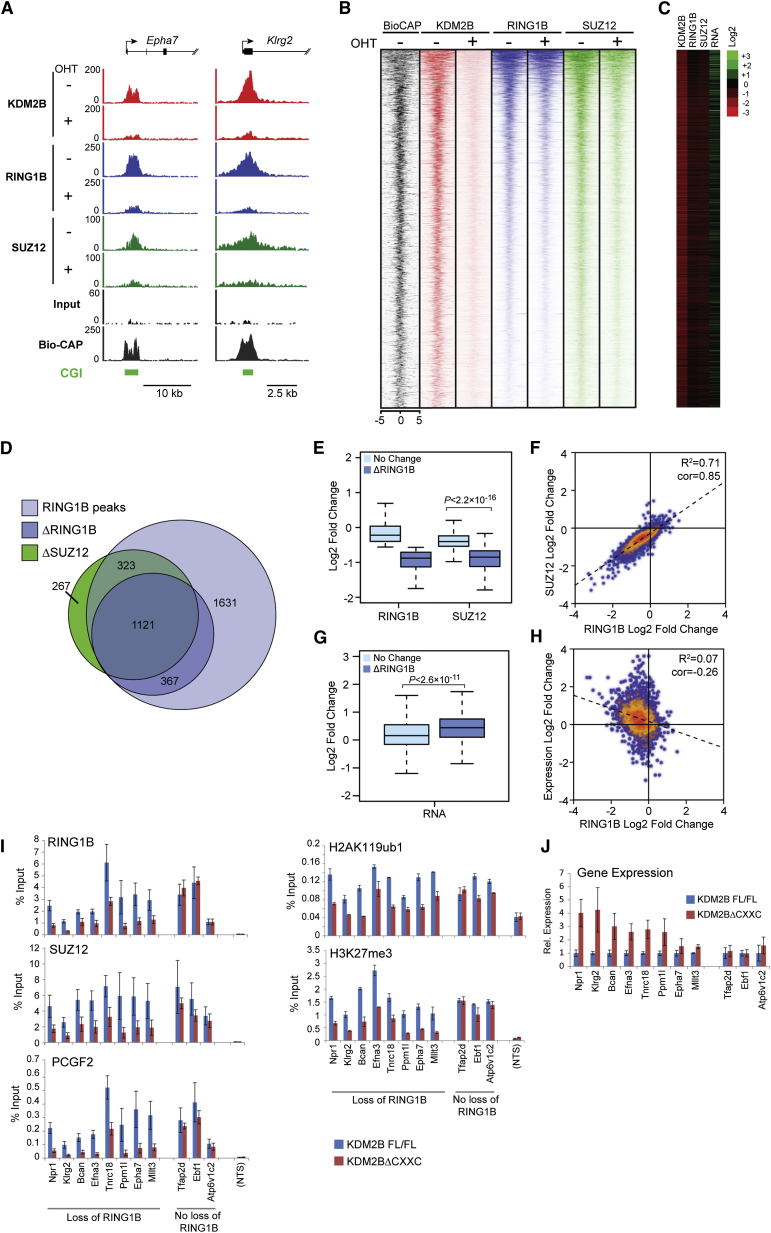
Figure 6.
Failure to Target the PCGF1/PRC1 Complex Leads to a Loss of H2AK119ub1, PRC2, and H3K27me3
(A) Snapshots of ChIP-seq traces for KDM2B, RING1B, and SUZ12 in the Kdm2bfl/fl cells prior to (−OHT) and after 72 hr (+OHT) of tamoxifen treatment at the Epha7 and Klrg2 genes.
(B) Heat map analysis of RING1B peaks (n = 3,488), showing ChIP-seq data for KDM2B, RING1B, and SUZ12 covering a 10 kb region centered over the RING1B peak −OHT and after 72 hr +OHT.
(C) Log2 fold changes in normalized read counts comparing the ChIP-seq and RNA-seq signal −OHT and after 72 hr +OHT.
(D) A Venn diagram showing all RING1B peaks (light blue), intersected with RING1B or SUZ12 peaks that have a greater than 1.5-fold reduction in RING1B/SUZ12 occupancy after 72 hr +OHT treatment (ΔRING1B [dark blue] and ΔSUZ12 [green]).
(E) A box and whisker plot indicating the log2 fold change in RING1B and SUZ12 occupancy at sites that have RING1B changes of greater than 1.5-fold (ΔRING1B) or less than 1.5-fold (No Change) following 72 hr +OHT treatment. The significance of the changes in SUZ12 occupancy at these sites is indicated above the plot.
(F) A scatter plot comparing the log2 fold change of RING1B and SUZ12 at RING1B sites in the Kdm2bfl/fl cells −OHT and after 72 hr +OHT.
(G) A box and whisker plot indicating log2 fold change in gene expression at sites described in (E).
(H) A scatter plot comparing the log2 fold change of gene expression to the fold change in RING1B occupancy at sites that show RING1B alterations.
(I) ChIP analysis for polycomb factors and modifications at regions showing loss of RING1B, no significant loss of RING1B, and a nontarget site (NTS). All ChIP experiments were performed in biological triplicate with error bars showing SEM.
(J) Gene expression analysis for the target genes analyzed by ChIP in (I). RT-PCR was performed in biological triplicate. Error bars show SEM.