Abstract
Metabolomics is a systems biology tool providing small molecule signatures of disease etiology. In order to estimate the biologic variability of the human serum metabolome, this study calculated intraclass correlation coefficients (ICCs) for 178 stably-detected metabolites measured by untargeted chromatography/mass spectrometry. We studied a subsample of 60 participants (57% males, 70% Caucasians, aged 73.77±5.3 years) in the Atherosclerosis Risk in Communities (ARIC) Study who provided two fasting serum samples 4–6 weeks apart. The median ICC across all metabolites was 0.60, and 82% of metabolites had at least fair variability (i.e., ICC>= 0.40). There was variation in the medium-term variability among metabolites, with those in the pathways of amino acid and lipid metabolism showing relatively high ICCs, and those in the carbohydrate pathway showing relatively low ICCs. The results of this study provide a valuable resource for future study design and outcome interpretation of mass spectrometry-based metabolomic studies in epidemiology.
Introduction
Metabolomics is an emerging approach to quantitate large numbers of low molecular weight molecules in a biological sample (Lewis et al., 2008), and it allows for global characterization of metabolic networks in an untargeted manner (Wang et al., 2011). Of the current metabolomic studies, most have used a single measure of metabolites for data analysis (Cheng et al., 2012; Suhre et al., 2011; Wang et al., 2011), with the assumption that individual metabolomic measurements are consistent at least over a medium-term period. However, the metabolome is dynamic and sensitive to external stimuli. There may be considerable day-to-day variations that are separate from technical laboratory phenomena or measurement error. Thus, a single measure of a metabolomic profile may have minimal relation to health beyond its physiologic effects over a medium-term period (e.g., one month). Low medium-term variability may also lead to a situation where a metabolomic profile related to disease may be missed (i.e., a false negative finding).
There are little data concerning medium-term, or week-to-week, biologic variability associated with metabolomic biomarkers (Evans et al., 2003; Floegel et al., 2011), although a number of population studies have collected and preserved a multitude of samples appropriate for untargeted metabolomic measurements (Cheng et al., 2012; Sreekumar et al., 2009; Suhre et al., 2010; 2011). The present report describes the intraclass correlation coefficients (ICCs) (Fleiss, 1986) and the coefficients of variation for the stably detected metabolites from a convenience subsample of 60 participants in the Atherosclerosis Risk in Communities (ARIC) Study (Wagenknecht et al., 2009). The study reported here investigated the biological variability of the serum metabolome measured by an untargeted mass spectrometry based protocol over a period of 4–6 weeks apart.
Materials and Methods
Study population and data collection
The ARIC Study is an observational biracial cohort study comprising 15,792 adults aged 45–64 years at baseline from four U.S. communities: Forsyth County, NC; Jackson, MS; suburban Minneapolis, MN; and Washington County, MD (Investigators, 1989). As an ancillary study, the ARIC Carotid MRI Study consisted of 2066 ARIC participants and was aimed to identify novel cellular, metabolic, and genomic correlates of atherosclerotic plaque in the carotid arterial wall. In order to quantify the variation in flow cytometry measurements, each field center recruited 15 volunteers, and they repeated the entire clinic visit at 4–6 weeks of their original visit in 2005–2006 (Catellier et al., 2008). These 60 participants formed the population of current medium-term biologic variability study. Blood samples were collected under standardized conditions (Investigators, 1987), and the participants were assigned alternate phantom IDs for data collection to ensure blinding of sample handling and laboratory measurements. The average age of the participants was 73.77±5.3 years, and 57% of the participants were male. African Americans made up 30% of the sample, while Caucasians made up the remainder (Table 1). This project was approved by the institutional review boards at each site, and written informed consents were provided by all study participants.
Table 1.
Characteristics of the Study Participants
| All (n=60) | Male (n=34) | Female (n=26) | |
|---|---|---|---|
| Age (years) | 73.77±5.3 | 74.21±5.0 | 73.19±5.7 |
| Caucasians (%) | 42 (70.0) | 25 (73.5) | 17 (65.4) |
| Centers (%) | |||
| Forsyth County | 7 (11.7) | 5 (14.7) | 2 (7.7) |
| Jackson | 18 (30.0) | 9 (26.5) | 9 (34.6) |
| Minneapolis | 15 (25.0) | 8 (23.5) | 7 (26.9) |
| Washington County | 20 (33.3) | 12 (35.3) | 8 (30.8) |
statistics: mean±standard error for continuous variables and n (%) for categorical variables.
Assessment of metabolites
Metabolite profiling was completed in June 2010 using serum samples which had been stored at −80°C since collection in 2004–2005. Metabolite levels in the serum were detected and quantified by Metabolon (Durham, USA). An untargeted, gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry based metabolomic quantification protocol was used in detecting and analyzing serum samples (60 pairs of samples for this variability study). This approach identifies and quantifies named compounds whose chemical identity is known, as well as additional unnamed compounds that do not currently have a chemical standard, and these unnamed compounds were tagged beginning with “X” and followed by numbers, such as “X-12345”.
The samples were prepared according to the manufacturer's protocol (Evans et al., 2009; Ohta et al., 2009) and the assay procedures have been described previously (Zheng et al., 2013a). Briefly, the analytical platform used here incorporated two separate ultrahigh performance liquid chromatography/tandem mass spectrometry (UHPLC/MS/MS2) injections; one injection was optimized for basic species, and the other was optimized for acidic species. The resulting MS/MS2 data were searched against an in-house generated authentic standard library that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments, as well as their associated MS/MS spectra for all molecules in the library. The instrument variability in our study was 4%, which was determined by the median relative standard deviation (%RSD) of the injection standards, and the overall process variability was 10% by calculation %RSD of all endogenous metabolites present in 100% of the technical replicate samples. Samples were run over four instrument run days (i.e., 30 samples per day). The day groups were selected serially from the shipment box order, and each metabolite was corrected in run-day blocks by registering the medians to equal one and normalizing each data point proportionately.
Statistical analysis
Metabolites meeting the following criteria were included in this analysis: 1) at least 12 pairs of measureable values across the 60 paired samples; 2) at least moderate blind duplicate repeatability (repeatability coefficient >0.6) calculated from 70 duplicate samples from ARIC participants at the baseline examination (Supplementary Data Text; supplementary material is available online at www.liebertonline.com/omi). Metabolites with low blind duplicate repeatability indicated relatively high measurement variance or instability during sample processing were excluded in order to focus on the components of biologic variances among those metabolites that are stably-detected. There were 178 metabolites, including 106 named and 72 unnamed metabolites, meeting both inclusion criteria. Medium-term variabilities information for all detected metabolites, regardless of their blind duplicate repeatability, is shown in Supplementary Data Table S1.
To assess the medium-term variability of the serum metabolites, the ICCs and 95% confidence intervals were calculated using a repeated-measures mixed ANOVA model (Weir, 2005), with metabolomic platform measurement treated as a fixed effect and participants considered as a random effect. The ICC was calculated as: σb2 / (σb2+σw2), where σb2 is the between-individual variance and σw2 is the within-individual variance over time. SAS procedure PROC GLM was used to partition the total variability into components due to between participant and within participant sources. The ICCs were categorized as: >0.75 excellent variability, 0.40–0.75 fair to good variability, and <0.40 poor variability (Fleiss, 1986). An ICC is a relative measure of variability, and it has been successfully applied in previous analyte studies (Bijari et al., 2011; Ma et al., 1995). The coefficients of variation, calculated by standard deviation/mean, were also presented as a dispersion measurement of the metabolite distribution for both the between- and within-individual components.
The level of a metabolite within a sample may be below the detection limit of the technology. For this medium-term variability study, each metabolite was analyzed in two ways: without and with imputation. Without imputation, only pairs with measureable values in both samples were included in the analysis and the ICC is labeled as ICC_noimp. For the imputation, values missing/below the detected limits (m/bdl) were assigned the lowest detected value for that metabolite in all samples and the ICC is labeled as ICC_imp. To explore the influence of outliers on the variability estimates, a third ICC (i.e., ICC_imp+no_outlier) was calculated for those metabolites with one or more outlier pairs. An outlier pair was defined if the difference between the paired measures is more than 3 standard deviations from the mean of the difference between paired measures among all paired observations for that metabolite.
All statistical analyses were performed in SAS version 9.2 (SAS Institute, Cary, NC).
Results
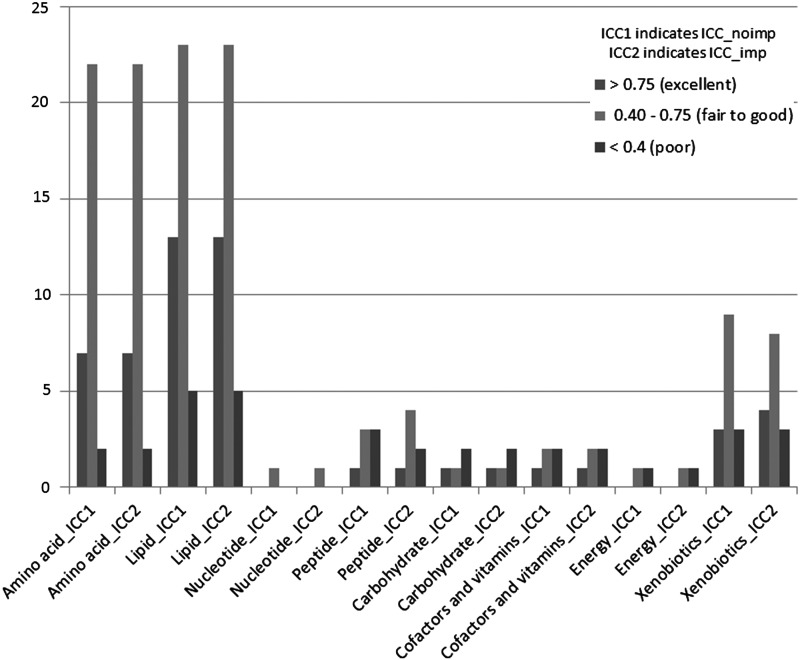
Among the 106 named metabolites, the majority were in the pathways of lipid (N=41) and amino acid (N=31) metabolism. The median ICC_noimp and ICC_imp for all analyzed metabolites were 0.60 and 0.63, respectively; for all named metabolites they were 0.57 and 0.60, respectively; for all unnamed metabolites they were 0.66 and 0.67, respectively. Based on the ICC_noimp, 51 (29%) metabolites had excellent variability, 94 (53%) had fair to good variability, and 33 (19%) had poor variability. Overall, 82% of metabolites had at least fair variability. For the ICC_imp, the distribution of variability groups was very similar (31%, 53%, and 16%, respectively). The distribution of variability groups across annotated pathways is shown in Figure 1. In general, the metabolites in amino acid and lipid pathways had higher variability than the metabolites in other pathways.
FIG. 1.
Distribution of variability groups across pathways among 106 stably-detected named metabolites.
The coefficients of variance and ICCs (ICC_noimp and ICC_imp) of metabolites in the amino acid and lipid pathways are shown in Table 2 and Table 3, respectively. Among metabolites in the amino acid pathway, the medium-term variability was lowest for 3-methylhistidine (ICC_noimp: 0.07 and ICC_imp: 0.15) and highest for indoleacetate (both ICC_noimp and ICC_imp: 0.90), with a median ICC (both ICC_noimp and ICC_imp) of 0.64. Among metabolites in the lipid pathway, variability was lowest for hyodeoxycholate (ICC_noimp: 0.22 and ICC_imp: 0.34) and highest for pregnen-diol disulfate (both ICC_noimp and ICC_imp: 0.99), with a median ICC_noimp of 0.65 and a median ICC_imp of 0.60. Table 4 shows the coefficients of variance and ICCs of metabolites in the remaining pathways (i.e., peptide, xenobiotics, nucleotide, carbohydrate, cofactors and vitamins, and energy).
Table 2.
Coefficients of Variation, and Intraclass Correlations of Measurements for 31 Metabolites in the Amino Acid Pathway
| No imputation method* | Imputation method* | |||||||
|---|---|---|---|---|---|---|---|---|
| Metabolites | Description | N of pairs without m/bdl value | CVw | CVb | ICC (95% CI) | CVw | CVb | ICC (95% CI) |
| N-Acetyl-beta-alanine | Alanine metabolism | 60 | 0.35 | 0.26 | 0.35 (0.16, 0.59) | 0.35 | 0.26 | 0.35 (0.16, 0.59) |
| 2-Aminobutyrate | Butanoate metabolism | 60 | 0.18 | 0.21 | 0.57 (0.40, 0.73) | 0.18 | 0.21 | 0.57 (0.40, 0.73) |
| Creatine | Creatine metabolism | 60 | 0.27 | 0.47 | 0.75 (0.63, 0.85) | 0.27 | 0.47 | 0.75 (0.63, 0.85) |
| 2-Hydroxybutyrate (AHB) | Cysteine metabolism | 60 | 0.25 | 0.27 | 0.56 (0.38, 0.72) | 0.25 | 0.27 | 0.56 (0.38, 0.72) |
| Glutamate | Glutamate metabolism | 60 | 0.27 | 0.23 | 0.42 (0.24, 0.63) | 0.27 | 0.23 | 0.42 (0.24, 0.63) |
| Pyroglutamine | 60 | 0.26 | 0.59 | 0.83 (0.74, 0.90) | 0.26 | 0.59 | 0.83 (0.74, 0.90) | |
| 5-Oxoproline | 60 | 0.10 | 0.17 | 0.74 (0.60, 0.83) | 0.10 | 0.17 | 0.74 (0.60, 0.83) | |
| Betaine | Glycine, serine and threonine metabolism | 60 | 0.14 | 0.19 | 0.64 (0.49, 0.78) | 0.14 | 0.19 | 0.64 (0.49, 0.78) |
| N-Acetylglycine | 44 | 0.58 | 0.58 | 0.50 (0.30, 0.71) | 0.58 | 0.61 | 0.53 (0.35, 0.70) | |
| 3-Methylhistidine | Histidine metabolism | 50 | 0.53 | 0.14 | 0.07 (0.00, 0.84) | 0.60 | 0.25 | 0.15 (0.02, 0.55) |
| Glutaroyl carnitine | Lysine metabolism | 60 | 0.16 | 0.22 | 0.66 (0.51, 0.79) | 0.16 | 0.22 | 0.66 (0.51, 0.79) |
| Pipecolate | 60 | 0.77 | 0.65 | 0.42 (0.23, 0.63) | 0.77 | 0.65 | 0.42 (0.23, 0.63) | |
| 3-(4-Hydroxyphenyl)lactate | Phenylalanine & tyrosine metabolism | 60 | 0.14 | 0.24 | 0.75 (0.62, 0.84) | 0.14 | 0.24 | 0.75 (0.62, 0.84) |
| 3-Phenylpropionate (hydrocinnamate) | 42 | 0.54 | 0.54 | 0.50 (0.29, 0.71) | 0.58 | 0.71 | 0.60 (0.44, 0.75) | |
| p-Cresol sulfate | 60 | 0.34 | 0.36 | 0.53 (0.35, 0.70) | 0.34 | 0.36 | 0.53 (0.35, 0.70) | |
| Phenol sulfate | 60 | 0.54 | 1.14 | 0.81 (0.71, 0.89) | 0.54 | 1.14 | 0.81 (0.71, 0.89) | |
| Phenylacetylglutamine | 60 | 0.32 | 0.53 | 0.74 (0.61, 0.83) | 0.32 | 0.53 | 0.74 (0.61, 0.83) | |
| Phenyllactate (PLA) | 48 | 0.50 | 0.53 | 0.54 (0.34, 0.72) | 0.55 | 0.56 | 0.51 (0.33, 0.69) | |
| 3-Indoxyl sulfate | Tryptophan metabolism | 60 | 0.28 | 0.36 | 0.63 (0.47, 0.76) | 0.28 | 0.36 | 0.63 (0.47, 0.76) |
| Indoleacetate | 60 | 0.20 | 0.61 | 0.90 (0.84, 0.94) | 0.20 | 0.61 | 0.90 (0.84, 0.94) | |
| Indolelactate | 53 | 0.26 | 0.28 | 0.53 (0.34, 0.71) | 0.28 | 0.34 | 0.59 (0.43, 0.74) | |
| Indolepropionate | 54 | 0.42 | 0.58 | 0.65 (0.48, 0.78) | 0.43 | 0.64 | 0.68 (0.54, 0.80) | |
| Kynurenine | 60 | 0.15 | 0.16 | 0.53 (0.36, 0.70) | 0.15 | 0.16 | 0.53 (0.36, 0.70) | |
| Tryptophan betaine | 57 | 0.39 | 0.94 | 0.86 (0.77, 0.91) | 0.39 | 0.98 | 0.86 (0.78, 0.91) | |
| Homostachydrine | Urea cycle; arginine-, proline-, metabolism | 58 | 0.31 | 0.45 | 0.68 (0.53, 0.80) | 0.32 | 0.46 | 0.68 (0.53, 0.80) |
| N-Acetylornithine | 60 | 0.31 | 0.50 | 0.72 (0.58, 0.82) | 0.31 | 0.50 | 0.72 (0.58, 0.82) | |
| N-Methyl proline | 47 | 0.63 | 1.22 | 0.79 (0.66, 0.88) | 0.65 | 1.26 | 0.79 (0.68, 0.87) | |
| Proline | 60 | 0.11 | 0.23 | 0.80 (0.70, 0.88) | 0.11 | 0.23 | 0.80 (0.70, 0.88) | |
| Urea | 60 | 0.18 | 0.17 | 0.49 (0.31, 0.68) | 0.18 | 0.17 | 0.49 (0.31, 0.68) | |
| alpha-Hydroxyisovalerate | Valine, leucine and isoleucine metabolism | 60 | 0.21 | 0.44 | 0.81 (0.71, 0.89) | 0.21 | 0.44 | 0.81 (0.71, 0.89) |
| isobutyrylcarnitine | 60 | 0.32 | 0.36 | 0.56 (0.39, 0.72) | 0.32 | 0.36 | 0.56 (0.39, 0.72) | |
No imputation method included only the pairs with detected values (without missing/bdl values) in both samples; imputation method included all 60 pairs with the missing/bdl values assigned with the lowest detected value for that metabolite in all samples.
ICC indicated intraclass correlation coefficient, CVw indicated the coefficient of within-person variance, and CVb indicated the coefficient of between-person variance. Coefficient of variation=standard deviation/mean.
Table 3.
Coefficients of Variation and Intraclass Correlations of Measurements for 41 Metabolites in the Lipid Pathway
| No imputation method* | Imputation method* | |||||||
|---|---|---|---|---|---|---|---|---|
| Metabolites | Description | N of pairs without m/bdl value | CVw | CVb | ICC (95% CI) | CVw | CVb | ICC (95% CI) |
| Cholate | Bile acid metabolism | 25 | 2.15 | 1.53 | 0.34 (0.10, 0.71) | 2.69 | 2.41 | 0.45 (0.26, 0.65) |
| Deoxycholate | 17 | 1.27 | 1.00 | 0.38 (0.10, 0.78) | 1.31 | 1.02 | 0.38 (0.19, 0.60) | |
| Hyodeoxycholate | 22 | 1.03 | 0.54 | 0.22 (0.03, 0.74) | 1.64 | 1.18 | 0.34 (0.16, 0.58) | |
| Ursodeoxycholate | 37 | 0.72 | 1.49 | 0.81 (0.67, 0.90) | 0.85 | 1.84 | 0.82 (0.73, 0.89) | |
| Glycochenodeoxycholate | 59 | 0.94 | 1.09 | 0.57 (0.40, 0.73) | 0.97 | 1.12 | 0.57 (0.40, 0.73) | |
| Glycocholate | 47 | 1.11 | 1.34 | 0.59 (0.40, 0.76) | 1.22 | 1.50 | 0.60 (0.44, 0.75) | |
| Glycocholenate sulfate | 59 | 0.25 | 0.41 | 0.72 (0.59, 0.83) | 0.26 | 0.40 | 0.71 (0.57, 0.82) | |
| Glycolithocholate sulfate | 58 | 0.57 | 0.79 | 0.66 (0.50, 0.79) | 0.57 | 0.78 | 0.65 (0.50, 0.78) | |
| Taurochenodeoxycholate | 37 | 0.79 | 0.48 | 0.27 (0.07, 0.63) | 1.14 | 0.84 | 0.35 (0.17, 0.59) | |
| Taurocholenate sulfate | 44 | 0.41 | 0.35 | 0.42 (0.22, 0.67) | 0.53 | 0.52 | 0.49 (0.31, 0.68) | |
| Taurodeoxycholate | 29 | 1.24 | 1.15 | 0.46 (0.21, 0.73) | 1.35 | 1.38 | 0.51 (0.33, 0.69) | |
| Taurolithocholate 3-sulfate | 39 | 0.67 | 0.68 | 0.51 (0.29, 0.72) | 0.65 | 0.68 | 0.52 (0.34, 0.69) | |
| 3-Dehydrocarnitine | Carnitine metabolism | 60 | 0.12 | 0.45 | 0.94 (0.90, 0.96) | 0.12 | 0.45 | 0.94 (0.90, 0.96) |
| Carnitine | 60 | 0.08 | 0.17 | 0.82 (0.72, 0.89) | 0.08 | 0.17 | 0.82 (0.72, 0.89) | |
| Decanoylcarnitine | 60 | 0.29 | 0.66 | 0.83 (0.74, 0.90) | 0.29 | 0.66 | 0.83 (0.74, 0.90) | |
| Deoxycarnitine | 59 | 0.09 | 0.17 | 0.77 (0.65, 0.86) | 0.09 | 0.17 | 0.77 (0.65, 0.86) | |
| Laurylcarnitine | 60 | 0.34 | 0.37 | 0.54 (0.37, 0.71) | 0.34 | 0.37 | 0.54 (0.37, 0.71) | |
| Octanoylcarnitine | 60 | 0.38 | 0.85 | 0.84 (0.75, 0.90) | 0.38 | 0.85 | 0.84 (0.75, 0.90) | |
| 3-Hydroxybutyrate (BHBA) | Ketone bodies | 60 | 0.70 | 0.50 | 0.34 (0.16, 0.58) | 0.70 | 0.50 | 0.34 (0.16, 0.58) |
| 3-Carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF) | Fatty acid, dicarboxylate | 60 | 0.47 | 0.80 | 0.74 (0.62, 0.84) | 0.47 | 0.80 | 0.74 (0.62, 0.84) |
| Octadecanedioate | 51 | 0.32 | 0.28 | 0.44 (0.24, 0.66) | 0.39 | 0.30 | 0.37 (0.19, 0.60) | |
| 2-Hydroxyoctanoate | Fatty acid, monohydroxy | 25 | 0.18 | 0.27 | 0.71 (0.48, 0.86) | 0.32 | 0.32 | 0.50 (0.32, 0.68) |
| 5-Dodecenoate (12:1n7) | Unsaturated Fatty acid | 58 | 0.25 | 0.25 | 0.48 (0.30, 0.67) | 0.25 | 0.25 | 0.50 (0.32, 0.68) |
| 10-Nonadecenoate (19:1n9) | 60 | 0.32 | 0.30 | 0.48 (0.29, 0.67) | 0.32 | 0.30 | 0.48 (0.29, 0.67) | |
| Eicosenoate (20:1n9 or 11) | 60 | 0.32 | 0.31 | 0.50 (0.31, 0.68) | 0.32 | 0.31 | 0.50 (0.31, 0.68) | |
| Myristoleate (14:1n5) | 60 | 0.33 | 0.33 | 0.50 (0.31, 0.68) | 0.33 | 0.33 | 0.50 (0.31, 0.68) | |
| Palmitoleate (16:1n7) | 60 | 0.31 | 0.34 | 0.54 (0.37, 0.71) | 0.31 | 0.34 | 0.54 (0.37, 0.71) | |
| Stearidonate (18:4n3) | 59 | 0.51 | 0.54 | 0.53 (0.35, 0.70) | 0.52 | 0.56 | 0.53 (0.36, 0.70) | |
| Dihomo-linoleate (20:2n6) | 60 | 0.30 | 0.31 | 0.52 (0.34, 0.69) | 0.30 | 0.31 | 0.52 (0.34, 0.69) | |
| Eicosapentaenoate (EPA; 20:5n3) | 60 | 0.20 | 0.59 | 0.89 (0.83, 0.93) | 0.20 | 0.59 | 0.89 (0.83, 0.93) | |
| Docosapentaenoate (n6 DPA; 22:5n6) | 59 | 0.21 | 0.31 | 0.69 (0.54, 0.80) | 0.21 | 0.32 | 0.69 (0.55, 0.81) | |
| Cortisol | Steroids | 60 | 0.27 | 0.24 | 0.44 (0.25, 0.64) | 0.27 | 0.24 | 0.44 (0.25, 0.64) |
| 21-Hydroxypregnenolone disulfate | 37 | 0.22 | 1.22 | 0.97 (0.94, 0.98) | 0.23 | 1.27 | 0.97 (0.95, 0.98) | |
| 4-Androsten-3beta,17beta-diol disulfate 2 | 58 | 0.17 | 0.69 | 0.94 (0.90, 0.96) | 0.17 | 0.71 | 0.94 (0.91, 0.97) | |
| 5alpha-Androstan-3beta,17beta-diol disulfate | 49 | 0.23 | 0.63 | 0.88 (0.81, 0.93) | 0.24 | 0.77 | 0.91 (0.85, 0.94) | |
| 5alpha-Pregnan-3beta,20alpha-diol disulfate | 33 | 0.44 | 0.61 | 0.65 (0.44, 0.82) | 0.46 | 0.76 | 0.73 (0.60, 0.83) | |
| Andro steroid monosulfate 2 | 50 | 0.82 | 1.14 | 0.66 (0.49, 0.80) | 0.85 | 1.27 | 0.69 (0.55, 0.81) | |
| Androsterone sulfate | 58 | 0.28 | 0.44 | 0.70 (0.56, 0.82) | 0.29 | 0.47 | 0.72 (0.59, 0.83) | |
| Dehydroisoandrosterone sulfate (DHEA-S) | 60 | 0.20 | 0.93 | 0.96 (0.93, 0.97) | 0.20 | 0.93 | 0.96 (0.93, 0.97) | |
| Pregn steroid monosulfate | 60 | 0.29 | 1.30 | 0.95 (0.92, 0.97) | 0.29 | 1.30 | 0.95 (0.92, 0.97) | |
| Pregnen-diol disulfate | 59 | 0.18 | 1.56 | 0.99 (0.98, 0.99) | 0.18 | 1.58 | 0.99 (0.98, 0.99) | |
No imputation method included only the pairs with detected values (without missing/bdl values) in both samples; imputation method included all 60 pairs with the missing/bdl values assigned with the lowest detected value for that metabolite in all samples.
ICC indicates intraclass correlation coefficient, CVw indicates the coefficient of within-person variance, and CVb indicates the coefficient of between-person variance. Coefficient of variation=standard deviation/mean.
Table 4.
Coefficients of Variation and Intraclass Correlations of Measurements for Metabolites in the Peptide, Xenobiotics, Nucleotide, Carbohydrate, Cofactors and Vitamins, and Energy Pathways
| No imputation method* | Imputation method* | |||||||
|---|---|---|---|---|---|---|---|---|
| Metabolites | Description | N of pairs without m/bdl value | CVw | CVb | ICC (95% CI) | CVw | CVb | ICC (95% CI) |
| Peptide | ||||||||
| gamma-Glutamylalanine | Gamma-glutamyl | 60 | 0.29 | 0.31 | 0.53 (0.35, 0.70) | 0.29 | 0.31 | 0.53 (0.35, 0.70) |
| gamma-Glutamylglutamate | 14 | 0.26 | 0.04 | 0.03 (0.00, 1.00) | 0.39 | – | – | |
| gamma-Glutamyltyrosine | 59 | 0.17 | 0.18 | 0.52 (0.34, 0.70) | 0.18 | 0.19 | 0.54 (0.36, 0.71) | |
| gamma-Glutamylglutamine | 60 | 0.14 | 0.17 | 0.59 (0.42, 0.74) | 0.14 | 0.17 | 0.59 (0.42, 0.74) | |
| gamma-Glutamylthreonine | 51 | 0.30 | 0.23 | 0.39 (0.19, 0.63) | 0.31 | 0.27 | 0.42 (0.24, 0.63) | |
| Aspartylphenylalanine | Dipeptide | 52 | 0.55 | 0.38 | 0.33 (0.14, 0.59) | 0.56 | 0.45 | 0.39 (0.21, 0.61) |
| Pro-hydroxy-pro | 60 | 0.26 | 0.52 | 0.80 (0.70, 0.88) | 0.26 | 0.52 | 0.80 (0.70, 0.88) | |
| Xenobiotics | ||||||||
| 1-Methylurate | Caffeine metabolism | 31 | 0.43 | 0.66 | 0.70 (0.50, 0.85) | 0.49 | 0.86 | 0.76 (0.63, 0.85) |
| 3-Methylxanthine | 27 | 0.38 | 0.42 | 0.55 (0.29, 0.78) | 0.47 | 0.60 | 0.63 (0.46, 0.76) | |
| 1,3,7-Trimethylurate | 13 | 0.40 | 0.57 | 0.67 (0.34, 0.89) | 0.33 | 0.60 | 0.78 (0.66, 0.86) | |
| Caffeine | 49 | 0.67 | 0.74 | 0.55 (0.35, 0.73) | 0.76 | 0.94 | 0.61 (0.44, 0.75) | |
| Theophylline | 41 | 0.55 | 2.94 | 0.97 (0.94, 0.98) | 0.65 | 3.52 | 0.97 (0.95, 0.98) | |
| 1,7-Dimethylurate | 27 | 0.44 | 0.42 | 0.47 (0.22, 0.74) | 0.56 | 0.71 | 0.61 (0.45, 0.76) | |
| Theobromine | 59 | 0.79 | 0.40 | 0.20 (0.05, 0.54) | 0.79 | 0.41 | 0.22 (0.06, 0.53) | |
| 4-Vinylphenol sulfate | Benzoate metabolism | 55 | 1.56 | 0.57 | 0.12 (0.01, 0.62) | 1.63 | 0.70 | 0.15 (0.03, 0.55) |
| Catechol sulfate | 60 | 0.38 | 0.44 | 0.57 (0.40, 0.73) | 0.38 | 0.44 | 0.57 (0.40, 0.73) | |
| Hippurate | 60 | 0.98 | 0.57 | 0.25 (0.09, 0.54) | 0.98 | 0.57 | 0.25 (0.09, 0.54) | |
| 2-Hydroxyhippurate (salicylurate) | 41 | 1.26 | 2.81 | 0.83 (0.72, 0.91) | 1.49 | 3.29 | 0.83 (0.74, 0.89) | |
| 4-Hydroxyhippurate | 43 | 0.34 | 0.43 | 0.62 (0.43, 0.78) | 0.38 | 0.53 | 0.66 (0.51, 0.79) | |
| Salicylate | Drug | 55 | 3.06 | 5.51 | 0.76 (0.64, 0.86) | 2.53 | 4.39 | 0.75 (0.62, 0.84) |
| Piperine | Food component/Plant | 56 | 1.36 | 1.26 | 0.46 (0.27, 0.66) | 1.39 | 1.34 | 0.48 (0.30, 0.67) |
| Thymol sulfate | 25 | 0.48 | 0.43 | 0.45 (0.18, 0.74) | 0.92 | 0.90 | 0.49 (0.31, 0.68) | |
| Nucleotide | ||||||||
| Uridine | Pyrimidine metabolism, uracil containing | 60 | 0.10 | 0.13 | 0.64 (0.48, 0.77) | 0.10 | 0.13 | 0.64 (0.48, 0.77) |
| Carbohydrate | ||||||||
| Gluconate | Nucleotide sugars, pentose metabolism | 59 | 0.27 | 0.15 | 0.23 (0.07, 0.54) | 0.27 | 0.16 | 0.26 (0.09, 0.54) |
| Mannitol | Fructose, mannose, galactose, starch, and sucrose metabolism | 47 | 13.80 | 11.80 | 0.42 (0.22, 0.66) | 13.60 | 12.30 | 0.45 (0.27, 0.65) |
| Glycerate | Glycolysis, gluconeogenesis, pyruvate metabolism | 60 | 0.29 | 0.20 | 0.32 (0.14, 0.57) | 0.29 | 0.20 | 0.32 (0.14, 0.57) |
| 1,5-Anhydroglucitol (1,5-AG) | 60 | 0.13 | 0.34 | 0.87 (0.79, 0.92) | 0.13 | 0.34 | 0.87 (0.79, 0.92) | |
| Cofactors and vitamins | ||||||||
| Biliverdin | Hemoglobin and porphyrin metabolism | 52 | 0.47 | 0.34 | 0.35 (0.16, 0.60) | 0.51 | 0.38 | 0.36 (0.18, 0.59) |
| Pantothenate | Pantothenate and CoA metabolism | 50 | 0.24 | 0.48 | 0.80 (0.69, 0.88) | 0.26 | 0.59 | 0.84 (0.75, 0.90) |
| Threonate | Ascorbate and aldarate metabolism | 60 | 0.24 | 0.30 | 0.61 (0.44, 0.75) | 0.24 | 0.30 | 0.61 (0.44, 0.75) |
| Pyridoxate | Vitamin B6 metabolism | 60 | 31.90 | 12.20 | 0.13 (0.02, 0.58) | 31.90 | 12.20 | 0.13 (0.02, 0.58) |
| gamma-CEHC | Metabolite of Vitamin E | 48 | 0.33 | 0.29 | 0.44 (0.23, 0.66) | 0.35 | 0.41 | 0.58 (0.41, 0.73) |
| Energy | ||||||||
| Malate | Krebs cycle | 53 | 0.31 | 0.19 | 0.27 (0.09, 0.57) | 0.32 | 0.22 | 0.31 (0.13, 0.57) |
| Succinylcarnitine | 59 | 0.17 | 0.24 | 0.67 (0.51, 0.79) | 0.17 | 0.23 | 0.64 (0.49, 0.78) | |
No imputation method included only the pairs with detected values (without missing/bdl values) in both samples; imputation method included all 60 pairs with the missing/bdl values assigned with the lowest detected value for that metabolite in all samples.
ICC indicated intraclass correlation coefficient, CVw indicated the coefficient of within-person variance, and CVb indicated the coefficient of between-person variance. Coefficient of variation=standard deviation/mean.
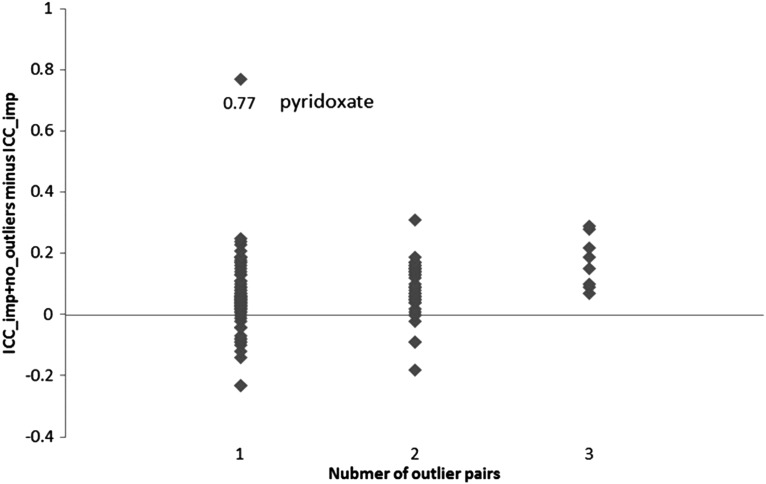
There were 136 metabolites that had at least one outlier pair of measures (8 had three pairs, 40 had two pairs, and the rest had one pair). The median difference between ICC_imp and ICC_imp+no_outlier was 0.06, with a range of −0.23 (theophylline and X - 07765_201) to 0.77 (pyridoxate) (Fig. 2). The difference between ICC_imp and ICC_imp+no_outlier was largest for the metabolite pyridoxate (difference=0.77), which had only one extreme outlier pair. The detailed information of ICC_imp+no_outlier for each metabolite is shown in Supplementary Data Table 1.
FIG. 2.
The distribution of difference between ICC_imp and ICC_imp+no_outlier among 136 metabolites that had at least one outlier pair of observations.
Discussion
Applications of untargeted metabolomics to the study of human disease have begun to emerge (Pirman et al., 2013; Wetmore et al., 2010; Wikoff et al., 2007; Zheng et al., 2013a; 2013b), but few studies have investigated the medium-term variability of the metabolomic measurements. We report that approximately 80% of the 178 stably-detected metabolites had at least fair medium-term variability (i.e., ICC≥0.40). Previous similar studies focused on selected metabolites of interests (Floegel et al., 2011), were from urine samples (Saude et al., 2007), or explored the long-term (i.e., ∼1 year) variability (Sampson et al., 2013; Townsend et al., 2013). The data presented here are to our knowledge the first medium-term variability study of the untargeted human serum metabolome, and it provides valuable information for the design and interpretation of future metabolomic, epidemiologic, and clinical studies.
The variability results for several metabolites, such as bile acids, deserve discussion as examples. Bile acids are commonly measured as routine tests for the screening and diagnosis of hepatobiliary disease (Steiner et al., 2011). In this study, the median ICC_noimp and median ICC_imp of metabolites in the bile acids pathway were 0.49 and 0.52, respectively. In general, these metabolites had considerable between-individual and within-individual variances among the endogenous metabolites. This phenomenon may partially result from activity of gut microflora (Swann et al., 2011). The levels of (glycine- and taurine-) conjugated bile acids mainly vary depending on food intake (Steiner et al., 2011) and the conjugated bile acids in this report had generally fair variability. The levels of the unconjugated bile acids are influenced by diurnal changes independent of food intake (Steiner et al., 2011). In this study, even though the samples were collected at the same time of day, the unconjugated bile acids had generally poor variability except for ursodeoxycholate. Previous studies relating bile acid profiles to disease have yielded inconsistent results (Abrams et al., 1982; Bennion and Grundy, 1977; Brufau et al., 2010; Steiner et al., 2011), and we speculate the relatively low over-time ICC may partially contribute to this phenomenon.
The second example is metabolites in the sex steroids pathway, for which the medium-term variability were generally good or excellent. Compared to a previous report in menstruating females (Shultz et al., 2011), the results from our males and postmenopausal females showed higher variability, suggesting that a single sample may be enough to measure their association with disease among an elderly population. We further explored the difference in variability across genders (Supplementary Data Table S2). Progestagens, such as pregn steroid monosulfate, 21-hydroxypregnenolone disulfate, and pregnen-diol disulfate, had higher ICCs, while 5α-pregnan-3β,20α-diol disulfate had lower ICCs in males than that in females. In general, androgens (andro steroid monosulfate 2, androsterone sulfate, 5α-androstan-3β,17β-diol disulfate and 4-androsten-3β,17β-diol disulfate 2, dehydroisoandrosterone sulfate (DHEA-S) in this study) had higher ICCs in females than that in males.
We identified two previous studies that reported over-time variability of human untargeted serum metabolomic measurements (Sampson et al., 2013; Townsend et al., 2013). The median unimputed ICC of 0.60 from 178 metabolites collected 4–6 weeks apart reported here is higher than the median variability of 0.43 from 385 metabolites measured on nonfasting samples from 60 Chinese females over a 1-year period (Sampson et al., 2013). It is similar to that of 0.58 from 210 metabolites (Townsend et al., 2013) over ∼1.5 years reported by Townsend et al. in a sample of 80 white Americans. The metabolites in the amino acid and lipid pathways have higher variability than the metabolites in other pathways (Fig. 1), and this may be expected because amino acids and lipids are relatively endogenous and genetically regulated compared to the metabolites in carbohydrate and xenobiotic pathways that are more likely to be influenced by the dietary intake and other environmental factors. This phenomenon may partially explain the observation that prominent findings in metabolomic biomarker discovery are often amino acids and lipids (Menni et al., 2013; Newgard et al., 2009; Wang et al., 2011).
There are a large number of factors that may influence the over-time variability of metabolites, including the time between measurements, selection of the metabolites, and the age, race, gender, life style, and overall health status of the study sample (Chambless et al., 1992). Similar to the previous studies (Floegel et al., 2011; Sampson et al., 2013), a limitation of the current report is the relatively small sample size, and restricted capability to assess these potential influences on the variability of metabolites. The study has been conducted among elderly ARIC participants in the U.S.; special attention should be paid when we generalize results to other populations.
There may be bias in studies of association between a single metabolite measurement and disease when the within-individual variance of the metabolite, including biology variance and measurement error, is large compared to the between-individual variance (i.e., ICC <0.5) (Floegel et al., 2011; Verghese et al., 2011). In simple linear or logistic regressions, it leads to bias toward the null hypothesis; that is, the size of relationship between the metabolite and the outcome under consideration is underestimated (Fuller, 1987). There are two possible ways to solve this problem (Chambless et al., 1992). One is to decrease the measurement variation by refining laboratory processes or making multiple measurements. The second solution is to adjust the estimate of relationships between a metabolite and disease for the amount of within-individual variation in the independent variable (e.g., the metabolite). For simple regressions this is accomplished by replacing the ordinary estimate 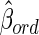 with
with 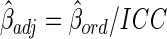 (Chambless et al., 1992). Therefore, for metabolites with low ICCs as the potential predictor variable of interest, the above equation suggests the way to adjust for ICC in estimating the relationship between a metabolite and disease. However, the true ICC of a metabolite over a certain period is seldom known.
(Chambless et al., 1992). Therefore, for metabolites with low ICCs as the potential predictor variable of interest, the above equation suggests the way to adjust for ICC in estimating the relationship between a metabolite and disease. However, the true ICC of a metabolite over a certain period is seldom known.
Conclusions
We report the medium-term variability for untargeted metabolomic measurements in the human serum from 60 individuals from four U.S. communities having two fasting samples collected 4–6 weeks apart. The median ICC was 0.60, and imputation of values below the detection limit had little impact on this estimate, and as expected, the impact of outliers in ICC varies among metabolites. There was variation among metabolites in this estimate with those in the pathways of amino acid and lipid metabolism showing higher ICCs, and those in the carbohydrate pathway showing lower ICCs. The results of this study serve as a resource to aid future study design and result interpretation.
Supplementary Material
Abbreviations Used
- ARIC
Atherosclerosis Risk in Communities
- DHEA-S
dehydroisoandrosterone sulfate
- ICCs
intraclass correlation coefficients
Acknowledgments
The authors thank the staff and participants of the Atherosclerosis Risk in Communities (ARIC) Study for their important contributions. The ARIC Study is a collaborative study supported by the National Heart, Lung, and Blood Institute, National Institutes of Health, Contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN2682011-00008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C. The metabolomics research was sponsored by the National Human Genome Research Institute (3U01HG004402-02S1). YZ and BY are supported in part by a training fellowship from the Burroughs Wellcome Fund (Grant 1008200).
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Abrams JJ, Ginsberg H, and Grundy SM. (1982). Metabolism of cholesterol and plasma triglycerides in nonketotic diabetes mellitus. Diabetes 31, 903–910 [DOI] [PubMed] [Google Scholar]
- Bennion LJ, and Grundy SM. (1977). Effects of diabetes mellitus on cholesterol metabolism in man. N Engl J Med 296, 1365–1371 [DOI] [PubMed] [Google Scholar]
- Bijari PB, Antiga L, Wasserman BA, and Steinman DA. (2011). Scan-Rescan reproducibility of carotid bifurcation geometry from routine contrast-enhanced MR angiography. J Magn Reson Imaging 33, 482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brufau G, Stellaard F, Prado K, et al. (2010). Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology 52, 1455–1464 [DOI] [PubMed] [Google Scholar]
- Catellier DJ, Aleksic N, Folsom AR, and Boerwinkle E. (2008). Atherosclerosis Risk in Communities (ARIC) Carotid MRI flow cytometry study of monocyte and platelet markers: Intraindividual variability and reliability. Clin Chem 54, 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambless LE, McMahon RP, Brown SA, Patsch W, Heiss G, and Shen YL. (1992). Short-term intraindividual variability in lipoprotein measurements: The Atherosclerosis Risk in Communities (ARIC) Study. Am J Epidemiol 136, 1069–1081 [DOI] [PubMed] [Google Scholar]
- Cheng S, Rhee EP, Larson MG, et al. (2012). Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 125, 2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, Dehaven CD, Barrett T, Mitchell M, and Milgram E. (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81, 6656–6667 [DOI] [PubMed] [Google Scholar]
- Evans DA, Bennett DA, Wilson RS, et al. (2003). Incidence of Alzheimer disease in a biracial urban community: Relation to apolipoprotein E allele status. Arch Neurol 60, 185–189 [DOI] [PubMed] [Google Scholar]
- Fleiss J. (1986). The Design and Analysis of Clinical Experiments. John Wiley & Sons, New York, NY [Google Scholar]
- Floegel A, Drogan D, Wang-Sattler R, et al. (2011). Reliability of serum metabolite concentrations over a 4-month period using a targeted metabolomic approach. PLoS One 6, e21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W. (1987). Measurement Error Models. John Wiley, New York [Google Scholar]
- Investigators TA. (1987). Atherosclerosis Risk in Communities Study. Operations manual. No. 9: Hemostasis determinations, version 1.0. National Heart, Lung, and Blood Institute., Bethesda, MD [Google Scholar]
- Investigators TA. (1989). The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol 129, 687–702 [PubMed] [Google Scholar]
- Lewis GD, Asnani A, and Gerszten RE. (2008). Application of metabolomics to cardiovascular biomarker and pathway discovery. J Am Coll Cardiol 52, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Folsom AR, Eckfeldt JH, Lewis L, and Chambless LE. (1995). Short- and long-term repeatability of fatty acid composition of human plasma phospholipids and cholesterol esters. The Atherosclerosis Risk in Communities (ARIC) Study Investigators. Am J Clin Nutr 62, 572–578 [DOI] [PubMed] [Google Scholar]
- Menni C, Fauman E, Erte I, et al. (2013). Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62, 4270–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9, 311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Masutomi N, Tsutsui N, et al. , (2009). Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol 37, 521–535 [DOI] [PubMed] [Google Scholar]
- Pirman DA, Efuet E, Ding XP, et al. (2013). Changes in cancer cell metabolism revealed by direct sample analysis with MALDI mass spectrometry. PLoS One 8, e61379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JN, Boca SM, Shu XO, et al. (2013). Metabolomics in epidemiology: Sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev 22, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saude E, Adamko D, Rowe B, Marrie T, and Sykes B. (2007). Variation of metabolites in normal human urine. Metabolomics 3, 439–451 [Google Scholar]
- Shultz SJ, Wideman L, Montgomery MM, and Levine BJ. (2011). Some sex hormone profiles are consistent over time in normal menstruating women: Implications for sports injury epidemiology. Br J Sports Med 45, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Steiner C, Othman A, Saely CH, et al. , (2011). Bile acid metabolites in serum: Intraindividual variation and associations with coronary heart disease, metabolic syndrome and diabetes mellitus. PLoS One 6, e25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Meisinger C, Doring A, et al. , (2010). Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS One 5, e13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, et al. , (2011). Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 [DOI] [PMC free article] [PubMed]
- Swann JR, Want EJ, Geier FM, et al. , (2011). Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA 108Suppl 1, 4523–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend MK, Clish CB, Kraft P, et al. (2013). Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem 59, 1657–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese PB, Castellano JM, and Holtzman DM. (2011). Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol 10, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht L, Wasserman B, Chambless L, et al. (2009). Correlates of carotid plaque presence and composition as measured by MRI: The Atherosclerosis Risk in Communities Study. Circ Cardiovasc Imaging 2, 314–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, et al. (2011). Metabolite profiles and the risk of developing diabetes. Nat Med 17, 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JP. (2005). Quantifying test-retest reliability using the intraclass correlation coefficient and the SEM. J Strength Cond Res 19, 231–240 [DOI] [PubMed] [Google Scholar]
- Wetmore DR, Joseloff E, Pilewski J, et al. (2010). Metabolomic profiling reveals biochemical pathways and biomarkers associated with pathogenesis in cystic fibrosis cells. J Biol Chem 285, 30516–30522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Gangoiti JA, Barshop BA, and Siuzdak G. (2007). Metabolomics identifies perturbations in human disorders of propionate metabolism. Clin Chem 53, 2169–2176 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yu B, Alexander D, et al. (2013a). Associations between metabolomic compounds and incident heart failure among African Americans: The ARIC Study. Am J Epidemiol 178, 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Yu B, Alexander D, et al. , (2013b). Metabolomics and incident hypertension among blacks: The atherosclerosis risk in communities study. Hypertension 62, 398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.