both serum and tissue glycoproteins in an efficient and high-throughput manner, we indigenously developed and employed lectin microarray integrated on a microfluidic lab-on-a-chip platform. We analyzed serum and gastric biopsy samples from 8 normal, 15 chronic Type-B gastritis, 10 chronic Type-C gastritis, and 6 gastric adenocarcinoma patients and found that the glycoprofile obtained from tissue samples was more distinctive than that of the sera samples. We were able to establish signature glycoprofile for the three disease groups, that were absent in healthy normal individuals. In addition, our findings elucidated certain novel signature glycan expression in chronic gastritis and gastric cancer. In silico analysis showed that glycoprofile of chronic gastritis and gastric adenocarcinoma formed close clusters, confirming the previously hypothesized linkage between them. This signature can be explored further as gastric cancer marker to develop novel analytical tools and obtain in-depth understanding of the disease prognosis.
INTRODUCTION
Gastric cancer has a vital contribution in worldwide cancer related death, even it happens to be the second most fatal form of cancer in USA with a 5-year survival rate of less than 4% at late stage.1, 2, 3 Ninety-five percent of malignant gastric tumors are reportedly gastric adenocarcinoma.4 To date, pathological analyses have related most of the gastric adenocarcinomas to gastritis, which is characterized by the inflammation of the epithelial lining of stomach. According to Correa's hypothesis, the development of gastric cancer is a multistep process consisting of a series of sequential steps from gastritis to malignant transformation through chronic active gastritis, chronic atrophic gastritis, intestinal metaplasia, dysplasia, and finally invasive type adenocarcinoma.4, 5, 6 Progression of gastritis occurs gradually, within years to decades.7 Unfortunately, 80% cases of gastric cancer are discovered only at the advanced stage because of the lack of markers characterizing early stages.8 Hence, there exists a need to recognize vulnerable changes which occur in these gastritis for accurate diagnosis of the gastric state. Therefore, it has great importance for evaluating the risk factors in gastric cancer, thus it is important to design and develop a novel biomarker screening technique which would be helpful to a large number of cancer patients and pave the way for personalized medicine.
Towards finding a cancer marker, the carbohydrate decoration or glycosylation pattern of cell surface adhesion glycoproteins, have been closely associated with progression of metastasis. Recent evidences suggest that activity of glycosyltrasferases change with the progression of gastric cancer and associated gastritis. These changes lead to differential expression of N- and O-glycans on the membrane-anchored and secreted glycoproteins.9 Thus, while progressing through several stages of gastric cancer, gastric mucosa promotes stage-specific altered glycan structures on cell surface as well as secretory glycoproteins. These structures could thus be potential biomarkers for different stages of cancer progression.10 Previous researchers have identified several serum glycoprotein biomarkers for gastric cancer (CA19-9, CA72-4, serum pepsinogen I, and α-feto protein). Yet the continuing low survival rate of adenocarcinoma patients shows that the discovery of novel glycan-based biomarkers for different phases of gastric cancer is essential. Moreover, only 1% of total serum glycoproteins are secreted at late stage of cancer and show altered glycosylation pattern10 and other nonrelated secreted glycoproteins mask the specific signatures of gastritis and gastric cancer. Conversely, surface glycoproteins from tissue biopsies could be potential sources of disease specific glycoproteins, which can efficiently be used in the establishment of the marker glycosylation profile or glyco-code for gastric carcinogenesis. To explore this unfathomed domain, in this report, we have analyzed sera and gastric tissue biopsies belonging to four clinical groups, namely normal healthy, chronic atrophic Type B gastritis, chronic Type C gastritis, and adenocarcinoma, to generate the corresponding disease specific glyco-code.
However, diverse complex glycan structures and their varieties of interactions with proteins pose great challenges for the development of disease specific glyco-code. Existing technologies for characterization of the glyco-codes include chromatography and mass spectroscopy. These techniques are inappropriate for high-throughput and systematic evaluation of protein glycosylation pattern.11 In comparison, members of a special class of carbohydrate-binding proteins, lectins, are capable of recognizing specific sugar moieties. For this reason, lectin microarray is now considered as a versatile platform for multiplexed analysis of the glycosylation pattern of patho-biological samples.12, 13, 14, 15, 16, 17, 18, 19 Moreover, one could also intergrate lectin microarrays with a microfluidics device to develop a high-throughput and low sample-volume requiring assay system.20, 21, 22, 23, 24 Lectin microarray is also more suitable and cost effective than the enzyme-linked immunosorbent assay (ELISA)-based screening as the former eliminates the use of specific monoclonal antibodies.25, 26, 27 We have previously investigated the effect of fluid flow on the reaction kinetics between two lectins in a microfluidics-integrated microarray system and showed that microfluidic platform augments lectin-glycan interactions and increases the sensitivity due to the confined reaction environment and controlled advection.22 The study also indicated the optimum design of the fluidic circuit as well as the hydrodynamic flow condition.24 Following the work of Zhao et al., who used lectin microarrays to profile pancreatitis and pancreatic cancer sera,3 in the present study, we combined the same microfluidic platform over several lectins to overcome the sensitivity issues and temporal limitations of lectin microarray (Figure 1) in order to efficiently establish glyco-codes specific to different stage of gastric cancer.
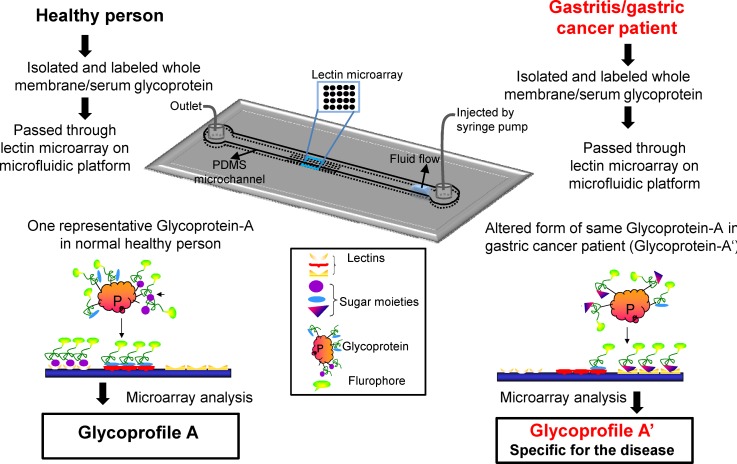
Figure 1.
Illustration of the principles of the experimental models used for lab on a chip based glycoprofiling. (a) Microfluidic channel with lectin microarray at bottom surface. (b) Representative cartoon of fluorescent labeled glycoproteins (isolated from sera or tissue biopsies) bind to different lectin microarrays within the microfluidic platform (c). (d) Fluorescent image of lectin microarray after binding of glycoproteins.
Here, we fabricated microfluidic architecture with a microarray of 17 different lectins at bottom surface of the channel. The lectins were chosen on the basis of their affinity to sugar moieties, involved in cancer and related chronic diseases.15 Next, we isolated sera and cell surface glycoproteins from the patients of the four clinical groups (mentioned above) and quantitatively analyzed glycoprofile for bound fraction of proteins at different lectin spots in our customized microarray on microfluidic platform. The quantitative and statistical inferences of the glycan profile provided us the glyco-code for the four gastric clinical groups. Specially, the expression pattern of GlcNAc-β(1-4)GlcNAc, Galβ(1-3)GalNAc, Neu5Ac, Gcα2,3Galβ1,3(Neu5Acα2,6)GalNAc, N-Ac-β-D-Glucosaminyl oligo, T-antigen, sialyl-T-antigen, Tn-antigen on the surface glycan distinguished the Type-B gastritis group from adenocarcinoma groups, whereas other N-glycans and O-glycans expression pattern distinguished Type-C gastritis from healthy normal. Moreover, glycosylation pattern in Type-B gastritis was more closely to gastric adenocarcinoma than to healthy normal and Type-C gastritis. The glyco-codes obtained from serum and tissue biopsy of each diseased group indicated the ideal specimen sources and comparative effectiveness of invasive and non-invasive screening. The present approach thus provided, for the first time, a simple yet efficient lab-on-a-chip device for detection of altered glycosylation pattern during gastric cancer progression. The development of the glyco-codes may be worth considering for screening the risk factor and relating to the prognosis of adenocarcinoma which would be of great interest from a clinical perspective. In future, as we could envisage, this novel exploration of microfluidics-based lectin microarray could inspire other devices towards complete understanding of the process of transformation from gastritis to gastric adenocarcinoma.
EXPERIMENTAL METHODS
Sample collections and processing
Investigated tissue biopsy samples and their serum samples were collected from patients at Kolkata Medical College Hospital (KMC), Kolkata from April 2009 to June 2012. Biopsy samples of antral mucosa with normal macroscopic appearances obtained at upper gastrointestinal endoscopy from the patients were assessed for the characteristic feature of the four studied clinical groups.28 Before inclusion in our study, all samples were analyzed by a histopathologist for the characteristic appearance of the four clinical groups. Total 39 patients were selected including gastric cancer (Adenocarcinoma of stage I/II/III) patient (n = 6), chronic active and atrophic Type B gastritis patient (n = 15), Type-C chronic gastritis (n = 10), and healthy control (n = 8) for the comparative study. The control group of subjects with histologically normal antral mucosa was also analyzed before incorporating into the study. All tissue and blood samples were collected with informed consent form from all individuals with permission granted by the institutional ethical committee of the hospital. See Table S1 in supplemental material for the summary of patient demographic characteristics and tumor status.29 All tissue and serum samples were labeled with a unique identifier to protect the confidentiality of the patient and were stored in PBS at −80 °C until processing. All the tissue biopsies were cut into two pieces—one for glycoprofiling and other for histopathological analysis. For glycoprofiling of the tissue membrane glycoprotein, whole membrane protein was isolated by two-phase solvent system method using the Membrane Protein Extraction Kit (Biovision, San Francisco, USA). Serum samples were immunodepleted by ProteomeLab IgY-12 proteome partitioning kit (Beckman Coulter, Fullerton, CA). Isolated glycoproteins were labeled with FITC and kept at −80 °C until using. See supplemental material for detailed protocol of sample processing.29
Production of the lectin microarray based microfluidic platform
The lectin microarray was produced as previously described with little modifications.22 Briefly, 0.5 mg/ml solutions of 17 types of different lectins in PBS (pH 7.4) supplemented with 10% glycerol were used as spotting solutions. Among these selected lectins, some had overlapping specificity and some had unique specificity. See Table S2 in supplemental material for the list of lectins and their affinity.29 These 17 lectins were spotted onto amino-propyltriethoxysilane (APTES)-glutaraldehyde coated glass slides in octuplicate using a noncontact microarray spotter Omnigrid® (Genomic Solution, USA) with minimum spot spacing of 100 μm. The glass slides were incubated overnight at 25 °C to allow lectin immobilization. Poly(dimethyl siloxane) (PDMS) microchannels were fabricated through soft lithographic techniques.30 The PDMS microfluidic channels were gently press bonded against the microarray patterned substrate to make the PDMS-glass microchannels. Inlet and outlet fluidic connections were made by inserting 20-gauge blunt-end needles in the punched holes and by silicone tubing. See supplemental material for detail fabrication protocol of microchannel.29
Lectin microarray hybridization
Before glycoprotein hybridization with the lectin microarray, microarrays containing PDMS-glass microchannels were washed thoroughly with probing buffer (25 mM Tris–HCl, pH 7.5, 140 mM NaCl (TBS) containing 2.7 mM KCl, 1 mM CaCl2, 1 mM MnCl2, and 1% Triton X-100), blocked with blocking reagent N102 (NOF Co.) at 20 °C for 1 h. The concentration, volume, and flow rate of each FITC labeled samples were optimized from our previous study to provide maximum fluorescence without saturation of binding sites of immobilized lectins within minimum time.22 For getting high-throughput glycoprofiling, we made the concentration of each membrane protein sample to 250 μg/ml. A volume of 200 μl of each of these samples was flown through separate fabricated microchannels at 50 μl/min flow rate for 4 min. After that, the unbound proteins were washed out with probing buffer flow at the same flow rate for 5 min. Fluorescence images of microarray spots were obtained with a fluorescence microscope (Olympus IX71) equipped with optical filters and a charge-coupled camera cooled (ProgRes® MFcool, Jenoptic Jena) to 0 °C. The fluorescent images were analyzed, and mean signal intensity of at least five most similar spots among octuplicate spots for each lectin was calculated by custom made program using MATLAB, after background subtraction.
Data analysis and clustering
Normalization of microarray data is required to adjust the data from each microarray to account for possible systematic variation in factors such as microarray quality, imaging stability, sample preparation reproducibility, and labeling efficiency. All the microarray spot intensities were normalized by mean normalization method as represented in Scheme S-1 and the corresponding mathematical expression in Eg1.16 The mean signal intensity of a single lectin in individual groups was multiplied by a normalization factor N for each sample, which was calculated by N = 1/μ, where μ is the final mean intensity value of that lectin in control group. Following the use of clustering and principal component analysis (PCA) by Zhao et al.,3 the mean normalized and log transformed data were analyzed by two multivariable analyses; average linkage hierarchical clustering (HC) (using Gene Cluster 3.0 and Java TreeView) and PCA (using Multi Variate Statistical Package (MVSP), UK) were used to provide graphical representations of the relationships among the samples. Unsupervised clustering method was used to achieve the hierarchical relationship of the 39 samples of four clinical groups on the basis of similarities in their glycan expression pattern. In PCA approach, using MVSP software samples were placed as points in a two dimensional Biplot (containing vectors superimposed over the scatter data points) without supervision, on the basis of normalized abundance of glycosylation and their covariance of log transformed data. The direction of an arrow of a particular lectin points toward the samples show maximum binding with the corresponding lectin. Moreover, the length of the vectors signify level of binding.
RESULT AND DISCUSSION
Histopathological diagnosis of the samples
Primary idea about the clinical groups of collected gastric tissue biopsies was obtained from the endoscopic observations and pathological tests. Prior to glycoprofiling, the clinical groups of each tissue biopsy were confirmed by histopathological analysis. Biopsies thus obtained were processed as described in material and methods section. The pathological classification of the patients was carried out from the gastroendoscopic and histopathological appearance (Figure S1).28, 29, 31, 32 Each biopsy was classified by searching the following characteristic appearances: (1) Presence of neutrophils in the mucosa (indicate active gastritis); (2) presence of lymphocytic and plasmacytic inflammatory reactions (indicative features of chronic gastritis); (3) presence of Helicobacer pylori; (4) presence of glandular atrophy also indicative of atrophic gastritis and intestinal metaplasia; and (5) presence of lesions and its topography in the oxyntic mucosa of the body and fundus, in antrum or both locations. Biopsies with the absence of all the above features and appearance of normal intact gastric squamous mucosal epithelium were classified as normal healthy control (Figure S1A).29 See supplementary material29 for the detailed histopathological characteristics of the four clinical groups.33 By means of the histopathological characteristics, 39 biopsies were grouped into four groups namely healthy control (n = 8), H. pylori associated chronic gastritis Type-B (n = 15), Type-C chronic gastritis (n = 10), and gastric adenocarcinoma (n = 6).
Lectin microarray based microfluidic platform efficient in glycoprofiling of glycoprotein
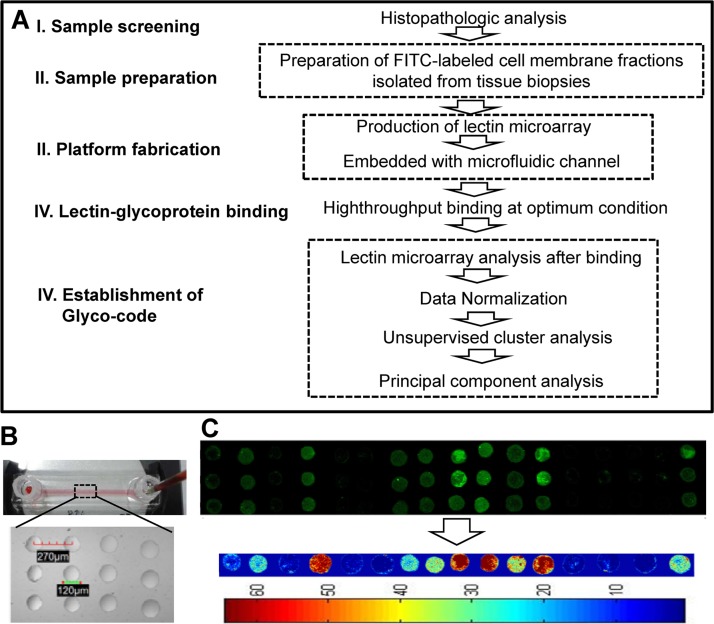
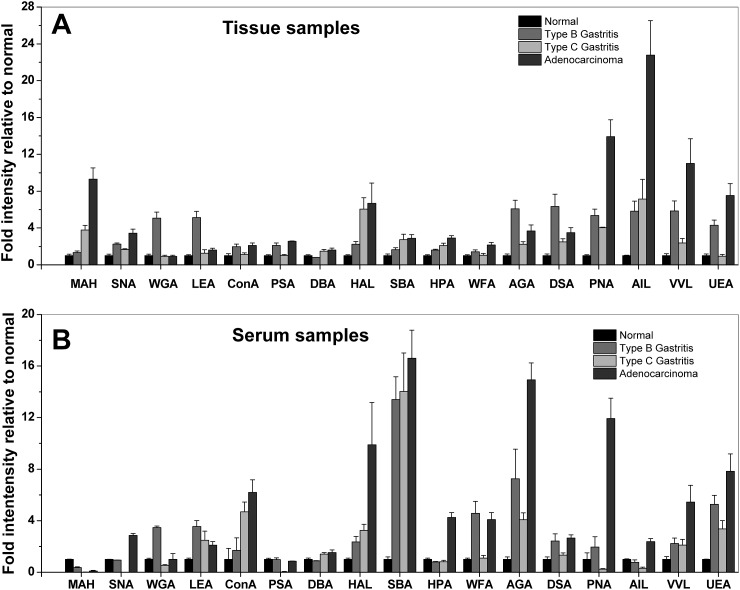
Glycoprofiling study of membrane glycoproteins through microfluidic platform based lectin microarrays was performed as per the scheme depicted in Figure 2a. Briefly, tissue and serum samples were processed as they were collected. Plasma membrane proteins of 10–100 μg were obtained from 10 mg tissue by two phase solvent system method, whereas the retained concentrated immunodepleted serum fraction was about 8% of total serum proteins. Prior to injection into the microfluidic channel, the concentration of the FITC-labeled samples was adjusted to 50 μg/ml. For the lectin microarray, 17 selected lectins were spotted on APTES-gluteraldehyde derivatized glass slides in octuplicate by non-contact microarray spotter.22 The bright-field images of the lectin spots (Figure 2b) within the microchannels clearly showed that the spots were in uniform circular shape. The average spot diameter and inter-spot distance were 120 μm and 270 μm, respectively. PDMS microchannel was pasted on the spotted area in such a way that the microarray remains at the bottom wall of the channels. The optimum flow rate and hybridization time for maximum binding were just as reported previously.22, 34 Therefore, based on this optimization, the labeled glycoprotein was hybridized with lectin microarray by infusing the protein solution into the microchannel at 50 μl/min flow rate for 4 min. Particularly in our approach, the integrated microfluidic channel, here, enables efficient delivery of the labeled glycoproteins to the lectin spot area. In contrast, comparatively slow and diffusion-limited conventional macro-scale microarray devices, the advantages of micro-scale transport process have been employed to increase the mass transfer rate to the lectin spot. Combinedly, the augmented mass transfer rate and high surface to volume ratio at the microconfined reaction reduce the required concentration of lectin and required time for maximum hybridization; both were one order magnitude lower than the conventional macro-scale system approach, signifying the overall increase of two order of sensitivity.22 Further, the easy, fast, and simple methodology allows efficient handling of multiple samples and also multiple times of same sample at a time. Moreover, microarray allows high density of experimental points available from a single chip run that augments the accuracy of the analysis. The amount of specific sugar moieties on the glycoproteins was quantified from the fluorescent intensity of corresponding lectin spots, fluorescent labeled glycoproteins bound to them. We observed that there was some intensity difference between the replicate spots; this might have occurred because of heterogeneous distribution of the lectin molecules during long time incubation of the immobilization process. And this demands further experimental optimization to reduce the heterogeneity. However, here we calculated the average intensity from the fluorescence signal intensity of at least five most similar spots among the octuplicate spots of each lectin by custom written program in MatLab (MathWorks, version 7.8) after performing proper background subtraction that might reduce the ambiguity (Figure 2c). The final normalized mean intensities, i.e., fold increase relative to normal corresponding to each lectin and disease types were plotted in Figure 3 (detail calculation and normalization method explained in average normalization methods in supplementary section).29 The variances between the samples of similar clinical type in Table S3 represent that the variance is acceptable. In order to demonstrate the capability of the platform in distinguishing among the cell surface glycan repertoires of four clinical groups, the increase of fold intensity relative to normal for each of the 17 lectins (Table S2)29 was plotted in Figure 3a. Similarly, the platform was used to establish the signature glycan contents of the four types of clinical sera (Figure 3b). The signal intensities of tissue samples following hybridization were significantly higher than that of serum samples, though the initial protein concentrations of both serum and tissue sample were the same (Figure 3). In Table S4, we represent the variance of fold change among the three diseased clinical groups for each lectin obtained from both tissue and serum samples.29 The highlighted values represent the higher value of variance between the corresponding tissue and serum sample for each lectin. This representation shows that most of the used lectins clearly separate the three disease types in tissue samples but not in serum samples. Moreover, Figure 3b represents that the many lectin binding does not significantly vary in the four clinical groups. Therefore, it is quite ambiguous to use the serum profile in disease specific glycoprofiling or screening. The result suggests that the microfluidic platform can efficiently distinguish the glyco-codes of the four clinical groups obtained from tissue samples. However, the expression of glycan in tissue glycoprotein is more than that in secreted glycoprotein and that may limit the glycoprofiling efficiency of the platform from serum samples. Glycoproteomic analysis of tissue samples using lectin microarray normally has been used to identify the glyco-biomarkers for cancer like choriocarcinoma, pancreatic carcinoma, and colon carcinoma.11, 35, 36 Here within the microfluidic platform, lectin microarray method became sufficiently efficient in the first time establishment of glyco-code for the gastritis and gastric adenocarcinoma from gastric biopsies. Among these lectins, some have overlapping affinity and others have unique specificity, which allows the platform for efficient detection of differential expression of complex glycan structure on glycoproteins. On the basis of specificity, the 17 lectins were categorized into four major groups, namely, sialic acid, N-acetyl-D-galactosamine (GalNAc) and galactose (Gal), glucose/mannose and fucose specific.
Figure 2.
(a) Strategy used to screen the glycosylation patterns and establish the glyco-code using samples of normal, chronic Type B, Type C gastritis, and gastric cancer sera and biopsies. (b) Representative microfluidic platform with fluidic setup. Inset: bright field image of lectin microarray on the ground surface of the channel. (c) Representative image of lectin microarray showing comparison of bound fraction of labeled glycoprotein across all the 17 lectins in triplicate. Matlab processed intensity contour picture of the above fluorescent image.
Figure 3.
Glycan profiles of tissue and serum glycoproteins isolated from normal (n = 8), chronic Type-B (n = 15), Type-C gastritis (n = 10), gastric adenocarcinoma (n = 6) patients. Data represent in Y-axis are the final mean normalized intensity (fold intensity relative to normal) of all samples in a single clinical group. Error bars represent SD and the corresponding p-value for each clinical group combination are tabulated in Table S4.29
Sialic acid expression profile
In particular, the terminal sialic acid residues of surface N-glycan have an important role in cellular adhesion and migration. In N-glycan, sialic acids are normally linked with Gal, GlcNAc, and Glc generally through α 2, 3, and α 2, 6 linkages. Using MAH, SNA, and WGA lectin hybridization, differential expression of sialic acid was observed in the four clinical groups. As shown in Figure 3a, the binding of gastric cancer samples to MAH and SNA lectin was higher than other sample groups. As summarized in Table TABLE I., gastric adenocarcinoma exhibited more frequent expression of α 2, 3, and α 2, 6 linked-sialic acids connected with Gal, GalNAc, and Glc. The Type-B gastritis tissue samples were moderately bound to these two lectins. But maximum binding with WGA lectin (specific to Neu5Ac and GlcNAc-β(1-4)GlcNAcβ(1-4)GlcNAc) revealed that sialic acid and β 1, 4 was expressed higher than α 2, 6 linked GalNAc in Type-B gastritis surface glycan. Figure 3b represents that significant amount of sialic acids with α 2, 6 linked GalNAc and β 1, 4 linked GlcNAc expressions were only observed in adenocarcinoma derived serum. Due to the similar type of sialic acids expression, the healthy control, Type-B and Type-C gastritis serum sample could not be well characterized only on basis of sialic acids expression. Previous studies have shown that aberrant expression of terminal sialic acids, and in particular α 2, 3-linked sialic acids induces the cellular migration and invasive properties of cancer cells which directly leads to increase of metastatic potential.37 The sialic acid expression pattern suggests that gastric mucosa of Type-B gastritis may have potency to transform into adenocarcinoma. Moreover, the presence of β 1, 4 linked GlcNAc and sialic acid both in Type-B gastritis containing tissue surface and adenocarcinoma serum revealed that this residue is secreted to serum from the gastric mucosa during Type-B to adenocarcinoma transformation.
TABLE I.
Color scale of surface glycans expression in tissue biopsies of the four clinical groups. a
| Expressed sugar moieties | Lectins | Healthy normal | Type B gastritis | Gastric adenocarcinoma | Type C gastritis |
|---|---|---|---|---|---|
| Neu5Ac | MAH, SNA, WGA | * | * | *** | |
| Neu5Ac-α(2-6) GalNAc | MAH, SNA | * | * | *** | |
| N-Ac-β-D-Galactosamine | WFA, HPA | *** | ** | *** | |
| N-Ac-α-D-Galactosamine | DBA, HAL, SBA | * | * | *** | |
| GlcNAc-β(1-4)GlcNAc | WGA | * | * | *** | |
| Gal β(1-3) GalNAc | AGA | *** | *** | *** | |
| N-Ac-β-D-Glucosamine | WGA, LEA | * | * | *** | |
| N-Ac-β-D-Lactosamine | DSA | *** | *** | *** | |
| Branched manose/ glucose | ConA | * | * | *** | |
| α-Mannose/Glucose | ConA, PSA | * | * | *** | |
| T-antigen | PNA | * | ** | *** | |
| Sialyl-T-antigen | AIL | * | * | *** | |
| Tn-antigen | VVL | * | * | *** | |
| Fucose | UEA | * | * | *** |
Expression color scale: green: (baseline relative to control; 1 fold increase); gray: 1(a little high (1–2 fold) increase relative to control); yellow: (moderate (2–4 fold) increase relative to control); red: high (>4 fold) increase relative to control. Single asterisk (*), double asterisk (**)I, and triple asterisk (***), respectively, indicating the sugar moieties that have been earlier reported in gastritic cases, earlier reported but not in gastritic cases and maiden observation in gastritic cases in the present work.11, 35, 37, 40, 41, 42, 45, 46, 48, 49, 50, 51, 52, 53
GalNAc and Gal expression profile
A blood group related antigen, Thomsen-Friedenreich (T) antigen is expressed in a majority of carcinomas and is related to invasion and metastasis of cancer.38, 39 In our study, the basic core structure of N-acetyl-α-D-galactosamine (Tn antigen), linked with Ser/Thr (mucin type of linkages) was detected by PNA, AIL and VVL, and sialyl T-antigen were probed by PNA and AIL, respectively. Results showed that most of the gastric cancers have elevated levels of T, sialyl-T, and Tn-antigen in both tissue and serum samples, and sialyl-T antigen expression was most distinct in gastric cancer (Figure 3). This O-glycosylation is critical for lubrication, protection, and controlling the local environment of the cell surface. In the present study, Gal/GalNAc expression profile in different clinical groups was determined by 7 lectins, including lectins SBA and HPA specific for N-Ac-galactosamine, lectin DBA and HAL specific for N-Ac-α-D-galactosamine, and lectins WFA, AGA, and DSA specific for N-Ac-β-D-galactosamine. AGA and DSA also have affinity towards galactose residues linked with GalNAc. Significantly higher binding of adenocarcinoma and Type-C gastritis tissue samples with DBA, HAL, SBA, and HPA lectins underscored the presence and high activity of GalNAc and αGalNAc in gastric tissue than the other groups. These sugar moieties were found to be expressed in moderate amount in Type-B gastritis and normal tissues, as well. However, abundant expression of N-Ac-α-D-galactosamine residue was observed only in adenocarcinoma serum glycan, whilst others showed the same but in lesser quantities. By probing with WFA, AGA, and DSA lectin, it was found that N-Ac-β-D-galactosamine and galactose were expressed to their maximum in adenocarcinoma and Type-B-gastritis tissue biopsies, respectively. However, Type-B serum sample contained both the moieties. Literature suggests that the patients positive for sialyl-T antigen have significantly less survival years than those who are negative for the same. This implies that sialyl-T antigen has an important role in cancer progression.40 During cancerous transformation, the GalNAc-transferases localize to endoplasmic reticulum, which leads to overexpression of Tn-bearing protein in lamellipodium of the migrating cells. Tn-bearing has been reported to enhance migration and invasiveness of cancer cells.39 Similarly N-glycan with Gal/GalNAc glycoconjugates, which can exist in both membrane-bound and secretory forms, have significant roles in cellular adhesion and migration. N-glycan, in addition, regulates the H. pylori adhesion and colonization to the gastric mucosa. These Gal/GalNAc expression profiles suggest that irrespective of α and β form, GalNAc is over-expressed in the surface glycan, indicating its probable role in regulation of the metastatic potential of the tumor cells. Higher expression of αGalNAc may have role in Type C gastritis prognosis through mucin depletion and localization of chronic inflammatory cells. The higher expression of βGalNAc and galactose residues may be a result of H. pylori adhesion and colonization at the gastric mucosa in Type B gastritis.
N-acetyl-D-glucosamine (GlcNAc) expression profile
Once H. pylori infects the stomach, the body defense mechanism is initiated, and this response is facilitated by the de novo expression of α1,4-GlcNAc which inhibits the bacterial adhesion and growth in Type-B gastritis patients.41 But the role and expression levels of β1, 4-GlcNAc in this type of gastritis are not clearly known. Therefore, in order to investigate the glycosylation profile, two lectins WGA and LEA were employed. Result showed that it expressed maximum in Type-B tissue surface glycan, moderately expressed in Type-C gastritis and gastric adenocarcinoma patients, and in very less amount in normal healthy persons (Figure 3a). Binding of adenocarcinoma serum samples only with WGA but non-significantly in LEA suggests an overall low but similar amount of expression of β-GlcNAc in serum glycan of all three types of patients except normal (Figure 3b). Since WGA has overlapping affinity to both sialic acid and β-GlcNAc, the peak fluorescence intensity may have come from sialic acid rather than β-GlcNAc in adenocarcinoma serum samples. The exact role of this glycan in this disease prognosis is yet to be deciphered.
Glucose/mannose expression profile
Decreased mannosidase activity in many type of cancer leads to decreased trimming of high mannose structures, with a corresponding augmented branching of the high mannose core in cell tumor surface and secreted glycoproteins.17 In order to investigate the α-glucose/mannose expression profiles in these four clinical types were investigated by ConA and PSA lectins. Results depict that all tissue sample contained mannose and glucose residues but Type-B gastritis and adenocarcinoma patients expressed very high amount of branched α-mannoside; α-D-glucose residues in their surface glycan, while serum samples showed decremented yet identical amount of these sugar moieties irrespective of the disease types (Figure 3). The elevated α-mannose/glucose expression in gastric cancer surface glycans suggests a premature termination of the glycosylation pathway. This high mannose/glucose is synthesized early in the process in endoplasmic reticulum and later final trimming of the sugar residues occurs in Golgi.42, 43 Because of the secretory mechanism involving both ER (mostly contain high mannose containing immature N-glycans) and Golgi (mostly contain mannose trimmed complex N-glycans), high mannose oligosaccharides are not abundant in serum profiles.44
Fucose expression profile
An elevated level of fucosylation in N-glycans is associated with hepatoma and ovarian cancers.45 In the present investigation, the expression level of α-1, 2-linked fucose was determined by Ulex europaeus agglutinin (UEA). As expected, fucose (UEA) was present in very high amount in adenocarcinoma and comparatively lesser in type-B tissue samples. This fucosylation leads to modifications of the functions of glycoproteins, such as growth factor receptors, adhesion molecules, and extracellular matrices, which in turn may be associated with the potential for the development of malignancy.45 However, in all the three diseased sera, differential expressions of the fucose residue were unnoticeable.
All the glycan expression patterns are tabulated in Table TABLE I.. The above glycoprofiling of four types of samples clearly suggests that each group has a distinct pattern of glycan expression profiles and the individual profiles can be used as characteristic glyco-code of the corresponding disease. Here, we first time observed certain specific glycan expression including N-Ac-β-D-Galactosamine, Gal β(1-3) GalNAc, and N-Ac-β–D-Lactosamine on tissue surface of chronic type-B gastritis and gastric adenocarcinoma. Simultaneously, the result first time demonstrated the glycoprofile in chronic type-C gastritis tissue and serum glycoprotein. Moreover, it is also observed that tissue biopsies are enriched with surface glycan and the variations in their expression pattern are easily distinguishable. However, in blood serums, non-abundant proteins are less apparent in terms of glycan expressions and their variations. Although, serum glycoprofiles may differentiate the diseased samples from normal, these cannot distinguish among the three types of diseases because of their similar patterns of expression in these three diseased conditions. More comprehensive investigation towards the exact role of this aberrant glycosylation in the corresponding disease prognosis is required for detailed understanding of this disease.
Statistical clustering of glycoprofile of four clinical types
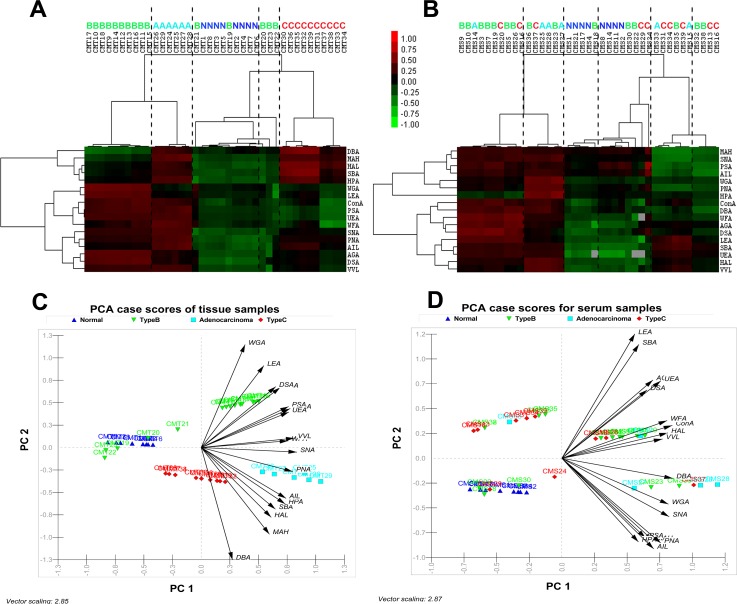
Statistical analysis of the lectin microarray data was executed to highlight the difference among the gastric disease specific glycan expression profiles in gastric biopsies and secreted serum glycoprotein. The statistical methods including HC and PCA were used to group and analyze all the glycoprofiles of different clinical conditions and to provide a graphical representation of relationships among the samples and diseases. In the HC approach, normalized data were log transformed and their pair-wise correlations were obtained by unsupervised clustering method to achieve the hierarchical relationship of the samples on basis of similarities in their glycan expression pattern. In combination with unsupervised plotting, supervised differential abundance approach was used to analyze 39 tissue and serum samples, each assayed in triplicates. As shown in Figure 4a, tissue samples obtained from clinically identical groups shared hierarchically closer relation and were grouped in same cluster due to the similar pattern of glycan expression on their surface. In contrast, serum samples from single clinical group seemed to be clustered with other clinical groups (Figure 4b). The result indicated that serum glycoprofile did not provide sufficient unique glyco-code for establishing as signature biomarker of these four types of clinical groups. Analysis of the HC plot obtained from statistical analysis of lectin microarray delivered many interesting findings. The clustering results for the glycan expression patterns of GlcNAc-β(1-4)GlcNAc, Galβ(1-3)GalNAc, Neu5Ac, Gcα2,3Galβ1,3(Neu5Acα2,6)GalNAc, N-Ac-β-D-Glucosaminyl oligo, T-antigen, sialyl-T-antigen, Tn-antigen generally distinguished the Type-B group from adenocarcinoma groups, whereas clustering due to other N-glycans and O-glycans expression pattern distinguished Type-C gastritis from healthy normal (Figure 4a). Despite the difference and disease pathophysiology, glycosylation pattern in Type-B gastritis was clustered more closely to gastric adenocarcinoma than to normal and Type-C gastritis. In addition, certain few Type-B gastritis samples were also found to be clustered with normal sample groups in HC plot, which might be due to heterogeneity of the individual patients.
Figure 4.
Unsupervised average linkage HC of the lectin array responses to tissue (a) and serum (b) samples of the four clinical groups. The contour plots also represent the glyco-codes of corresponding four clinical groups. Mean-normalized data were analyzed by Cluster 3.0. Positive: red, negative: green, black: zero and grey: missing data. Clustering method: complete linkage. The heat map with clustering was acquired using Java TreeView. The normalized lectin microarray responses to four groups were visualized by PCA. Tissue samples (c) and serum samples (d) were represented in corresponding Biplots (containing vectors superimposed over the scatter data points) to provide graphical representations of the relationships among the samples on the basis of their glycan expression. The direction of an arrow of a particular lectin points toward the samples show maximum binding with the corresponding lectin. Moreover, the length of the vectors signifies level of binding.
Simultaneously, in PCA based statistical approach, data corresponding 39 samples were placed as points in a two dimensional scatter plot without supervision on the basis of normalized abundance of glycosylation and their covariance of log transformed data, and each clinical group was indicated with different symbols (Figure 4c). Positions of the points and their close association, defined by two components namely PC1 and PC2, highlighted the similarity in glycan pattern. PCA based scatter plot showed that data corresponding to tissue samples were clearly segregated into the four clinical conditions by two principal components PC1 and PC2. The analysis demonstrated that the four clinical groups could be clearly distinguished depending on their glycan structures on tissue surface. However, in case of serum samples, no such clear distinction was observed (Figure 4d). This is well in coherence with the analytical data interpreted from HC plot. Additionally, the variation in the different surface glycan expression or binding with different lectins throughout glycosylation space was given by the direction and dimension of the superimposed vectors represented in the bi-plot as arrows (Figure 4d). The direction of an arrow of a particular lectin pointed toward the samples showed maximum binding with the corresponding lectin, and length of arrow was proportional of the level binding. The direction of the arrow indicates the direction of maximum change for that variable and its length is proportional to the rate of change.
Type-B gastritis tissue samples were clustered at the right-hand side of the plot representing higher binding with WGA LEA, DSA, AGA, PSA, and UEA lectins, whereas clustering of adenocarcinoma tissue samples at right-hand side was associated with high binds to PNA, MAH, SNA, ConA, WFA, UEA, VVL, AIL, HPA, PSA lectins (Figure 4c). Normal tissue samples were clustered at the left-hand side of the plot and corresponded to low bindings to all the lectins. Type-C gastritis clustered at middle but lower side of the plot was found to be associated with high LEA binding. Type-B, adenocarcinoma, and Type-C gastritis tissue samples were also clustered separately from normal samples (Figure 4c). However, similar to the observation in the HC plots, few Type-B tissue samples were co-clustered with normal tissue samples due to individual patient heterogeneity. Occurrence of such coherences in the data may happen due to two probable reasons. First and foremost reason could be a simple heterogeneity in individual patients, which often leads to ambiguous results in clinical result. Although unreported as yet, the second reason could be the inability of cells to manifest the infection with a change in glycosylation pattern probably because of a less severity of the infection. This may also signify a transition phase between normal and Type-B gastritis. However, concrete data affirming the fact are not currently available.
Furthermore, a linkage was shown to have developed between Type-B gastritis and gastric adenocarcinoma as per the HC plot, which was later confirmed by the principal component analysis. Further, the closeness between the clusters of Type-B gastritis and gastric adenocarcinoma was more apparent than the other type of gastritis. Glycoproteins obtained from these two clinical groups got bound with two uniquely different sets of lectins. Although different, these lectins correspond to common sugar residues such as Neu5Ac and branched manose/glucose and announced the increment in these residues in the membrane protein fraction of both the diseased tissues. Moreover, it has already been reported that such increments of the above sugar residues occur in cancer cells.17, 46, 47 Similar trends occurring in Type-B gastritis patients were a unique finding presented in this case. It is implicative that, there may also be certain correlation between these two disease subtypes. Earlier report suggests that although direct conversion of H. pylori infection to adenocarcinoma is atypical, adenocarcinoma may be a culminating stage of developing chronic gastritis through atrophic gastritis.7 However, in order to confirm the glycosylation changes appearing in lectin and glycoprotein, mapping through LC-MS and immunostaining should be performed for further authentication.
CONCLUSIONS
Lectin microarray based microfluidic platform provides a comparative analysis of invasive (from tissue biopsy) and non-invasive (serum sample) glyco-biomarker screening of chronic gastritis and stage specific gastric cancer in terms of accuracy and specificity. Results highlighted the potential utility of using the information about altered glycosylation patterns in diseased state from normal healthy group, rather than absolute protein levels, in screening of gastric disease states. Hence, it may be worth to consider in developing a glycan tissue marker for the risk factor screening of a disease like gastric cancer. Differential expressions and link of these glycans on their membrane proteins in cancer and normal tissue lend support to the theory that glycosylation changes may have clinical utility in establishing a specific glyco-code, like in distinguishing amongst cancer and chronic gastritis, cancer and normal tissues and chronic gastritis and normal tissues. Conclusively, it could be said that the study deciphers certain typical and novel glycosylation pattern in the glycoprotein of gastric cancer tissues. The experimentation would have been highly strenuous and time consuming if the high-throughput microfluidic-chip based lectin microarray system was not employed. This robust analytical platform helped us to investigate quickly and systematically the complex “glyco-codes” for different stages of gastric cancer and to combine glycomic information toward a significant understanding in futuristic use of the codes as an early and quick diagnosis tool for gastric cancer and detailed understanding of their role in the disease prognosis. Although, as shown in our study no effective glyco-pattern signature was deduced from serum samples, for other type of disease serum may serve as a good candidate. Finally, a more comprehensive investigation towards the exact role of this aberrant glycosylation in the corresponding disease prognosis is required for detailed understanding of gastric cancer and chronic gastritis.
ACKNOWLEDGMENTS
We would like to thank Ethical Committee of Kolkata Medical College Hospital for necessary clearance, and medical staff of Department of Surgical Gastroenterology, Kolkata Medical College Hospital, Kolkata, for consistence support in sample collections. We acknowledge the financial support of this work through the Department of Biotechnology, Government of India (Grant No. BT/PR9999/NNT/28/85/2007, dated 08/05/2008).
References
- American Cancer Society, Cancer Facts & Figures (American Cancer Society, 2005), pp. 1–60. [Google Scholar]
- Jemal A., Murray T., Ward E., Samuels A., Tiwari R. C., Ghafoor A., Feuer E. J., and Thun M. J., CA. Cancer J. Clin. 55, 10 (2005). 10.3322/canjclin.55.1.10 [DOI] [PubMed] [Google Scholar]
- Zhao J., Patwa T. H., Qiu W., Shedden K., Hinderer R., Misek D. E., Anderson M. A., Simeone D. M., and Lubman D. M., J. Proteome Res. 6, 1864 (2007). 10.1021/pr070062p [DOI] [PubMed] [Google Scholar]
- Schwartz G. K., Semin. Oncol. 23, 316 (1996); seen online at http://www.ncbi.nlm.nih.gov/pubmed/8658215. [PubMed] [Google Scholar]
- Dowall J. E., Willis P., Prescott R., Lamonby S., and Lynch D. A., J. Clin. Pathol. 53, 784 (2000). 10.1136/jcp.53.10.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M., Chen S. L., Karam S. M., Engstrand L., and Gordon J. I., Proc. Natl. Acad. Sci. U.S.A. 105, 4358 (2008). 10.1073/pnas.0800668105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipponen P. and Marshall B. J., Gastroenterol. Clin. 29, 579 (2000). 10.1016/S0889-8553(05)70131-X [DOI] [PubMed] [Google Scholar]
- Layke J. C. and Lopez P. P., Am. Fam. Physician 69, 1133 (2004); seen online at http://www.ncbi.nlm.nih.gov/pubmed/15023013. [PubMed] [Google Scholar]
- Kawaguchi T., Curr. Drug Targets: Cardiovasc. Haematol. Disord. 5, 39 (2005); seen online at http://www.ncbi.nlm.nih.gov/pubmed/15720223. [DOI] [PubMed] [Google Scholar]
- Narimatsu H., Sawaki H., Kuno A., Kaji H., Ito H., and Ikehara Y., FEBS J. 277, 95 (2010). 10.1111/j.1742-4658.2009.07430.x [DOI] [PubMed] [Google Scholar]
- Li Y., Wen T., Zhu M., Li L., Wei J., Wu X., Guo M., Liu S., Zhao H., Xia S., Huang W., Wang P., Wu Z., Zhao L., Shui W., Li Z., and Yin Z., Mol. Biosyst. 9, 1877 (2013). 10.1039/c3mb00013c [DOI] [PubMed] [Google Scholar]
- Olkhov R. V., Weissenborn M. J., Flitsch S. L., and Shaw A. M., Anal. Chem. 86, 621 (2014). 10.1021/ac402872t [DOI] [PubMed] [Google Scholar]
- Gao J., Liu D., and Wang Z., Anal. Chem. 82, 9240 (2010). 10.1021/ac1022309 [DOI] [PubMed] [Google Scholar]
- a Fry S., Afrough B., Lomax-Browne H. J., Timms J. F., Velentzis L. S., and Leathem A. J. C., Glycobiology 21, 1060 (2011). 10.1093/glycob/cwr045 [DOI] [PubMed] [Google Scholar]
- Rakus J. F. and Mahal L. K., Annu. Rev. Anal. Chem. 4, 367 (2011). 10.1146/annurev-anchem-061010-113951 [DOI] [PubMed] [Google Scholar]
- Tateno H., Kuno A., Itakura Y., and Hirabayashi J., in Methods in Enzymology, 1st ed. (Elsevier Inc., 2010), pp. 181–195. [DOI] [PubMed] [Google Scholar]
- Zhao J., Patwa T. H., Lubman D. M., and Simeone D. M., Curr. Opin. Mol. Ther. 10, 602 (2008); seen online at http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2920894. [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi J., Kuno A., and Tateno H., Electrophoresis 32, 1118 (2011). 10.1002/elps.201000650 [DOI] [PubMed] [Google Scholar]
- Dai Z., Zhou J., Qiu S.-J., Liu Y.-K., and Fan J., Electrophoresis 30, 2957 (2009). 10.1002/elps.200900064 [DOI] [PubMed] [Google Scholar]
- Situma C., Hashimoto M., and Soper S. A., Biomol. Eng. 23, 213 (2006). 10.1016/j.bioeng.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Wang L. and Li P. C. H., Anal. Chim. Acta 687, 12 (2011). 10.1016/j.aca.2010.11.056 [DOI] [PubMed] [Google Scholar]
- Roy B., Das T., Maiti T. K., and Chakraborty S., Anal. Chim. Acta 701, 6 (2011). 10.1016/j.aca.2011.05.049 [DOI] [PubMed] [Google Scholar]
- Yang S., Eshghi S. T., Chiu H., Devoe D. L., and Zhang H., Anal. Chem. 85, 10117 (2013). 10.1021/ac4013013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCalla S. E. and Tripathi A., Annu. Rev. Biomed. Eng. 13, 321 (2011). 10.1146/annurev-bioeng-070909-105312 [DOI] [PubMed] [Google Scholar]
- Gupta G., Surolia A., and Sampathkumar S.-G., OMICS 14, 419 (2010). 10.1089/omi.2009.0150 [DOI] [PubMed] [Google Scholar]
- Ng R. C. Y., Roberts A. N., Wilson R. G., Latner A. L., and Turner G. A., Br. J. Cancer 55, 249 (1987). 10.1038/bjc.1987.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanina O. E., Mecklenburg M., Nifantiev N. E., Pazynina G. V., and Bovin N. V., Lab Chip 3, 260 (2003). 10.1039/b305963d [DOI] [PubMed] [Google Scholar]
- Sepulveda A. R. and Patil M., Arch. Pathol. Lab. Med. 132, 1586 (2008); seen online at http://www.archivesofpathology.org/doi/abs/10.1043/1543-2165(2008)132%5B1586%3APATTPD%5D2.0.CO%3B2. [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4882778 for supporting experimental methods, notes, figures, and tables.
- Das T., Maiti T. K., and Chakraborty S., Lab Chip 8, 1308 (2008). 10.1039/b803925a [DOI] [PubMed] [Google Scholar]
- Huang J., Sridhar S., Chen Y., and Hunt R., Gastroenterology 114, 1169 (1998). 10.1016/S0016-5085(98)70422-6 [DOI] [PubMed] [Google Scholar]
- Rugge M. and Genta R. M., Hum. Pathol. 36, 228 (2005). 10.1016/j.humpath.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Sipponen P., Kekki M., Haapakoski J., Ihamäki T., and Siurala M., Int. J. Cancer 35, 173 (1985). 10.1002/ijc.2910350206 [DOI] [PubMed] [Google Scholar]
- Lee M.-Y., Kumar R. A., Sukumaran S. M., Hogg M. G., Clark D. S., and Dordick J. S., Proc. Natl. Acad. Sci. U.S.A. 105, 59 (2008). 10.1073/pnas.0708756105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Masuda K., Banno K., Kobayashi N., Umene K., Nogami Y., Tsuji K., Ueki A., Nomura H., Sato K., Tominaga E., Shimizu T., Saya H., and Aoki D., Oncol. Rep. 31, 1121 (2014). 10.3892/or.2014.2979 [DOI] [PubMed] [Google Scholar]
- Fry S., Afrough B., Leathem A., and Dwek M., in Metastasis Research Protocol (Humana Press, 2012), pp. 267–272. [Google Scholar]
- Wang F.-L., Cui S.-X., Sun L.-P., Qu X.-J., Xie Y.-Y., Zhou L., Mu Y.-L., Tang W., and Wang Y.-S., Cancer Detect. Prev. 32, 437 (2009). 10.1016/j.cdp.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Springer G. F., Science 224, 1198 (1984). 10.1126/science.6729450 [DOI] [PubMed] [Google Scholar]
- Gill D. J., Min K., Chia J., Chyi S., Steentoft C., Clausen H., Bard-chapeau E. A., and Bard F. A., Proc. Natl. Acad. Sci. U.S.A. 110, E3152 (2013). 10.1073/pnas.1305269110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya A., Kikuchi Y., Ando Y., and Abe R., Br. J. Surg. 82, 960 (1995). 10.1002/bjs.1800820734 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Lee H., Nakayama J., and Fukuda M., Glycobiology 19, 453 (2009). 10.1093/glycob/cwp004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- a de Leoz M. L., Young L. J. T., An H. J., Kronewitter S. R., Kim J., Miyamoto S., Borowsky A. D., Chew H. K., and Lebrilla C. B., Mol. Cell. Proteomics 10, M110.002717 (2011). 10.1074/mcp.M110.002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S. A., Dwek M. V., and Schumacher U., Functional and Molecular Glycobiology, 1st ed. (BIOS Scientific Publishers Ltd, Oxford, UK, 2002). [Google Scholar]
- Kronewitter S. R., An H. J., de Leoz M. L., Lebrilla C. B., Miyamoto S., and Leiserowitz G. S., Proteomics 9, 2986 (2009). 10.1002/pmic.200800760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ikeda Y., Miyoshi E., Yaginuma Y., Iahikawa M., and Taniguchi N., Int. J. Cancer 88, 914 (2000). [DOI] [PubMed] [Google Scholar]
- Durand G. and Seta N., Clin. Chem. 46, 795 (2000); seen online at http://www.clinchem.org/content/46/6/795.long. [PubMed] [Google Scholar]
- Jeong H.-H., Kim Y.-G., Jang S.-C., Yi H., and Lee C.-S., Lab Chip 12, 3290 (2012). 10.1039/c2lc40248c [DOI] [PubMed] [Google Scholar]
- Wang P., J. Cancer Mol. 1, 73 (2005). [Google Scholar]
- Pinho S. S., Carvalho S., Marcos-Pinto R., Magalhães A., Oliveira C., Gu J., Dinis-Ribeiro M., Carneiro F., Seruca R., and Reis C. A., Trends Mol. Med. 19, 664 (2013). 10.1016/j.molmed.2013.07.003 [DOI] [PubMed] [Google Scholar]
- Ozcan S., Barkauskas D. A., Renee Ruhaak L., Torres J., Cooke C. L., An H. J., Hua S., Williams C. C., Dimapasoc L. M., Han Kim J., Camorlinga-Ponce M., Rocke D., Lebrilla C. B., and Solnick J. V., Cancer Prev. Res. (Phila). 7, 226 (2014). 10.1158/1940-6207.CAPR-13-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meany D. L. and Chan D. W., Clin. Proteomics 8, 7 (2011). 10.1186/1559-0275-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-W., Ho C.-W., Huang H.-H., Chang S.-M., Popat S. D., Wang Y.-T., Wu M.-S., Chen Y.-J., and Lin C.-H., Proc. Natl. Acad. Sci. U.S.A. 106, 14581 (2009). 10.1073/pnas.0903286106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobata A. and Amano J., Immunol. Cell Biol. 83, 429 (2005). 10.1111/j.1440-1711.2005.01351.x [DOI] [PubMed] [Google Scholar]