Abstract
Background: The aim of this study was to determine the prevalence of virulence-associated genes and enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) analysis of Campylobacter spp. isolated from children with diarrhea in Iran. Methods: A total of 200 stool specimens were obtained from children under 5 years during July 2012 to July 2013. Detection of C. jejuni and C. coli was performed by standard biochemical and molecular methods. The presence of virulence-associated genes and genetic diversity of isolates was examined using PCR and ERIC-PCR analyses. Results: A total of 12 (6%) Campylobacter spp. were isolated from patients including 10 (4.5%) C. jejuni and 2 (1.5%) C.coli. The flaA, cadF and ciaB genes were present in 100% of isolates, while no plasmid of virB11 gene was present in their genome. The prevalence of invasion-associated marker was 100% among C. coli and was not detected in C. jejuni isolates. The distribution of both pldA and the genes associated with cytolethal distending toxin (CDT) was 58.3% in C. jejuni isolates. Seven distinct ERIC-PCR profiles were distinguished in three clusters using ERIC-PCR analysis. Genotyping analysis showed a relative correlation with geographic location of patients and virulence gene content of isolates. Conclusion: To our knowledge, this is the first molecular survey of Campylobacter spp. in Iran concerning genotyping and virulence gene content of both C. jejuni and C. coli. ERIC-PCR revealed appropriate discriminatory power for clustering C. jejuni isolates with identical virulence gene content. However, more studies are needed to clearly understand the pathogenesis properties of specific genotypes.
Key Words: Campylobacter jejuni, Campylobacter coli, Ddiarrhea, Virulence factors
INTRODUCTION
Campylobacter spp. especially C. jejuni and C. coli are considered as potential etiological agents that caused many undiagnosed cases of acute diarrhea in children in developing countries including Iran [1-3]. The purpose of our experiments was to determine the rate and molecular survey of C. jejuni and C. coli virulence and also their diversity that leads to the practical applications to elucidate Campylobacter colonization and the control of this organism. Recurrent exposure to these organisms might raise level of specific immunity correlated with age in developing countries. Therefore, children younger than 5 years of age are mainly affected by these organisms [4].
Above organisms are fastidious and need nutrient-rich-based medium and microaerobic atmosphere [5]. This reason may be the main cause that Campylobacter spp. are not applied in routine diagnostic programs of clinical laboratories in most developing countries. The pathogenicity of Campylobacter species is dependent on their ability to bind to the human intestinal cells and CadF protein. This protein, which is encoded by cadF gene, is responsible for Campylobacter binding to extracellular matrix of human intestinal cells [6]. Another gene, flaA, encodes a flagella protein which mediates motility, colonization, and invasion of gastrointestinal tract and it is essential for establishing human infection.
The ciaB (an invasion protein), virB11 (the IV secretory system), and pldA (an outer membrane phospholipase A) genes encode proteins associated with increased bacterial invasion on cultured epithelial cells; however, their exact roles in invasion have remained to be elucidated [7]. Cytolethal distending toxin (CDT) is encoded by three linked genes, including cdtA, cdtB, and cdtC. In epidemiology of infectious diseases, bacterial typing is of great value in source tracking studies. To analyze the genetic relatedness of C. jejuni, several molecular typing methods based on PCR have been developed. enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) has been shown high discriminatory power, good legibility, and ease of use in most epidemiological investigations.
In this study, we aimed to determine the prevalence of virulence-associated genes and ERIC-PCR analysis of C. jejuni and C. coli isolated from children with diarrhea. CDT is one of the most characterized virulence factors in Campylobacter spp. Pathogenesis induces cell cycle arrest in G2 phase and promotes DNA damage together with apoptotic death in human monocytic cells; therefore, its presence is supposed to be associated with the severity of the disease.
MATERIALS AND METHODS
Study design and data collection procedure. The study was started after obtaining ethical approval from Research and Publication ethics office of Tarbiat Modares University (Tehran, Iran). A total of 200 stool specimens were collected from children with acute diarrhea attended to two major Children’s Hospital Medical Center in Tehran from July 2012 to July 2013. All children under five years of age were included in this study. Children with persistent diarrhea or previous treatment with antibiotics in the last 5 days were excluded from the study. Acute diarrhea was defined as diarrhea which takes about 14 days or less. Demographic data were collected by a co-worker resident in hospital. All samples were transported to the laboratory in a modified Cary-Blair transport medium (5.00 g/L sodium chloride, 1.50 g/L sodium thioglycollate, 1.10 g/L disodium phosphate, and 0.09 g/L calcium chloride, pH 8.4 ± 0.2 at 25ºC( with reduced agar content (1.6 g/L). Identification of C. jejuni and C. coli was performed by the standard culture, Gram staining, and conventional biochemical tests and confirmed by molecular methods [8, 9].
Confirmation of presumptive Campylobacter species by duplex-PCR. Samples were incubated at 42°C for 48-72 h in microaerophilic conditions onto modified charcoal cefoperazone deoxycholate agar medium (10 g/L meat extract, 10 g/L peptone, 5 g/L sodium chloride, 4 g/L bacteriological charcoal, 3 g/L casein hydrolysate, 1 g/L sodium deoxycholate, 0.25 g/L iron (II) sulfate, 0.25 g/L sodium pyruvate, and 15 g/L agar, pH 7.4 ± 0.2 at 25º) plus campylobacter CCDA selective supplement (cefoperazone 3,200 mg/L). DNA templates were extracted by boiling method [10]. Confirmation of Campylobacter spp. was performed by PCR amplification of cadF gene. A duple-PCR was applied for simultaneous detection of hipO and asp genes, specific to C. jejuni and C. coli, respectively [11]. PCR was performed in a 25-μl reaction mixture, containing 10 ng DNA template, 2.5 μl PCR buffer 10×, 200 μM dNTP, 5 mM MgCl2, 0.1 μM each primer, 1 unit of Taq DNA polymerase, and deionized water. The primer sequences and their designation are shown in Table 1. The C. jejuni ATCC 29428 and C. coli ATCC 43478 were used as reference strains.
Table 1.
Primers, PCR conditions, and respective references
| Primers | Sequence (5 → 3') | Target |
PCR condition
|
Amplicon
(bp) |
References | ||
|---|---|---|---|---|---|---|---|
| Denaturin | Annealin | Extension | |||||
| cadFU cadFR |
TTGAAGGTAATTTAGATATG CTAATACCTAAAGTTGAAAC |
cadF | 94οC, 30 s | 43οC, 30 s | 72οC, 30 s | 400 | [11] |
| hipOU hipOR |
GAAGAGGTTTGGGTGGTG AGCTAGCTTCGCATAATAACTTG |
hipO | 94οC, 30 s | 53οC, 30 s | 72οC, 30 s | 735 | [11] |
| aspU aspR |
GGTATGATTTCTACAAAGCGAG ATAAAAGACTATCGTCGCGTG |
asp | 94οC, 30 s | 53οC, 30 s | 72οC, 30 s | 500 | [11] |
| flaAU flaAR |
TTTCGTATTAACACAAATGGTGC CTGTAGTAATCTTAAAACATTTTG |
flaA | 94οC, 45 s | 46οC, 45 s | 72οC, 60 s | 1743 | this study |
| cdtjAU cdtjAR |
AGGACTTGAACCTACTTTTC AGGTGGAGTAGTTAAAAACC |
Cj-cdtA | 94οC, 30 s | 55οC, 30 s | 72οC, 30 s | 631 | [31] |
| cdtj BU cdtjBR |
ATCTTTTAACCTTGCTTTTGC GCAAGCATTAAAATCGCAGC |
Cj-cdtB | 94οC, 30 s | 56οC, 30 s | 72οC, 30 s | 714 | [31] |
| cdtjCU cdtjCR |
TTTAGCCTTTGCAACTCCTA AAGGGGTAGCAGCTGTTAA |
Cj-cdtC | 94οC, 30 s | 55οC, 30 s | 72οC, 30 s | 524 | [31] |
| cdtCAU cdtCAR |
ATTGCCAAGGCTAAAATCTC GATAAAGTCTCCAAAACTGC |
Cc-cdtA | 94οC, 30 s | 55οC, 30 s | 72οC, 30 s | 329 | [31] |
| cdtCBU cdtCBR |
TTTAATGTATTATTTGCCGC TCATTGCCTATGCGTATG |
Cc-cdtB | 94οC, 30 s | 56οC, 30 s | 72οC, 30 s | 413 | [31] |
| cdtCCU cdtCCR |
TAGGGATATGCACGCAAAAG GCTTAATACAGTTACGATAG |
Cc-cdtC | 94οC, 30 s | 55οC, 30 s | 72οC, 30 s | 313 | [31] |
| ciaBU ciaBR |
TGCTAGCCATACTTAGGCGTTTT TTGATAATAGCGGACAATTTGAAA |
ciaB | 94οC, 30 s | 54οC, 30 s | 72οC, 30 s | 610 | this study |
| pldAU pldAR |
AAGCTTATGCGTTTTT TATAAGGCTTTCTCC |
PldA | 94οC, 30 s | 46οC, 30 s | 72οC, 30 s | 913 | [32] |
| iamAU iamAR |
GCGCAAAATATTATCACCC TTCACGACTACTATGCGG |
iam | 94οC, 30 s | 47οC, 30 s | 72οC, 30 s | 518 | [33] |
| virB11U virB11R |
GAACAGGAAGTGGAAAAACTAGC TCCCGCATTGGGCTATATG |
virB11 | 94οC, 30 s | 52οC, 30 s | 72οC, 120 s | 708 | [33] |
| ERICF ERICR | ATGTAAGCTCCTGGGGATTCA AAGTAAGTGACTGGGTGAGCG |
ERIC | 94οC, 30 s | 52οC, 30 s | 72οC, 300 s | variable | [34] |
Detection of virulence/invasion-associated genes. The presence of virulence/invasion-associated genes, including invasion-associated marker (iam), pldA, and ciaB, (responsible for Campylobacter invasion and attachment), virB11 (involved in Campylobacter virulence), and CDT were investigated using specific primers which specifically amplify within the coding region of each gene. The distribution of flaA gene (responsible for Campylobacter attachment) was examined by primers specifically designed according to the flaA locus sequence of C. jejuni strain (GenBank accession no. AF050186.1). Due to the species-allele-specification of CDT sequence, two separate primer pairs were used which were selected according to cdt locus sequence of C. jejuni and C. coli standard strains.
Enterobacterial repetitive intergenic consensus PCR. ERIC-PCR assay was performed according to the method introduced by Versalovic and colleagues [12]. The primer sequences, their designation, and amplification conditions are depicted in Table 1. ERIC- PCR amplification reactions were performed in a 25-μl reaction mixture, containing 10 ng genomic DNA, 2.5 μl reaction buffer 10×, 200 μM dNTP, 5 mM MgCl2, 0.2 μM each primer, and 1 unit Taq DNA polymerase. The reaction was placed in a DNA thermal cycler (Mini Bio-Rad, USA). ERIC-PCR patterns were analyzed based on the Dice similarity coefficient using GelClust software [13].
RESULTS
The study population was made up of 200 children with acute diarrhea, including 110 (55%) male and 90 (45%) female with the mean age of 27.4 months (2.3 years). Among 200 stool samples, Campylobacter spp., including 10 (4.5%) C. jejuni and 2 (1.5%) C. coli were isolated from 12 (6%) samples.
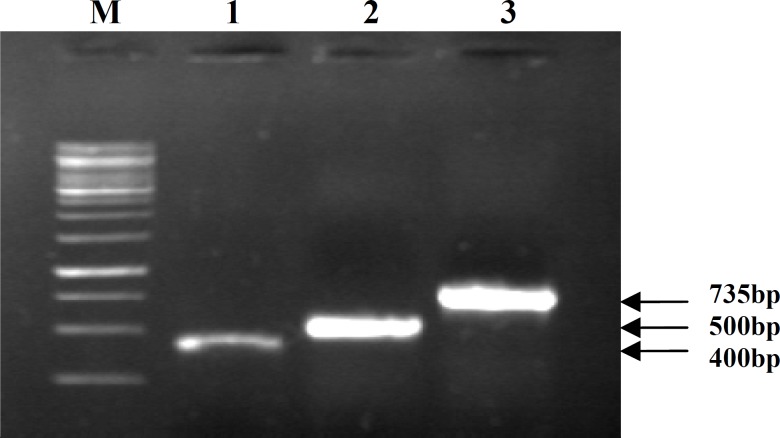
Duplex-PCR assay. The cadF gene was positive in 12 (100%) suspected cultures, and an amplification band of 400 bp was obtained for cadF+ isolates indicative of Campylobacter spp. Duplex-PCR for species identification indicated the presence of 10 (4.5%) C. jejuni and 2 (1.5%) C. coli isolates, respectively (Fig. 1).
Fig. 1.
PCR and Duplex- PCRfor species identification of Campylobacter spp. and C. jejuni/ C.coli isolates, respectively; Lane1, cadF gene; lane 2, asp gene; lane 3, hipO gene, and M, 1 kb DNA size marker
Prevalence of virulence-associated genes and cytolethal distending toxin. All of our Campylobacter isolates harbored flaA and ciaB genes. CDT encoding genes (cdtA, cdtB, and cdtC) involved in CDT production, and pldA was found in 7 (58.3%) of C. jejuni isolates corresponding to amplification bands of 631, 714, 524, and 913 bp, respectively. However, no CDT encoding genes were found among C. coli isolates. The plasmid virB11 gene was also absent among our Campylobacter spp. isolates. The prevalence of iam was 100% among C. coli, while no amplification was obtained for C. jejuni isolates (Table 2).
Table 2.
Prevalence of virulence and toxin genes in C. jejuni and C. coli isolates under study
| Species (No.) |
No. of PCR positive (%)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cad F | hip O | asp | vir B11 | cia B | iam | pld A | fla A | cdt A | cdtB | cdtC | |
| C. jejuni (10) | 10 (100) | 10 (100) | 0 (0) | 0 (0) | 10 (100) | 0 (0) | 7 (58.3) | 10 (100) | 7 (58.3) | 7 (58.3) | 7 (58.3) |
| C. coli (2) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) |
| Total (12) | 12 (100) | 10 (83.4) | 2 (16.6) | 0 (0) | 12 (100) | 2 (16.6) | 7 (58.3) | 12 (100) | 7 (58.3) | 7 (58.3) | 7 (58.3) |
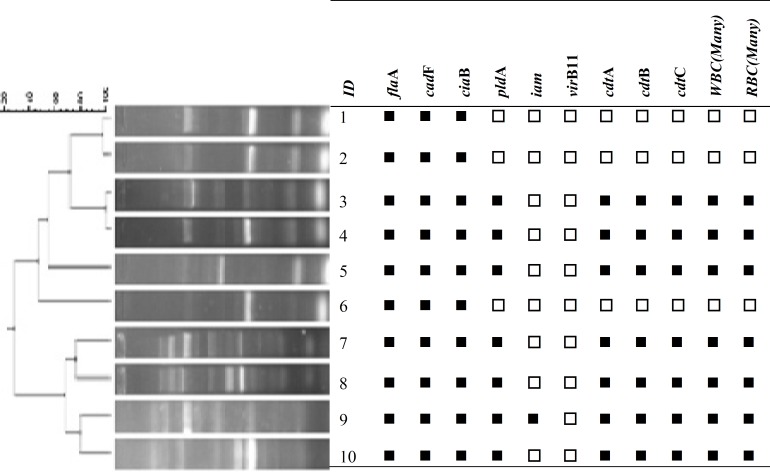
Enterobacterial repetitive intergenic consensus PCR. In total, 10 out of 12 Campylobacter isolates under study produced 7 distinguishable banding profiles by ERIC-PCR genomic fingerprinting, which were corresponded to 10 C. jejuni isolates. Dendrogram of ERIC-PCR was created with UPGMA algorithm, which revealed 3 major clusters with 4, 4, and 2 members. No obvious banding pattern was obtained for C. coli isolates even after multiple attempts (Fig. 2).
Fig. 2.
Dendrogram generated from the ERIC-PCR profiles of C. jejuni isolates from humans in relation to their Profile of virulence-associated genes. Strains 1, 2, 3, and 4 share 80% overall similarity, but strains 7, 8, 9, and 10 share 70% overall similarity. positive Negative
DISCUSSION
Campylobacteriosis is one of the most common bacterial causes of food-borne illness and leading cause of bacterial diarrheal disease in the world [9]. In the present study, the prevalence of Campylobacter spp. among diarrheic children was about 6%. In a few previous studies performed in Iran, the prevalence of Campylobacter spp. was reported to be 8%, 10.8%, and 8.7% in 2007, 2009, and 2011, respectively [3, 14, 15]. This observation shows the trend of Campylobacter infections to be almost unchanged through years in this country.
The prevalence of Campylobacter species among diarrheic children was reported to be 5.4% in Turkey [14], 7% in India [15] and 11.1% in Lebanon [16]. As reported by WHO [17], the incidence of Campylobacteriosis was 9.3 per 1,000 people in Europe America [17]. All isolates in the present study, either C. Jejuni or C .coli, harbored cadF, flaA, and ciaB genes. These genes are essential virulence factors involved in Campylobacter adhesion and colonization to human intestinal epithelial cells during human infection. The ubiquitous existence of the highly conserved cadF gene in 100% of Campylobacter spp. was previously reported by Konkel and coworkers [18] and was subsequently used by other investigators for successful detection of Campylobacter spp. [19, 20].
The prevalence of virulence-associated genes (ciaB and flaA) was reported to be 80-100% in different studies concerning Campylobacter spp. infections in children with moderate to severe diarrhea [21-23]. Similarly, these genes were also detected within all of our isolates. The ciaB and flaA are both involved in maximal invasion of human intestinal cells. This result can justify broadly existence of these gens in clinical Campylobacter spp.
The virB11 gene was not found in any of Campylobacter isolates under study. This finding is in agreement with the studies by other investigators who did not find virB11 gene among Campylobacter isolates of children from Brazil and Bangladesh [23, 24]. However, a few other studies indicated the prevalence of virB11 to be 10.7-22.7% among clinical isolates [21, 25]. This finding emphasizes the low prevalence of type IV secretion system apparatus in
Campylobacter spp., which can probably be due to plasmid basis of the gene.
In this study, the iam gene that codes for iam was detected in 100% of C. coli isolates, while none of C. jejuni isolates harbored the gene. Similar results have been also reported regarding the high prevalence of iam gene among C. coli as well as its absence or low distribution among C. jejuni isolates of children with diarrhea in Brazil [26]. However, several studies demonstrated no substantial difference in its occurrence among the two species. This result shows that iam frequency is controversial [21, 27, 28].
The distribution of both pldA and the genes associated with CDT production (cdtA, cdtB, and cdtC) was 58.3% in C. jejuni isolates, while none of the genes were detected among C. coli isolates. The CDT toxin induces cell cycle arrest in G2 phase and promotes DNA damage; therefore, its presence is supposed to be associated with the severity of the disease in C. jejuni. However, variations which may occur within cdt gene sequences and may affect their detection through amplification methods should not be ignored during interpretation of negative results in C. coli. Moreover, a direct correlation was observed in this study between detection of pldA gene and the presence of white and red blood cells in stool of patients, which may be due to contribution of pldA gene product in pathological changes in intestinal epithelium. In agreement with our results, Rizal and colleagues [21] reported the presence of cdt and pldA genes among 50% and 55% of C. jejuni isolates, respectively, while none of the genes were detected within their C. coli isolates. Seven distinct ERIC-PCR profiles were distinguished from 10 C. jejuni isolates which were located in three clusters by ERIC-PCR. Cluster analysis showed that all isolates (no. 4) within cluster I were isolated from patients who lived in the south of Tehran. Except one isolate, all others within cluster II (no. 3) have also isolated from the patients in a similar geographical location in Tehran (east and center). Moreover, all of the isolates in cluster I and II revealed identical virulence gene content. Two isolates within cluster III were isolated from west of Tehran, but no clear correlation could be determined between ERIC-PCR profile and virulence genes content of isolates within this cluster. Sahilah and colleagues [29] reported that no specific relationship could be extracted between ERIC-PCR analysis and virulence gene content of their C. jejuni isolates. However, Wardak et al. [30] showed that ERIC-PCR could clearly divide C. jejuni and C. coli into two clusters.
ERIC-PCR was unable to type our C. coli isolates which raises the question that to how extent is the typeability power of ERIC-PCR for C. coli strains. This emphasizes that more studies are needed to clearly understand the ability and role of this typing method in C. coli epidemiological studies. However, it is noteworthy that ERIC-PCR analysis has been proved as a well-documented molecular tool in epidemio-logical studies of C. jejuni strains.
To our knowledge, this is the first molecular survey of C. jejuni and C. Coli genotypes in Iran. Nevertheless, further studies are needed to more clearly understand the correlation between virulence-associated genes and specific genotypes of C. jejuni and C. coli clinical isolates.
ACKNOWLEDGMENTS
This work was supported by a grant from Research council of Tarbiat Modares University, and it is a part of Ph.D. thesis of Mahdi Ghorbanalizadgan in medical bacteriology branch. We thank M. Akbari and L. Kashi for their contributions to this study.
References
- 1.Sangaré L, Nikiéma A, Zimmermann S, Sanou I, Congo-Ouédraogo M, Diabaté A, et al. Campylobacter Spp Epidemiology and Antimicrobial Susceptibility in a Developing Country, Burkina Faso (West Africa) Afr J Clin Exp Microbiol. 2012 May;13(2):10–117. [Google Scholar]
- 2.Silva J, Leite D, Fernandes M, Mena C, Gibbs PA, Teixeira P. Campylobacter spp as a Foodborne Pathogen: A Review. Front Microbiol. 2011 Sep;2:200. doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jafari F, Garcia-Gil L, Salmanzadeh-Ahrabi S, Shokrzadeh L, Aslani MM, Pourhoseingholi MA, et al. Diagnosis and prevalence of enteropathogenic bacteria in children less than 5 years of age with acute diarrhea in Tehran children's hospitals. J Infect. 2009 Jan;58(1):21–7. doi: 10.1016/j.jinf.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Koenraad P, Rombouts F, Notermans S. Epidemiological aspects of thermophilic Campylobacter in water-related environments: a review. Water Environ Res. 1997 Jan;69(1):52–63. [Google Scholar]
- 5.Ica T, Caner V, Istanbullu O, Nguyen HD, Ahmed B, Call DR, et al. Characterization of mono- and mixed-culture Campylobacter jejuni biofilms. Appl Environ Microbiol. 2011 Dec;78(4):1033–1038. doi: 10.1128/AEM.07364-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayak R, Stewart TM, Nawaz MS. PCR identification of Campylobacter coli and Campylobacter jejuni by partial sequencing of virulence genes. Mol Cell Probes. 2005 Jun;19(3):187–193. doi: 10.1016/j.mcp.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Konkel ME, Kim BJ, Rivera‐Amill V, Garvis SG. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999 May;32(4):691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- 8.Isenberg HD. Clinical microbiology procedures handbook. 2rd ed. USA: ASM Press; 2007. [Google Scholar]
- 9.Ripabelli G, Tamburro M, Minelli F, Leone A, Sammarco ML. Prevalence of virulence-associated genes and cytolethal distending toxin production in Campylobacter spp isolated in Italy. Comp Immunol Microbiol Infect Dis. 2010 Jul;33(4):355–64. doi: 10.1016/j.cimid.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Mohran ZS, Arthur RR, Oyofo BA, Peruski LF, Wasfy MO, Ismail TF, et al. Differentiation of Campylobacter isolates on the basis of sensitivity to boiling in water as measured by PCR-detectable DNA. Appl Environ Microbiol. 1998 Jan;64(1):363–5. doi: 10.1128/aem.64.1.363-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al Amri A, Senok AC, Ismaeel AY, Al-Mahmeed AE, Botta GA. Multiplex PCR for direct identification of Campylobacter spp in human and chicken stools. J Med Microbiol. 2007 Oct;56(10):1350–5. doi: 10.1099/jmm.0.47220-0. [DOI] [PubMed] [Google Scholar]
- 12.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec;19(24):6823–31. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khakabimamaghani S, Najafi A, Ranjbar R, Raam M. GelClust: a software tool for gel electrophoresis images analysis and dendrogram generation. Comput Methods Programs Biomed. 2013 Aug;111(2):512–8. doi: 10.1016/j.cmpb.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Kayman T, Abay S, Hizlisoy H. Identification of Campylobacter spp. isolates with phenotypic methods and multiplex polymerase chain reaction and their antibiotic susceptibilities. Mikrobiyol Bul. 2013 Apr;47(2):230–9. doi: 10.5578/mb.4532. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee P, Ramamurthy T, Bhattacharya MK, Rajendran K, Mukhopadhyay AK. Campylobacter jejuni in hospitalized patients with diarrhea, Kolkata, India. Emerg Infectious Dis. 2013 Jul;19(7):1155–6. doi: 10.3201/eid1907.121278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dabboussi F, Alam S, Mallat H, Hlais S, Hamze M. Preliminary study on the prevalence of Campylobacter in childhood diarrhoea in north Lebanon. East Mediterr Health J. 2012 Dec;18(12):1225–8. [PubMed] [Google Scholar]
- 17.WHO and FAO. The global view of campylobacteriosis: report of an expert consultation. www.who.int/ foodsafety/publications/foodborne_disease/global_view_campylobacterosis/en/.2012. html.
- 18.Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol. 1999 Mar;37(3):510–7. doi: 10.1128/jcm.37.3.510-517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieczorek K, Denis E, Lynch O, Osek J. Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in Polish slaughterhouses. Food Microbiol. 2013 May;34(1):130–6. doi: 10.1016/j.fm.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Parahitiyawa N, Jin L, Leung W, Yam W, Samaranayake L. Microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev. 2009 Apr;22(1):46–64. doi: 10.1128/CMR.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizal A, Kumar A, Vidyarthi AS. Prevalence of pathogenic genes in Campylobacter jejuni isolates from poultry and human. Internet J Food Safety. 2010 Jan;12:29–34. [Google Scholar]
- 22.Hamidian M, Sanaei M, Azimi-Rad M, Tajbakhsh M, Dabiri H, Zali M-R. fla-typing, RAPD analysis, isolation rate and antimicrobial resistance profile of Campylobacter jejuni and Campylobacter coli of human origin collected from hospitals in Tehran, Iran. Ann Microbiol. 2011 Sep;61(2):315–21. [Google Scholar]
- 23.Quetz Jda S, Lima IF, Havt A, Prata MM, Cavalcante PA, Medeiros PH, et al. Campylobacter jejuni infection and virulence-associated genes in children with moderate to severe diarrhoea admitted to emergency rooms in northeastern Brazil. J Med Microbiol. 2012 Apr;61(Pt 4):507–13. doi: 10.1099/jmm.0.040600-0. [DOI] [PubMed] [Google Scholar]
- 24.Talukder KA, Aslam M, Islam Z, Azmi IJ, Dutta DK, Hossain S, et al. Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J Clin Microbiol. 2008 Apr;46(4):1485–88. doi: 10.1128/JCM.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003 Apr;52(4):345–8. doi: 10.1099/jmm.0.05056-0. [DOI] [PubMed] [Google Scholar]
- 26.Andrzejewska M, Klawe J, Szczepańska B, Śpica D. Occurrence of virulence genes among Campylobacter jejuni and Campylobacter coli isolates from domestic animals and children. Pol J Vet Sci. 2011;14(2):207–11. doi: 10.2478/v10181-011-0031-x. [DOI] [PubMed] [Google Scholar]
- 27.Rozynek E, Dzierzanowska-Fangrat K, Jozwiak P, Popowski J, Korsak D, Dzierzanowska D. Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J Med Microbiol. 2005 Jul;54(7):615–9. doi: 10.1099/jmm.0.45988-0. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho AC, Ruiz-Palacios GM, Ramos-Cervantes P, Cervantes L-E, Jiang X, Pickering LK. Molecular characterization of invasive and noninvasive Campylo- bacter jejuni and Campylobacter coli isolates. J Clin Microbiol. 2001Apr;39(4):1353–9. doi: 10.1128/JCM.39.4.1353-1359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahilah AM, Tuan Suraya TS, Noraida I, Ahmad Azuhairi A, Chai LC, Son R. Detection of virulence genes and enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) analysis among raw vegetables isolates of Campylobacter jejuni. Int Food Res J. 2010 Apr;17:681–90. [Google Scholar]
- 30.Wardak S, Jagielski M. Evaluation of genotypic and phenotypic methods for the differentiation of Campylo- bacter jejuni and Campylobacter coli clinical isolates from Poland PFGE, ERIC-PCR, PCR-flaA-RFLP and MLST. Med Dosw Mikrobiol. 2009;61(1):63–77. [PubMed] [Google Scholar]
- 31.Asakura M, Samosornsuk W, Taguchi M, Kobayashi K, Misawa N, Kusumoto M, et al. Comparative analysis of cytolethal distending toxin (cdt) genes among Campylobacter jejuni, C. coli, and C. fetus strains. Microb Pathog. 2007 May-Jun;42(5):174–83. doi: 10.1016/j.micpath.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Biswas D, Hannon SJ, Townsend HG, Potter A, Allan BJ. Genes coding for virulence determinants of Campylobacter jejuni in human clinical and cattle isolates from Alberta, Canada, and their potential role in colonization of poultry. Int Microbiol. 2011 Mar;14(1):25–32. doi: 10.2436/20.1501.01.132. [DOI] [PubMed] [Google Scholar]
- 33.Müller J, Schulze F, Müller W, Hänel I. PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Vet Microbiol. 2006 Mar;113(1-2):123–9. doi: 10.1016/j.vetmic.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Zulkifli Y, Alitheen N, Son R, Raha AR, Samuel L, Yeap SK, Nishibuchi M. Random amplified poly-morphic DNA-PCR and ERIC PCR analysis on Vibrio parahaemolyticus isolated from cockles in Padang, Indonesia. Int Food Res J. 2009 Apr;16(2):141–50. [Google Scholar]