Abstract
Significance: Skeletal muscle is a highly plastic tissue. Exercise evokes signaling pathways that strongly modify myofiber metabolism and physiological and contractile properties of skeletal muscle. Regular physical activity is beneficial for health and is highly recommended for the prevention of several chronic conditions. In this review, we have focused our attention on the pathways that are known to mediate physical training-induced plasticity. Recent Advances: An important role for redox signaling has recently been proposed in exercise-mediated muscle remodeling and peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) activation. Still more currently, autophagy has also been found to be involved in metabolic adaptation to exercise. Critical Issues: Both redox signaling and autophagy are processes with ambivalent effects; they can be detrimental and beneficial, depending on their delicate balance. As such, understanding their role in the chain of events induced by exercise and leading to skeletal muscle remodeling is a very complicated matter. Moreover, the study of the signaling induced by exercise is made even more difficult by the fact that exercise can be performed with several different modalities, with this having different repercussions on adaptation. Future Directions: Unraveling the complexity of the molecular signaling triggered by exercise on skeletal muscle is crucial in order to define the therapeutic potentiality of physical training and to identify new pharmacological compounds that are able to reproduce some beneficial effects of exercise. In evaluating the effect of new “exercise mimetics,” it will also be necessary to take into account the involvement of reactive oxygen species, reactive nitrogen species, and autophagy and their controversial effects. Antioxid. Redox Signal. 21, 154–176.
Introduction
Adult skeletal muscle is a highly plastic tissue, as it is able to change phenotype without changing genotype in response to external stimuli (60). Although muscle plasticity is limited by cell lineage determined during development, myofibers' physiological properties, metabolism, and size vary according to environmental stimuli. Changes in nutrient availability, paracrine/autocrine conditions, and intracellular oxygen availability influence adult skeletal muscle features. This tissue is also extremely adaptable to changes in contractile activity, and adaptation to exercise training has numerous beneficial effects on health. For this reason, physical activity is highly recommended for the prevention of several chronic conditions. In this review, we examine the signaling pathways triggered by exercise in skeletal muscle, with particular attention to those involving the peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α). We then focus on the role recently suggested for reactive species and for autophagy as mediators of molecular and metabolic response to contraction.
Myofiber Types
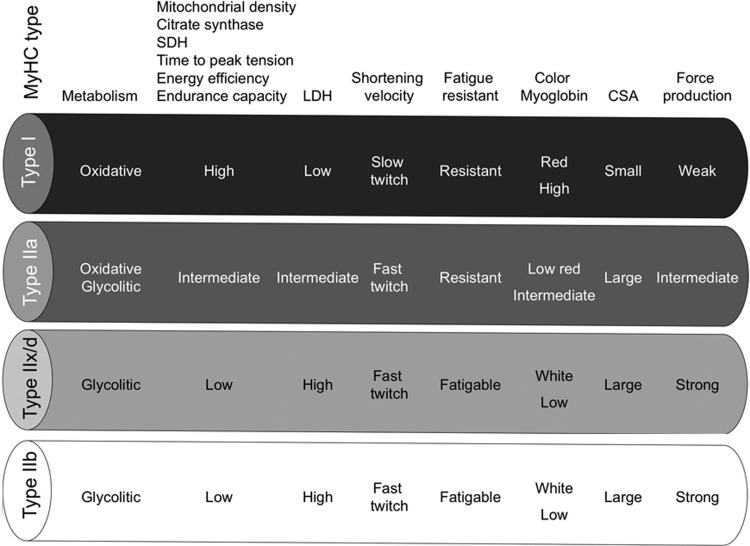
Mammalian skeletal muscles are composed of myofibers with various contractile properties (such as force production, endurance, twitch duration, and shortening velocity) and differing metabolism. Myofibers are mainly classified as slow-twitch or fast-twitch based on the maximal speed of shortening (166). The variability in physiological properties of myofibers strongly depends on the isoform of myosin heavy-chain (MyHC) expressed. Indeed, myosin ATPase activity determines the sliding velocity between actin and myosin, thereby shortening the velocity of the fiber (6). Myosin ATPase type I histochemical staining identifies slow-twitch fibers, while myosin ATPase type II (which has the highest ATPase activity) stains fast-twitch myofibers. Based on the expression of the predominant isoforms of MyHC protein expressed, myofibers are mainly classified as type I fibers, type IIx/d fibers, and type IIa fibers (166, 167) (Fig. 1).
FIG. 1.
Characteristics of mammalian skeletal muscle fiber types. The red color is associated with a high content of myoglobin. MyHC, myosin heavy-chain; SDH, succinate dehydrogenase; LDH, lactate dehydrogenase; CSA, cross-sectional area.
Type I fibers (slow-twitch fibers) contain the slow isoform of MyHC and slow isoforms of other contractile proteins. They have a predominatly oxidative metabolism. They are characterized by high mitochondrial content, high capillary density and express mainly glucose and fatty acid oxidative enzymes. Type I fibers are rich in myoglobin and are red colored. They develop a slow contractile force and are resistant to fatigue. They are involved in continuous tonic activity. Force production depends on the time the myosin head spends bound to actin, on the myosin head density and on the duty ratio (16).
Type IIx/d fibers (fast-twitch fibers) express a fast isoform of MyHC and fast isoforms of other contractile proteins and, therefore, develop a fast contractile force. Type IIx/d fibers mainly metabolize glucose by glycolysis and are characterized by low mitochondrial content and low capillary density. They are also poor in myoglobin and are white in appearence. Type IIx/d fibers express low glucose transporter 4 (GLUT4) and have low sensitivity to insulin that type I fibers. They are involved in phasic activity (103).
Type IIa fibers (fast-twitch fibers) have intermediate features. They have a mixed (oxidative/glycolytic) metabolism. They are fast-twitch fibers with a fast contractile force development, but mainly express oxidative enzymes. Although muscle endurance and resistance to fatigue rely on several cellular factors, there is a strict correlation between these properties and high oxidative capacity and high content of mitochondria of the fiber. Therefore, type IIa fibers are fast but they are more resistant to fatigue than type IIx/d fibers as they are more oxidative (60, 143).
Rodents also possess type IIb fibers that are more fast-twich and glycolytic than IIx/d fibers (Fig. 1). Many other contractile and structural proteins are also present in distinct isoforms whose expression is more or less tightly connected to fiber type. For example, the shortening velocity also depends on myosin light chain isoforms; thus, it might vary among fibers of the same MyHC type. Therefore, the classification reported earlier in four main fiber types is an oversimplification. Moreover, muscle also contains hybrid fibers with a combination of myosin transcripts (I-IIa-IIx/d-IIb).
The velocity of shortening and the fiber's twitch duration depend not only on myosin composition but also on the speed of Ca2+ release and uptake in the fiber. These, in turn, depend on the development of sarcoplasmic reticulum and on sequestering systems such as the sarcoplasmic reticulum Ca2+ ATPases (SERCAs) whose isoforms are differentially expressed in different fiber types (60, 142).
Skeletal Muscle Metabolism
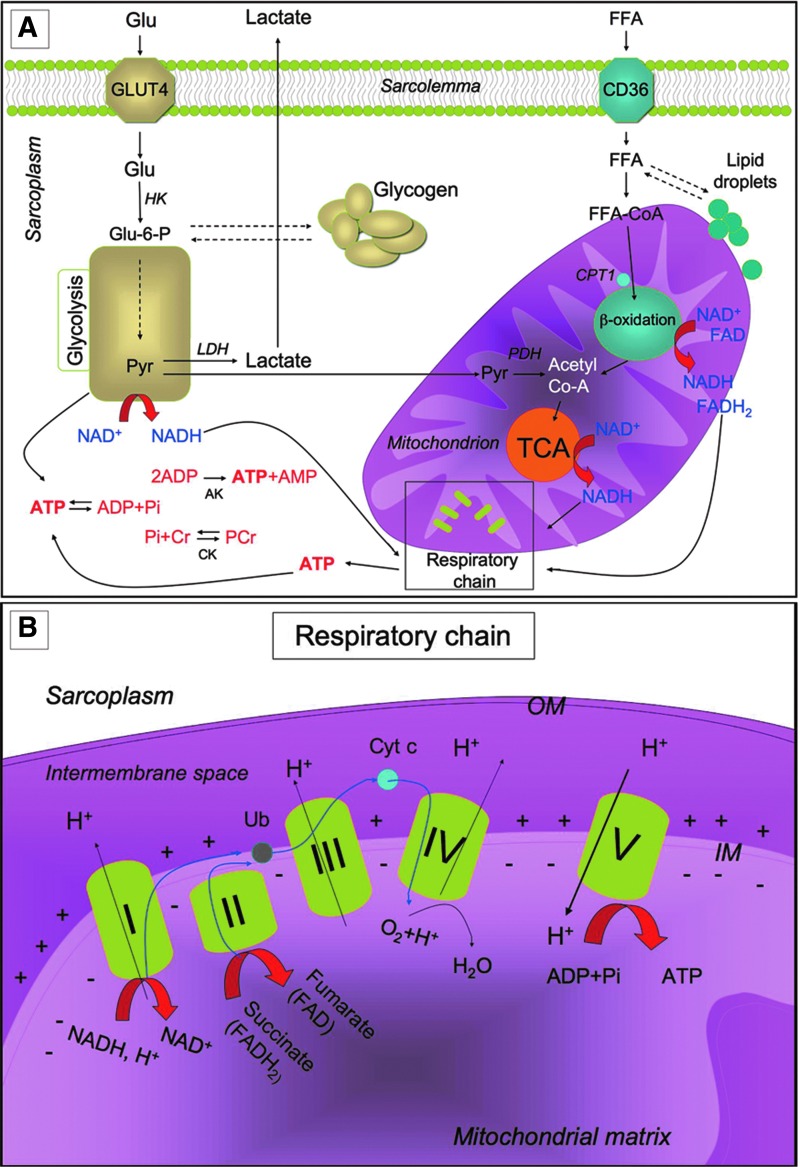
To enable contraction, the skeletal muscle needs a high amount of ATP, which is hydrolyzed by myosin ATPase and is also necessary to enable exchange of ions occurring during contraction (48). ATP is provided by a complex network of metabolic pathways that are briefly described in Figure 2. To obtain the huge amount of energy required during exercise, glucose uptake from the bloodstream increases while, at rest, glucose can be stored as glycogen. Skeletal muscle is the main storage site for glucose in the mammalian body. Glycogen is distributed in distinct localizations within the myofibers, depending on myofiber type and on training status (139). Lipids can also be stored as triglycerides in the myofiber, although excessive lipid accumulation is deleterious and might cause lipotoxicity and inflammation. Energy requirement during exercise induces the lipolysis of lipid droplets with the liberation of free fatty acids (FFAs) and the catabolism of glycogen with the release of glucose-1-phosphate, both of which are further catabolized to produce ATP (Fig. 2).
FIG. 2.
Metabolic pathways for ATP production in skeletal myofibers. (A) Skeletal muscles require a high amount of ATP for contraction. The main sources of energy are Glu and FFA. Glu uptake into the sarcoplasm from blood occurs, among other things, through the GLUT4. Once in the cytosol, Glu is phosphorylated by HK and forms Glu-6-P. One molecule of Glu-6-P can be converted into two molecules of Pyr through glycolysis, a metabolic anaerobic pathway involving 10 enzymes (the enzyme phosphofructokinase is an important control point in the glycolytic pathway). Depending on the energy needs, Glu-6-P can also be stored as glycogen. In anaerobic conditions Pyr is reduced to lactate by LDH. Alternatively, in aerobic conditions, Pyr might be transferred into the mitochondria matrix, where it is decarboxylated into acetyl-CoA by the PDH complex. Acetyl-CoA is then metabolized through the TCA cycle. The first enzyme acting in the TCA cycle is the citrate synthase that forms citrate from acetyl-CoA and oxaloacetate. The TCA cycle produces reducing equivalents (NADH, FADH2) and CO2. In addition to Pyr, another important source of acetyl-CoA is the β-oxidation of FFA. FFA enter the myofiber through a passive flip-flop or through a protein-mediated mechanism such as the FAT/CD36. In the cytosol, FFA undergo esterification and form triglycerides stored as lipid droplets that are surrounded by mitochondria. Alternatively, at the mitochondrial OM, they can be condensed with CoA to form FFA-CoA and, through the CPT1, they can cross the mitochondrial IM and reach the mitochondrial matrix where they undergo β-oxidation. β-oxidation is a cycle of four reactions. Each cycle produces a molecule of acetyl-CoA which, in turn, enters the TCA cycle. Along with acetyl-CoA, during β-oxidation, FADH2 and NADH are also formed. In skeletal muscles, at rest, excess of ATP produced is stored as PCr. ATP is converted into ADP and Pi by ATPase, and the Pi is used to convert Cr in PCr whose amount is roughly 10 times higher than the amount of ATP. During intense activity, PCr can anaerobically donate a phosphate group to ADP and form ATP for quick regeneration of ATP. PCr is, therefore, a rapid system to supply energy during contraction. The reversible phosphorylation of Cr is catalyzed by several CK. Once ATP also produced by PCr is consumed, the AK (myokinase) catalyzes the formation of ATP and AMP from two ADP molecules. During exercise, the amount of ATP produced by the myofiber increases enormously. However, the stores of ATP that can be detected in the myofiber are not as high, as ATP is stored in the form of PCr. (B) Reducing equivalents (NADH and FADH2) generated mainly during TCA, β-oxidation, and glycolysis are oxidized by the complexes of the respiratory chain (Complex I, II, III, and IV) in the oxidative phosphorylation pathway. Electrons are transferred from NADH and FADH2 to oxygen (which is reduced to H2O) by means of the enzyme complexes and by the electron carriers Ub and Cyt c of the respiratory chain. The energy released by reducing equivalent oxidation as electrons pass from one complex to the next is used to pump protons (H+) across the IM into the intermembrane space. This creates an electrochemical proton gradient across the IM, which is highly energetic. Protons can flow along this gradient through ATP synthase (ATPase or complex V); this backflow releases the energy of the proton gradient, which is used by ATP synthase to phosphorylate ADP and to form ATP. This phosphorylation of ADP is called oxidative, as it is coupled to the presence of oxygen that enables the oxidation of reducing equivalents. By this mechanism, nutrients are oxidated and their energy is stored in usable energy as ATP. ATP is also produced in a lower amount during glycolysis. OM, outer membrane; Glu, glucose; FFAs, free fatty acids; Glu-6-P, glucose-6-phosphate; Pyr, pyruvate; PDH, pyruvate dehydrogenase; TCA, tricarboxylic acid; FAT/CD36, fatty acyl translocase; CPT, carnitine palmitoyltransferase; CK, creatine kinases; AK, adenylate kinase; Ub, ubiquinon; IM, inner membrane; Cr, creatine; Cyt c, cytochrome c; GLUT4, glucose transporter 4; HK, hexokinase; PCr, phosphocreatine. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Exercise-Induced Adaptation
Skeletal muscle is extremely adaptable to environmental changes and is characterized by a high metabolic flexibility: It is able to rapidly modify the rate of ATP synthesis, the blood flow, and the kind of substrate used, depending on needs (103, 105). Skeletal muscle is also extremely adaptable to changes in contractile activity: Physical exercise strongly modifies metabolic potential, morphology, and physiology of skeletal muscle, thus producing a strong beneficial effect on health (96, 144). All pathways of ATP generation are active during exercise, but the relative contribution of each is determined by the intensity and duration of contraction. Indeed, exercise might be performed with different modalities, thus producing different effects on muscles (39).
Physical exercise might be grossly classified as “endurance training” and “resistance training.” Endurance training is based on endurance and is aerobic, while resistance training is based on strength. Endurance exercise (e.g., performed by marathon runners, swimmers, and cyclists) is generally characterized by high-frequency, long duration, and low power output. Resistance exercise (e.g., body building and throwing events) is, in general, characterized by low frequency, high resistance, high intensity, and short duration. Along with the modality of exercise, other parameters such as duration, frequency, and intensity of the exercise influence the effect of physical training on the muscle (131).
Exercise triggers a metabolic and structural remodeling in skeletal muscle, thus leading to changes in contractile properties and to increased angiogenesis in order to reduce muscle fatigue. These adaptations improve skeletal muscle performance (13). The specific features of skeletal muscle adaptation to exercise depend on the modality of exercise performed (60). Resistance exercise acts mainly by increasing muscle mass and strength (see “Exercise and skeletal muscle mass” section). On the other hand, endurance exercise stimulates mitochondrial biogenesis and expression of mitochondrial respiration and FFA β-oxidation genes, thereby providing a phenotypic adaptation toward a more oxidative phenotype. Submaximal aerobic activities increase insulin-independent glucose uptake and utilization in skeletal muscle, along with insulin sensitivity and redistribution of GLUT4 to the plasma membrane (125). With regard to the contractile properties, endurance exercise promotes fiber type transformation toward the slow-twitch contractile apparatus by inducing a dramatic modification of gene expression and physiological properties of the myofiber. The muscle used frequently needs to be more energy efficient, with both longer twitches and slower MyHC types contributing to higher energy efficiency (60).
Exercise provides numerous beneficial effects on skeletal muscle and, in general, on health (12). Although both exercise modalities are beneficial for health, endurance exercise is more effective for preventing cardiovascular diseases; while resistance training (mostly inducing muscle hypertrophy) is more effective for the maintenance of muscle mass contrasting atrophy and age-related muscle wasting (15). Hypertension, coronary heart disease, and cardiovascular risk profile mainly benefit from endurance exercise, which also increases angiogenesis and capillarization and protects from inflammation. The combination of both modalities of exercise increases bone mineral density and greatly improves insulin sensitivity, thereby protecting from type 2 diabetes. Exercise remains the primary preventive approach against obesity, glucose intolerance, and metabolic disease (31, 39, 64, 67).
Signaling in Skeletal Muscle Exercise-Induced Adaptation
The adaptation of myofibers and the benefits of regular exercise are mediated by a network of molecular and metabolic pathways that are activated by muscle contraction. Changes in contractile activity are sensed by intracellular sensors, which trigger intracellular signaling cascades. These are, in turn, converted into a transcriptional modulation that induces a metabolic reprogramming and a change of physiological properties of myofibers. Although these signaling mechanisms are not fully clarified, the transcription factors induced by exercise include nuclear factor of activated T cells (NFAT), myocyte enhancer factor 2 (MEF2), myogenic differentiation factor (myoD), myogenin, and PPARs. These transcription factors induce/repress genes coding for the fast and slow isoforms of various contractile proteins and for metabolic enzymes mediating a modulation of fiber-type specification and a remodeling of skeletal muscle. The transcriptional status (repressed or activated) of specific genes might also be modulated by exercise-induced epigenetic modifications (such as DNA methylation, phosphorylation, acetylation, and histone modifications) triggering chromatin remodeling (54, 123). The transcriptional modifications induced by exercise are deeply reviewed elsewhere (60, 103). Interestingly, skeletal muscle might also act as a secretory organ during exercise. It releases into circulation cytokines and peptides called “myokines” (such as IL-6 and irisin), which act both on other organs and on skeletal muscle (141).
Peroxisome proliferator-activated receptor γ coactivator-1α
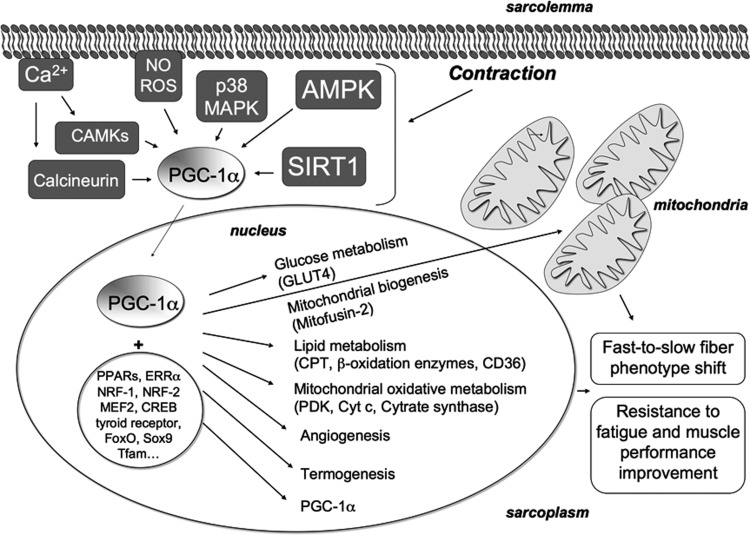
Although the molecular mechanisms of the adaptive response to exercise remain to be fully elucidated, PGC-1α is currently considered a major regulator of phenotypic adaptation induced by exercise. PGC-1α has been identified as a transcriptional coactivator of peroxisome proliferator-activated receptor γ (PPARγ) in brown fat cells (152). PGC-1α and its homolog PGC-1β are also co-activators for PPARα and PPARδ (involved in adipocyte differentiation and thermogenesis), and for a variety of transcription factors other than PPARs (63, 101, 105, 124, 151, 152, 199). PGC-1α promotes up-regulation of itself by an interaction with MEF2 on its own promoter (63) (Fig. 3).
FIG. 3.
Signaling pathways triggered by contraction and involving PGC-1α. PGC-1α is a major regulator of skeletal muscle remodeling induced by exercise. Changes in contractile activity are sensed by intracellular sensors involved in PGC-1α activation. These include Ca2+-dependent calcineurin and CAMKs, NO, ROS, p38 MAPK, AMPK, and SIRT1. Once activated, PGC-1α co-activates a variety of transcription factors and nuclear receptors such as PPARs, ERR-α, NRF-1, NRF-2, MEF2, CREB, thyroid receptor, FoxO, Sox9, and Tfam, which upregulate genes coding for mitochondrial proteins such as Mitofusin-2, PDK, Cyt c, and Cytrate synthase, thereby inducing mitochondrial biogenesis. PGC-1α also induces the transcription of genes encoding proteins that are involved in lipid metabolism (e.g., CPT, β-oxidation enzymes, and CD36), in angiogenesis, and in termogenesis. Moreover, PGC-1α triggers transcription of itself by interacting with MEF2 on its own promoter. By interacting with MEF2 (which binds the promoter of the gene encoding GLUT4), it also induces GLUT4 overexpression and enhances insulin sensitivity. The final effects of PGC-1α are fast-to-slow myofiber phenotype shift and muscle performance improvement. ERR-α, estrogen-related receptor-α; AMPK, AMP-activated protein kinase; CREB, cAMP-response element-binding protein; FoxO, forkhead box; Sox9, sex determining region Y-box 9; Tfam, transcription factor A mitochondrial; MEF2, myocyte enhancer factor 2; NRF=nuclear respiratory factor; p38 MAPK, p38 mitogen-activated protein kinase; PDK, pyruvate dehydrogenase kinase; PGC-1α=peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α; ROS, reactive oxygen species; SIRT1, sirtuin 1.
PGC-1α expression in skeletal muscle is greatly induced by a single bout of exercise and also after prolonged physical activity (5, 118, 146, 180, 181, 197). Exercise seems to regulate not only the level of PGC-1α but also the translocation of the protein to the nucleus (197). PGC-1α is the master regulator of mitochondrial biogenesis; its overexpression increases mitochondrial content. PGC-1α promotes mitochondrial oxidative metabolism, glucose and lipid metabolism, and energy homeostasis and enhances angiogenesis (124, 199). There are also strong indications that PGC-1α plays a key role in fiber-type specificity; indeed, PGC-1α is involved in slow phenotype specification and is mainly expressed in slow muscles (102, 103) (Fig. 3). Moreover, transgenic mice overexpressing PGC-1α in skeletal muscles develop slower muscles and display a muscle phenotype similar to that of aerobically trained mice—high levels of mitochondrial enzymes, high respiratory capacity, increased resistance to fatigue, and thus improved exercise performance (19, 102, 190). On the other hand, PGC-1α knock-out mice have reduced oxidative capacity and muscle performance, but no clear influence on MyHC fiber type. By contrast, the muscle-specific PGC-1α knock-out animals display a slow-to-fast shift in MyHC fiber type, as well as reduced oxidative capacity and impaired muscle function (62, 190). However, on exercise, muscle-specific knock-out mice exhibit normal running activity and normal fast-to-slow fiber type transformation; while mitochondrial biogenesis and angiogenesis appears to depend on PGC-1α as well as on endurance exercise (49).
Since PGC-1α enhances GLUT4 expression and insulin sensitivity, it has been proposed that the improved glucose transport and insulin sensitivity occurring during exercise might be, in part, mediated by PGC-1α. However, data from transgenic and knock-out mice are highly controversial as discussed by Lira et al. (49, 103, 131).
PGC-1α activity is regulated by transcriptional regulation and also by post-translational modifications: phosphorylation, sumolation, and deacetylation as well as methylation and ubiquitination. A single bout of exercise or endurance exercise induces PGC-1α deacetylation in skeletal muscle, and this correlates with the up-regulation of PGC-1α target genes. Both phosphorylation and deacetylation of PGC-1α have been suggested to be necessary for PGC-1α ability to induce up-regulation of mitochondrial genes and of PGC-1α gene itself (21, 74). The signal transduction pathways induced by contraction sensors include those mediated by AMP-activated protein kinase (AMPK), sirtuin 1 (SIRT1), protein kinase C, changes in intracellular Ca2+ concentration, p38 mitogen-activated protein kinase (p38 MAPK), nitric oxide (NO•), reactive oxygen species (ROS), redox balance, and hypoxia-inducible factor-1 (HIF-1). Several of these signaling pathways may contribute to exercise-induced PGC-1α activation (60) (Fig. 3).
AMP-activated protein kinase
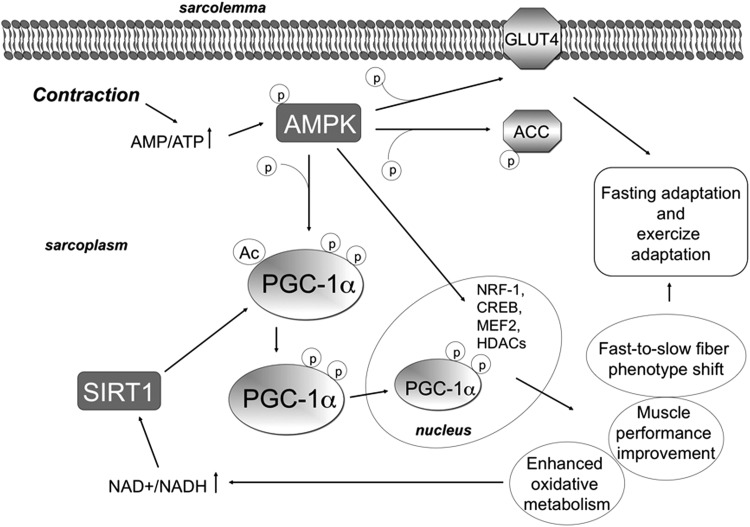
It has been shown, both in tissue culture and in vivo, that PGC-1α could be regulated by AMPK (4, 71, 72). AMPK is a serine/threonine kinase that regulates metabolic adaptation of skeletal muscle in response to an altered cellular energy status. Being a metabolic sensor for energy deprivation, AMPK plays a key role in metabolic flexibility of skeletal muscle, which is critical for the energy homeostasis of the whole organism. AMPK is activated allosterically by increased AMP/ATP and creatine/phosphocreatine (Cr/PCr) ratios that are caused not only by fasting or glucose deprivation, but also by cellular stress and oxidative stress (80). In order to rebuild energy stores in the cell, AMPK triggers signaling pathways that induce the transcriptional up-regulation of enzymes involved in lipid, glucose, and mitochondrial oxidative metabolism and in mitochondrial biogenesis (8, 74, 105, 192) (Fig. 4). Since AMPK activation acts to spare ATP, it inhibits anabolic pathways such as glycogen synthesis and protein synthesis and induces catabolic pathways (23).
FIG. 4.
Modulation of PGC-1α by AMPK and SIRT1. The serine/threonine kinase AMPK is activated by increased AMP/ATP ratio. AMPK is a heterotrimer with a catalytic α subunit and two (β and γ) regulatory subunits. Exercise requires a high amount of ATP, thereby leading to AMP/ATP ratio increase, which enhances AMPK phosphorylation and AMPK enzymatic activity. AMPK acts by phosphorylating and modulating some transcription factors such as NRF-1, CREB, and MEF2, as well as HDACs, and also by phosphorylating metabolic enzymes. For example, it induces lipid metabolism by phosphorylating and inactivating ACC. In the skeletal muscle, AMPK activation also triggers Glu uptake by enhancing GLUT4 translocation to the sarcolemma. Moreover, AMPK phosphorylates PGC-1α; this is needed for its deacetylation by SIRT1. These modifications enable PGC-1α to migrate into the nucleus, where it plays its role as a transcription factor, thereby triggering numerous effects. ACC, acetyl-CoA carboxylase; HDAC, histone deacetylase.
In addition to fasting, AMPK is also activated in the skeletal muscle by exercise. Intense contraction requires high amounts of ATP, thereby leading to AMP/ATP ratio increase. Exercise enhances AMPK phosphorylation and AMPK enzymatic activity in an intensity-dependent manner (65, 192, 195). Strikingly, the pharmacologic activator of AMPK, 5-aminoimidazole-4-carboxamide-1-β-4-ribofuranoside (AICAR) increases the amount of mitochondria; promotes mitochondrial oxidative enzymes, GLUT4, hexokinase (HK) II, FFA β-oxidation, and PGC-1α up-regulation both in vitro and in vivo (176, 193). The AMPK agonist AICAR is able to enhance running performances of mice in the absence of exercise training (133). Pharmacological compounds that activate beneficial endurance exercise-induced signalings are defined as “exercise mimetics.” The PPARβ/δ agonist GW1516 is a part of this category (89, 179). In addition, resveratrol and chitooligosaccaride have recently been included in this class of compounds (78, 127). “Exercise mimetics” are also beneficial to dystrophin-deficient skeletal muscle, with some having been tested as a therapeutic for type 2 diabetes (75, 78).
It has been proposed that AMPK may be necessary for PGC-1α activity. AMPK is able to directly phosphorylate PGC-1α in Thr 177 and Ser 538, thus being necessary for its deacetylation by SIRT1. Indeed, the down-regulation of AMPK inhibits PGC-1α deacetylation induced by exercise (21). AMPK also acts by modulating metabolic enzymes such as acetyl-CoA carboxylase (ACC) and GLUT4 (Fig. 4). Moreover, AMPK phosphorylates some transcription factors acting on PGC-1α and mitochondrial gene expression (Fig. 4). Similar to exercise, thyroid hormones increase energy demand and oxygen consumption, hyperthyroid conditions resulting in increased glucose uptake, FFA β-oxidation, and oxidative capacity. Interestingly, it has been shown that electrical stimulation plus thyroid hormone treatment trigger mitochondrial biogenesis in skeletal muscle through the phosphorylation of AMPK by liver kinase B1 (LKB1) and its associated proteins. AMPK, in turn, increases the phosphorylation of cAMP-response element-binding protein (CREB) and the expression of CREB-responsive mitochondrial genes such as those coding for PGC-1α and cytochrome c (Cyt c) (17, 182). However, while it has been shown that fast-to-slow fiber-type shift during exercise depends on AMPK, AMPK inhibition does not impair PGC-1α and mitochondrial induction, as reviewed by Lira et al. (103). Despite this discrepancy, most data strongly suggest that skeletal muscle adaptation induced by endurance exercise may be mediated by AMPK-induced PGC-1α. Moreover, this incongruity might be explained by the fact that, in addition to AMPK, other pathways (described in the next paragraphs) have been proposed as being induced by exercise and as activating PGC-1α.
Sirtuins
As stated earlier, PGC-1α may also be regulated by deacetylation. Most probably, PGC-1α deacetylation occurs by means of the protein deacetylase SIRT1. It has been suggested that AMPK-mediated PGC-1α phosphorylation is necessary for PGC-1α deacetylation by SIRT1 (20, 21) (Fig. 4). The regulation of the sirtuin family of deacetylases is NAD+ dependent (168). AMPK might activate SIRT1 by increasing the NAD+/NADH ratio through the induction of oxidative metabolism and also by promoting the expression of nicotinamide phosphoribosyltransferase (NAMPT) in NAD+ biogenesis (20, 21, 47). SIRT1 activity is associated with enhanced mitochondrial function and exercise performance. After exercise or fasting, dynamic NAD+/NADH ratio changes occur as a consequence of increased oxidative metabolism, which determines a reoxidation of NADH to NAD+. It has been demonstrated that endurance exercise is able to stimulate SIRT1 by promoting the expression NAMPT in NAD+ biogenesis. However, it is also possible that the changes in sirtuins and NAD+ metabolism occur in parallel with exercise-induced skeletal muscle remodeling, rather than being a causative factor of the same. Recently, the role of SIRT1 in PGC-1α deacetylation and mitochondrial biogenesis on endurance exercise has been confuted: It has been suggested that another deacetylating agent (general control of amino-acid synthesis [GCN5]), and not SIRT1, is involved in PGC-1α deacetylation (145).
Independently of PGC-1α deacetylation, enhanced sirtuin activity is associated with skeletal muscle remodeling on exercise (50, 94). By sensing the NAD+/NADH ratio and deacetylating lysine residues on enzymes and transcription factors, sirtuins couple the alterations in the cellular redox state with the adaptive changes in gene expression and metabolism. Interestingly, exercise-induced increase of SIRT3 is associated with improved mitochondrial function and enhanced skeletal muscle insulin sensitivity (20, 94).
p38 Mitogen-activated protein kinase
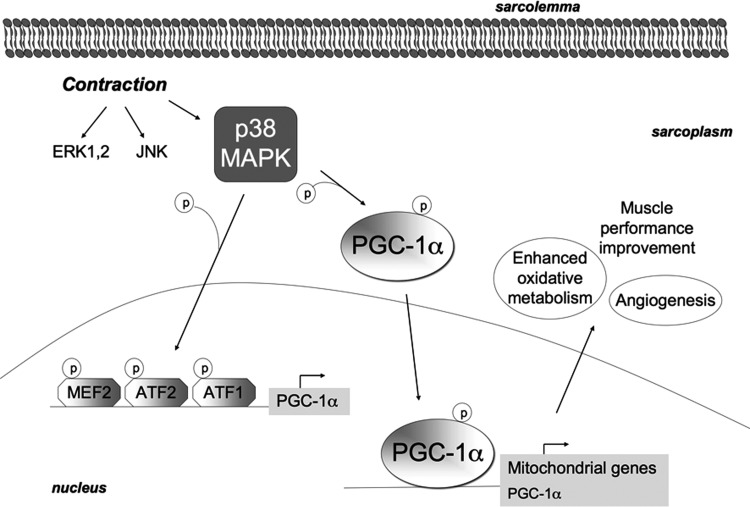
Acute exercise activates some MAPKs (Fig. 5): It has been suggested that PGC-1α activation and the following skeletal muscle metabolic adaptation triggered by exercise might be mediated by p38 MAPK, which phosphorylates PGC-1α and enables its translocation to the nucleus (Fig. 5). Both PGC-1α phosphorylation and deacetylation seem to be necessary for PGC-1α nuclear translocation (151, 197). Moreover, p38 MAPK phosphorylates some transcription factors directly inducing up-regulation of PGC-1α and, in turn, of mitochondrial genes (2, 22) (Fig. 5). p38 MAPK is necessary for PGC-1α-mediated angiogenesis due to endurance exercise and to motor nerve stimulation in mice (147). PGC-1α-mediated metabolic adaptation to exercise specifically depends on the p38γ MAPK isoform, which induces mitochondrial biogenesis and PGC-1α overexpression. The key role of p38γ MAPK in exercise-induced metabolic adaptation is strongly supported. By contrast, exercise-induced contractile adaptation was unaffected in p38γ MAPK muscle-specific knock-out mice, and, therefore, p38 MAPK may not be necessary for fast-to-slow fiber type switch, which might be otherwise regulated.
FIG. 5.
MAPKs are activated by exercise. During acute exercise, three MAPKs are activated: specifically, ERK1/2, JNK, and p38 MAPK. p38 MAPK is able to phosphorylate PGC-1α and to favor its translocation to the nucleus, where it acts as a transcription factor for mitochondrial genes as well as for itself. In addition, p38 MAPK phosphorylates and activates the transcription factors MEF2, ATF1, and ATF2, which bind to the PGC-1α promoter, thereby inducing up-regulation of PGC-1α and, in turn, of mitochondrial genes. It has also been proposed that p38 MAPK might regulate PGC-1α induction by endurance exercise mainly by mediating its nuclear translocation; while PGC-1α up-regulation might occur as a secondary effect of PGC-1α translocation and activity. The increased activity of PGC-1α-induced by p38 MAPK mainly mediates angiogenesis and metabolic adaptation to exercise. ATF, activating transcription factor; ERK, extracellular-signal-regulated kinase; JNK, c-Jun N-terminal kinase.
Ca2+ signaling
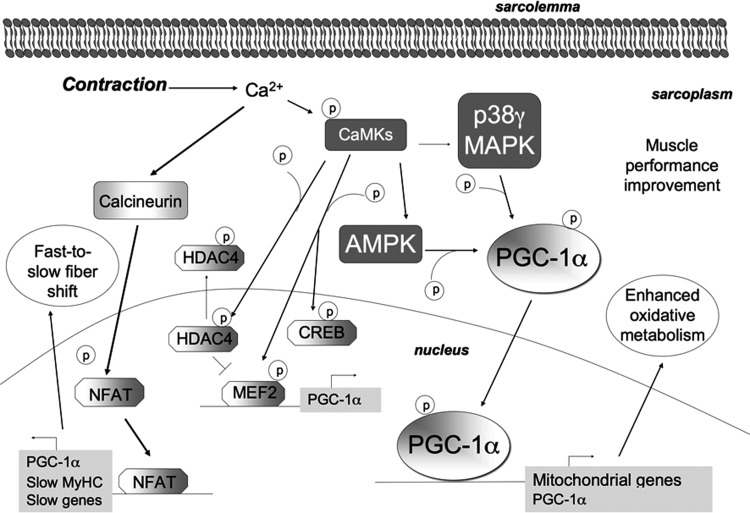
Several in vitro, ex vivo, and in vivo experiments strongly suggest that the concentration of calcium in the myofiber has a key role in PGC-1α regulation during contraction. The Ca2+ ionophore caffeine or electrical stimulation induces PGC-1α mRNA up-regulation, with this effect being mediated by Ca2+/calmodulin-dependent calcineurin and Ca2+/calmodulin-dependent kinases (CaMKs) (93) (Fig. 6).
FIG. 6.
Exercise adaptation is mediated by Ca2+ signaling. During exercise, Ca2+ concentration increases inside the myofiber and activates CaMKs. Calcineurin is a phosphatase that primarily dephosphorylates and activates NFAT. NFAT mainly promotes the transcription of PGC-1α and of slow genes (such as MyHC), thus most probably mediating the fast-to-slow myofiber transition. CaMKII is the main isoform of CaMKs in human skeletal muscle. On Ca2+ increase due to exercise, CaMKs become phosphorylated in an intensity-dependent manner; they, along with p38 MAPK, target PGC-1α and mitochondrial biogenesis. In addition, it is considered that CaMKs might also phosphorylate CREB, AMPK, and HDACs. CaMKs phosphorylate and directly activate MEF2, which promotes the transcription of PGC-1α. Nuclear HDAC4 inhibits MEF2. By phosphorylating HDAC4, CaMKs induce HDAC4 export out of the nucleus, thereby releasing MEF2. CaMKs, Ca2+/calmodulin-dependent kinases; NFAT, nuclear factor of activated T-cells.
Calcineurin is a serine/threonine phosphatase that is Ca2+/calmodulin dependent and activated during skeletal muscle contraction. Although the role of calcineurin in exercise-triggered muscle remodeling and in PGC-1α overexpression has not been fully demonstrated, its activity has been associated with slow myofiber gene expression. Calcineurin dephosphorylates and activates the NFAT transcription factor, which is involved in slow-twitch gene expression (30, 171). Accordingly, calcineurin overexpression in mice induces genes coding for slow-twitch proteins such as myoglobin, GLUT4, pyruvate dehydrogenase kinase (PDK), and mitochondrial enzymes and is associated with enhanced endurance exercise performance (79, 134). Calcineurin induces PGC-1α transcription, and inhibition of the calcineurin/NFAT axis reduces type I fiber gene expression and reduces fast-to-slow fiber type transition (199). Available data strongly suggest that, while p38 MAPK/PGC-1α controls mitochondria biogenesis and angiogenesis in response to endurance exercise, calcineurin/NFAT axis controls fiber-type shift.
CaMKs are also considered as being involved in the activation of slow oxidative gene expression in myocytes, although their role in adaptation due to exercise needs further elucidation (29, 38, 103). CaMKs' overexpression is associated with increased PGC-1α gene expression, mitochondrial biogenesis, glucose and lipid uptake and oxidation, skeletal muscle plasticity, and reduced fatigability (37). CAMKs might act along with p38 MAPK in activating PGC-1α and mitochondrial biogenesis (29, 198) (Fig. 6).
Exercise induces histone deacetylase 4 (HDAC4) and HDAC5 export out of the nucleus. HDAC4 usually represses MEF2 and, consequently, PGC-1α transcription. The activation of CaMKII leads to phosphorylation and nuclear exclusion of HDAC4, which releases MEF2, thereby enabling the transcription of the genes encoding PGC-1α and GLUT4 (104). It has been postulated that CAMKs might be upstream kinases for AMPK, p38 MAPK, and HADC4 during contraction (103) (Fig. 6).
ROS and NO•
The skeletal muscle generates a complex set of ROS and reactive nitrogen species (RNS), both at rest and during contractile activity (61). The primary ROS produced by mitochondria is the superoxide anion (O2•−). Through various reactions, it gives rise to other ROS, mainly hydrogen peroxide (H2O2) and hydroxyl radical (OH•) (Fig. 7A). The main RNS is NO• that is generated by different isoforms of nitric oxide synthases (NOS). O2•− can react rapidly with NO• and produce more dangerous RNS (e.g., peroxynitrite [ONOO−]) (43, 148) (Fig. 7A).
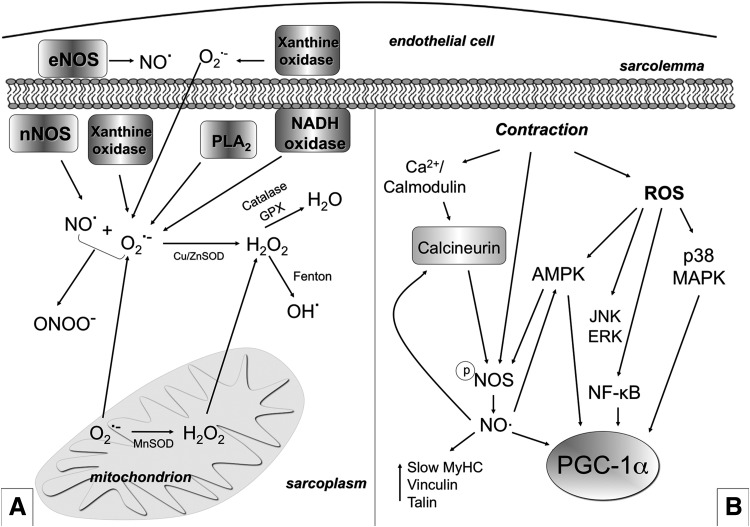
FIG. 7.
ROS and RNS production and their role in exercise-mediated skeletal muscle plasticity. (A) Several ROS and RNS are produced in skeletal muscle both at rest and during contraction. NO• is produced by NOS (eNOS in the endothelial cells and nNOS in the muscle fibers). The O2•− is generated into mitochondria in at least five sites (three of which being well characterized) within the mitochondrial respiratory chain through incomplete reduction of oxygen in the electron transport system. O2•− can also be generated as a specific product of some enzymes into mitochondria. Moreover, NADH oxidase, xanthine oxidase, and PLA2 are other O2•− generators. O2•− can undergo spontaneous dismutation or dismutation catalyzed by MnSOD (in the matrix) and Cu/ZnSOD (in the intermembrane space and in the cytosol) and form H2O2. H2O2 is cytotoxic; however, it is poorly reactive and is considered a relatively weak oxidizing agent. H2O2 is unable to oxidize DNA or lipids directly, but it can inactivate some enzymes. H2O2 might be de-tossificated in H2O by catalase or GPX or be transformed, through the Fenton reaction, into OH• which is, by contrast, highly reactive and able to damage most macromolecules, including DNA, proteins, and lipids. O2•− can react with NO• and produce ONOO−. ONOO− is able to rapidly cross membranes and is a strong oxidant agent that can lead to DNA damage and nitration of proteins. Sub-intracellular measurements and detection of ROS are usually prone to artifacts, while O2•− intracellular catabolism is extremely complex and controversial. Therefore, the earlier description is an oversimplification. Skeletal muscle has a well-developed system that prevents potentially deleterious effects of ROS (such as catalases, SODs, glutathione, GPX, peroxiredoxins, and thioredoxins). The abundance of scavengers abrogates free-radical chain reaction propagation under physiological conditions. If redox homeostasis is disrupted, the cell becomes damaged, thus leading to a pathological condition. (B) ROS and NO• mediate a contraction-induced adaptive response to exercise. NO• and ROS mediate the up-regulation of PGC-1α, GLUT4, mitochondrial genes, and slow genes. Moreover, ROS increase Glu uptake by triggering p38 MAPK (and also ERK) phosphorylation and activation, which possibly phosphorylates and activates PGC-1α. ROS and RNS trigger PGC-1α phosphorylation and Glu uptake also through AMPK. Moreover, ROS induce NF-κB-mediated transcription of PGC-α. Contraction also induces NO• production by NOS. NO•, in turn, activates calcineurin/NFAT induction of fast-to-slow phenotype. NOS is a Ca2+/calmodulin-dependent enzyme that should be dephosphorylated in order to produce NO•, and this might depend on calcineurin. NOS is also regulated by AMPK. NO•, nitric oxide; O2•−, superoxide anion; H2O2, hydrogen peroxide; GPX, glutatione peroxidase; NF-κB, nuclear factor-kappa B; NOS, nitric oxide synthases; OH•, hydroxyl radical; ONOO−, peroxynitrite; PLA2, phospholipase A2; RNS, reactive nitrogen species; SOD, superoxide dismutase.
Mitochondria are the predominant site for ROS generation in cells (43). Other potential sites for ROS and RNS generation in skeletal muscle are the sarcoplasmic reticulum, the transverse tubule, and the sarcolemma, all of which contain an NAD(P)H-dependent oxidase that generates O2•−. More specifically, in skeletal muscle, this enzyme appears to use NADH as a substrate (148). Other sources of oxidative stress during physical exercise are the inflammatory response mediated by neutrophils, the activity of xanthine oxidase, and the phospholipase A2 (PLA2) (56, 186) (Fig. 7A).
The most ROS and RNS can irreversibly modify and damage proteins, DNA, and lipids; lipid peroxidation is, in turn, a source of new free radicals (43). However, the traditional view by which ROS are a by-product of oxidative metabolism with only negative effects on cellular components has been now ruled out. ROS have a physiological function, as they are necessary for regulating several key biological processes. It has been hypothesized that their beneficial or detrimental effects depends on their concentration. At a very low concentration, ROS would induce biological processes such as proliferation and differentiation. At a slightly higher concentration, they would mediate the response of cells to ROS by inducing antioxidant genes. If ROS concentration becomes too high, they would then induce cell death (36, 178). Low and physiological levels of ROS are required for force production in skeletal muscle. By contrast, high levels of ROS promote contractile dysfunction, resulting in muscle weakness and fatigue (150). In order to maintain the redox balance, the levels of ROS, their generation, and elimination are finely tuned: Skeletal muscle has a well-developed system of scavengers that prevent deleterious effects of ROS (Fig. 7A).
Reactive species and muscle plasticity
The elevated metabolic rate associated with physical exercise and muscle contraction increases mitochondrial oxygen utilization in muscle tissue and, consequently, increases ROS generation. It has been recently proposed that both contraction-induced ROS generation and NO• play an important physiological function in the regulation of both muscle force production and contraction-induced adaptive response of muscle fibers to exercise training (81, 103, 140, 150, 174).
Since ROS's seemingly contradictory beneficial and detrimental effects are probably due to the difference in both the magnitude and the temporal pattern of ROS generation, it is plausible that moderate exercise can produce low levels of ROS and activate signaling pathways, leading to cellular adaptation and protection against stress. By contrast, maximal, or near maximal, bouts of high-intensity physical exercise and high levels of ROS production may result in chronic activation of signaling pathways that promote cell damage and, potentially, cell death (1).
Of particular interest is the role of NO• signaling in the adaptive responses to exercise. More specifically, it has been clearly shown that NO• and its derivatives play a role in the modulation of cellular metabolism and in fast-to-slow fiber-type transformation (138). NO• seems to mediate the up-regulation of PGC-1α, GLUT4, and mitochondrial proteins, thus modulating mitochondrial function and biogenesis (122) (Fig. 7B). This hypothesis is supported by several data indicating that NO• production increases in skeletal muscle during contractile activity (140, 173); that low levels of NO• induce mitochondrial biogenesis, PGC-1α and GLUT4 expression in cultured muscle cells (138); and that NO• is required for the up-regulation of slow MyHC induced by overload (169). It has also been observed that administration to humans of inorganic nitrate (which can be converted into NO• in the body) significantly improves energy metabolism during exercise (24). Moreover, a genetic deletion of NOS or their pharmacological inhibition prevents PGC-1α induction that is triggered by endurance exercise (188). It has also been demonstrated that NOS activity mediates sarcomere addition during remobilization of muscle and increases expression of the structural proteins talin and vinculin during cyclic stretching of skeletal muscle (85), thus further proving a role for NO• in the skeletal muscle phenotypic adaptation to exercise. H2O2 also seems to be important for plasticity of skeletal muscle and for PGC-1α induction (174). As such, cultured muscle myotubes treated with H2O2 show induction of PGC-1α, and the antioxidant N-acetyl-l-cysteine (NAC) inhibits this up-regulation (71). Moreover, recent findings suggest that ROS are involved in the regulation of glucose uptake by muscles (95, 163) and that ROS scavengers and generic antioxidants are able to modify glucose uptake (25). Interestingly, antioxidants inhibit the up-regulation of PGC-1α, mitochondrial proteins, and the increase of insulin sensitivity induced by endurance exercise or by electrical stimulation (55, 156).
PGC-1α activation induced by ROS also seems to trigger protection against excessive oxidative stress. This is possibly due to either the PGC-1α-induced up-regulation of antioxidant enzymes and/or to the fact that the increased number of mitochondria triggered by PGC-1α might enable lower levels of respiratory activity for each mitochondrion while maintaining the same amount of global ATP generation (155).
Interaction between reactive species and the other pathways involved in muscle plasticity
It has been suggested that ROS and RNS might play a role in the phenotypic adaptation induced by exercise by triggering p38 MAPK and AMPK, which are known to activate PGC-1α after contraction (70, 81, 103) (Fig. 7B). Exogenous ROS increase glucose uptake via p38 MAPK activation/phosphorylation; accordingly, the inhibition of p38 MAPK reduces ROS-induced glucose uptake (25). Moreover, contraction-induced RNS and ROS are able to increase glucose uptake via AMPK; while NAC and NOS inhibitors inhibit this effect. PGC-1α might be a part of a redox-sensitive pathway, which might also involve the ROS-sensitive nuclear factor-kappa B (NF-κB). In fact, human PGC-1α promoter contains several NF-κB binding sites and ROS might induce PGC-1α transcription through NF-κB (121) (Fig. 7B). ROS also mediate c-Jun N-terminal kinase (JNK) signaling activation triggered by acute exercise (141). In addition, it has recently been shown that the xanthine oxidase inhibitor allopurinol is able to attenuate skeletal muscle signaling after acute exercise (namely, the increased phosphorylation of p38 MAPK and extracellular-signal-regulated kinase [ERK]), although this inhibitor does not impair mitochondrial adaptations to endurance training (PGC-1α, GLUT4, and superoxide dismutase [SOD] increase) (87, 116, 189). Moreover, some authors suggest that NOS are activated by both Ca2+ and AMPK. As such, several studies have demonstrated that NO• is involved in Ca2+/calmodulin and AMPK-mediated effects and regulation of PGC-1α (103, 122).
In summary, data in literature strongly suggest that contraction stimulates a new molecular and metabolic profile, which is most probably mediated by reactive species. ROS scavengers and generic antioxidants are useful to reduce the damage due to excessive oxidative stress; however, they might also impair beneficial phenotypic adaptation to exercise in skeletal muscle such as glucose uptake (25, 36). The specific role that ROS/RNS play in mitochondrial biogenesis needs to be unraveled and further defined, as does the question as to whether PGC-1α expression in skeletal muscle depends on ROS/RNS.
Hypoxia-inducible factor-1
During exercise, oxygen consumption increases enormously; while the partial pressure of oxygen (PO2) in the contracting muscle decreases (60). This might activate the transcription factor HIF-1. In normoxic conditions, the subunit HIF-1α is hydroxylated by prolyl hydroxylase enzymes (PHD), which are sensors of cellular oxygen tension: Hydroxylation of HIF-1α finally results in its proteasomal degradation. When the intracellular PO2 decreases, pyruvate dehydrogenase (PDH) are inhibited and HIF-1α is no longer degraded, thus enabling HIF-1 accumulation (73, 170). It has been suggested that HIF-1-dependent processes are important for muscle adaptation to low oxygen tension occurring during endurance exercise. In fact, HIF-1 enhances the transcription of genes involved in angiogenesis, erythropoiesis, and metabolic pathways, thereby enabling more oxygen delivery to tissues; it also enhances the transcription of genes that are involved in oxygen-independent energy production such as glycolysis (45, 117). This is in accordance with the finding that HIF-1α mRNA and protein levels increase after acute exercise (86). On the other hand, other authors have found that training reduces HIF-1 expression (107); this suggests that HIF-1 is not responsible for the improvement of oxygen transport and utilization during exercise. Accordingly, it has recently been found that HIF-1 signaling and overall gene expression on prolonged exercise is different when compared with changes observed on hypoxia (86). Furtheremore, albeit controversial, it has also been proposed that HIF-1 may induce a slow-to-fast fiber type transformation which is in conflict with the induction of mitochondria and oxidative metabolism generally occurring on exercise (60, 86, 117).
Exercise and Skeletal Muscle Mass
As stated earlier, while endurance exercise acts by up-regulating mitochondrial metabolism and fiber-type transformation, the beneficial effects of resistance exercise mainly depend on its ability to increase muscle mass. Skeletal muscle mass depends on a delicate balance between protein synthesis and protein degradation: Resistance exercise influences both these processes by activating the phosphoinositide 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) signaling (52). The kinase mTOR exists in two independent complexes: mTOR complex 1 (mTORC1) and mTORC2 (Fig. 8). Raptor and Rictor are specific functional components of TORC1 and TORC2, respectively. mTORC1 controls protein translation by phosphorylating the eukaryotic translation initiation factor 4E-binding protein-1 (4E-BP1) and p70 ribosomal protein S6 kinase (p70S6K). p70S6K phosphorylates the ribosomal subunit S6 and up-regulates protein synthesis. mTORC2 prevents protein degradation by phosphorylating and inhibiting the forkhead box (FoxO) class of transcription factors. Indeed, FoxO transcription factors induce the expression of atrogin-1/muscle atrophy F-box (MAFbx) and muscle ring finger protein 1 (MuRF-1), two E3 ubiquitin ligases, which promote the ubiquitination and the proteasome-mediated degradation of critical sarcomeric proteins (9). The ubiquitin-proteasome system mediates muscle atrophy in several conditions, and the oxidative stress plays a key role in the regulation of the proteasome proteolytic activity (149). Mechanosensory regulation of protein synthesis is determined by high-force contractions that damage the sarcolemma and activate the membrane phospholipid phosphatidic acid, which, in turn, activates mTOR. During resistance exercise, mechanosensory regulation of protein synthesis also involves some transmembrane receptors called focal adhesion kinase (FAK) proteins, which transmit the contractile force through the skeletal muscle architecture and trigger protein synthesis by inducing mTOR activation (39, 84).
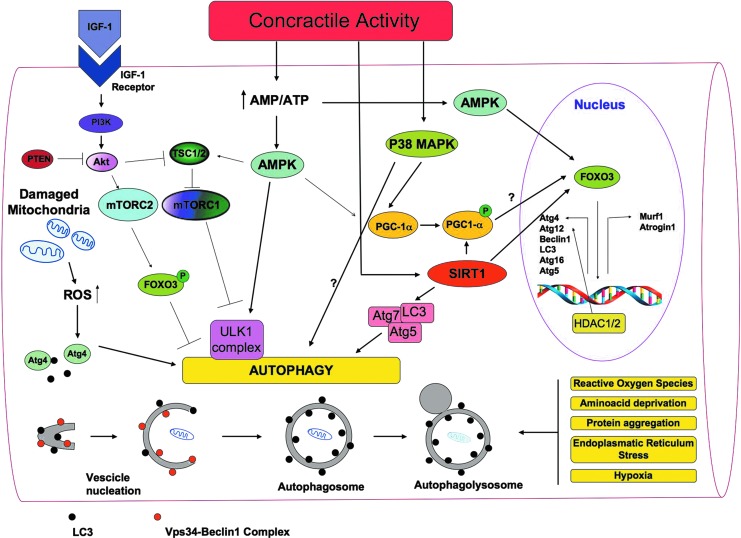
FIG. 8.
A general overview of the signaling molecules involved in the regulation of autophagy in skeletal muscles. Autophagy is a multi-step process that involves distinct phases during which part of the cytoplasm intracellular organelles are sequestered within characteristic double-membraned autophagic vacuoles (autophagosomes) that fuse with lysosomes and become autophagolysosomes. There, defective intracellular organelles and proteins are digested by a battery of lysosomal hydrolases. LC3 and the Vps34-Beclin1 complex are required, among many other proteins, for autophagosome formation. In skeletal muscles, ROS production influences different cell signaling pathways, including selective mitochondrial autophagy (mitophagy). Damaged mitochondria removal is particularly needed during exercise when the oxidative metabolism and the turnover of mitochondria increase. Moreover, ROS are mainly produced by mitochondria. The activation of the pivotal autophagy protein LC3 is mediated by the redox-sensitive Atg4 protease, which cleaves LC3. Growth factors such as IGF induce the PI3K/Akt signaling, which activates mTORC1 and mTORC2. mTORC1 (known to activate protein synthesis) also inhibits autophagy, as it inhibits the formation of the Atg1 (in humans ULK1) complex. ULK1 is a serine/threonine kinase that forms a complex with different regulatory proteins such as Atg13 and Atg17. Atg13 hyper-phosphorylation inhibits its association with Atg1, while the Atg1–Atg13 interaction enables the generation of autophagosomes. mTORC2 phosphorylates and inhibits FoxO transcription factors, thus inhibiting autophagy. Indeed, dephosphorylated FoxOs migrate into the nucleus and activate the transcription of genes that control muscle mass. FoxOs activate both ubiquitin-proteasome genes (atrogin-1 and MuRF-1) and autophagy-lysosome genes such as Beclin1, LC3, Atg4, Atg12, Atg16, and Atg5. PTEN inhibits the PI3K/Akt signaling pathway, and, therefore, it enables autophagy. Exercise, mitochondrial dysfunctions, starvation, and oxidative stress increase the intracellular AMP/ATP ratio, thus activating the energy stress sensor AMPK, which, in turn, promotes autophagy by inhibiting mTORC1 through the phosphorylation of TSC2. The TSC complex, consisting of TSC1 and TSC2 proteins, regulates the activity of the mTORC via Rheb, a small GTPase. In addition, AMPK induces autophagy by triggering ULK1 phosphorylation. By integrating signals from upstream sensors such as mTOR and AMPK, the ULK1 complex plays a central role in autophagy. AMPK activates FoxO transcription factors and leads to the expression of LC3 and Beclin1. Through a mechanism not yet known, p38 MAPK also seems to induce autophagy. In addition, SIRT1 can deacetylate Atg5, Atg7, and LC3, thus inducing autophagy; nuclear SIRT1 might induce the expression of autophagy genes through the activation of FoxOs. HDAC1 and 2 regulate muscle autophagy by controlling the expression of autophagy genes. During exercise, PGC-1α induction is mediated, among other things, by AMPK and p38 MAPK. Exercise up-regulates SIRT1 that removes acetyl groups from PGC-1α, enabling its translocation to the nucleus. PGC-1α induction has been associated with increased autophagy, although this hypothesis needs further investigation. IGF, insulin growth factor; LC3, microtubule-associated protein 1-light chain 3; mTOR, mammalian target of rapamycin; mTORC, mTOR complex; MuRF-1, muscle ring finger protein 1; PI3K, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homologue; TSC, tuberosus sclerosis complex; ULK1, unc-51-like kinase1; Vps34, vacuolar protein sorting 34. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The influence of exercise on muscle mass also involves muscle stem cells. As such, exercise induction of hypertrophy is accompanied by satellite cell fusion to myofibers. Mitochondria are considered as being involved in the regulation of myoblast proliferation/differentiation; therefore, PGC-1α-mediated mitochondrial biogenesis triggered by endurance exercise might possibly influence satellite cell fusion (113, 157). Interestingly, PGC-1α up-regulation occurs during differentiation (88, 130). Other signalings triggered by endurance exercise, such as p38 MAPK and Akt, contribute to satellite cell differentiation (1, 153, 183). Therefore, muscle wasting might be counteracted by endurance training through enhancement of myoblast differentiation and fusion. In addition, it has been suggested that PGC-1α might control muscle wasting pathways (18). It reduces the FoxO3-associated muscle atrophy, and mice overexpressing PGC-1α are protected from sarcopenia and have an increased lifespan (162, 191). Moreover, increasing mitochondrial oxidative metabolism and biogenesis protects from atrophy, and this might be achieved by endurance exercise-induced PGC-1α (75, 191). Regular submaximal aerobic activities have also been found to be beneficial for patients afflicted with Duchenne muscular dystrophy (DMD), while “exercise mimetics” decrease muscle inflammation and inhibit FoxO1 signaling (75). It has also been shown that exercise inhibits MuRF up-regulation due to diabetes and that this might mediate exercise's beneficial effects on this disease (26, 51).
Autophagy
Along with the ubiquitin-proteasome activity, another major proteolytic process mediates protein degradation in cells—the autophagy-lysosome system. At baseline levels, autophagy is a housekeeping mechanism cleaning cells of aberrant and dysfunctional molecules and organelles, thereby maintaining cell homeostasis. Autophagy is a multi-step process during which a part of the cytoplasm (including intracellular organelles) is sequestered within double-membraned autophagic vacuoles (autophagosomes), which then fuse to lysosomes and become autophagolysosomes (Fig. 8). By this mechanism, defective organelles and proteins are digested by lysosomal hydrolases (41, 126). Under stress conditions, autophagy increases and promotes temporary cellular adaptation to unfavorable conditions. It primarily favors survival during nutritional stress imposed by decreased nutrients; the degradation of intracellular material through autophagy becomes an alternative source of energy (83, 91).
More than 31 autophagy-related genes (ATG) have been identified. They encode Atg proteins, which form different complexes during the various steps of the autophagy process. In mammalian cells, the vacuolar protein sorting 34 (Vps34) complex regulates the initial steps of autophagosome formation. This complex is composed of the class III-PI3K Vps34, the kinase Vps15 (p150 in mammals), Atg14 (Barkor or mAtg14 in mammals), and Atg6/Vps30 (Beclin1 in mammals). Beclin1 interacts with several enhancing (Ambra1, Atg14/Barkor, and UVRAG) or inhibitory (Rubicon, B-cell lymphoma 2 [Bcl2], and BclX/l) factors that modulate its binding to Vps34 (44, 100, 119). In particular, the dissociation of Beclin1 from Bcl2 is essential for its autophagic activity. After the initial step, two ubiquitin-like conjugation systems are required for autophagosome formation completion: the Atg12-Atg5 and the microtubule-associated protein 1-light chain 3 (LC3)-phosphatidyl-ethanolamine (PE) systems. By means of the first conjugation system, the Atg12-Atg5-Atg16L complex is added to the elongating membrane of autophagosomes, thus enabling it to grow. With regard to the other conjugation system, after the activation by Atg7 and Atg3, the Atg4-cleaved LC3 interacts and conjugates with PE. PE-LC3 is the active form of LC3 (named LC3-II), which stably associates with the autophagosomal membrane. In the final stages of autophagy, the autophagosomes fuse with the lysosomes for degradation of their “cargo” (126).
Autophagy and skeletal muscle mass maintenance
The role of autophagy in the maintenance of muscle mass is controversial. While excessive autophagy is detrimental to skeletal muscle and contributes to muscle wasting, basal autophagy is required for the maintenance of skeletal muscle homeostasis and integrity (112, 161). The autophagy-lysosome system is activated in several atrophy conditions such as fasting, caloric restriction, cancer cachexia, aging, disuse, and denervation (114, 161). Conversely, the key role of autophagy in skeletal muscle homeostasis maintenance is supported by the fact that muscle-specific ablation of key autophagy proteins such as Atg7 or Atg5 produces myofiber degeneration and muscle weakness (10, 161). Accordingly, ablation of Vps15 and ablation of the autophagy regulator nutrient deprivation autophagy factor-1 (Naf1) leads to myopathic features in mice (10, 135). Notably, something that has been recently identified is the first human multisystemic disorder which is associated with defective autophagy; the Vici syndrome, which is caused by a mutation in the ATG ectopic P-granules autophagy-protein-5 (EPG5): Both Vici patients and EPG5 homolog knock-out mice display skeletal muscle-impaired autophagy (32). The impairment of autophagy also contributes to the pathogenesis of several genetic muscle diseases such as Bethlem and Ullrich dystrophies, where defective autophagy causes the accumulation of dysfunctional organelles into myofibers: Strikingly, the reactivation of autophagy rescues the myopathic phenotype (10, 53). Moreover, impaired autophagy and accumulation of damaged organelles have recently been found in DMD patients and in dystrophin-deficient mice (35). It has also been suggested that unbalanced autophagy might contribute to sarcopenia (10). A particular type of autophagy-defined chaperone-assisted selective autophagy (CASA) has also been found to be essential for muscle maintenance. CASA is required for the efficient disposal of damaged filamin, which would form aggregates in muscles. CASA differs from chaperone-mediated autophagy, as it also needs the ubiquitination of the chaperone substrate and the recruitment of the ubiquitin adaptor p62. CASA's relevance in skeletal muscle maintenance is also supported by the finding that the limb-girdle muscular dystrophy type 1D is associated to a mutation on a gene coding a co-chaperone interacting with the CASA complex (184).
On the other hand, the autophagy-lysosome system is activated in several atrophy conditions such as fasting, caloric restriction, cancer cachexia, aging, disuse, and denervation (114, 161). Moreover, the phenotype of some transgenic mice suggests that autophagy may favor muscle atrophy: The laminin-2 knock-out mouse model of muscular dystrophy displays an enhancement of autophagy whose inhibition significantly improves their dystrophic phenotype (10). Increased Vps34 activity and autophagy due to a mutation on the gene encoding Jumpy have been associated to a centronuclear myopathy (185), while skeletal muscle-specific ablation of Raptor (negative modulator of autophagy) results in dystrophy (7). The mutation of genes related to lysosomal functions are a primary cause of severe myopathies, including Danon and Pompe diseases, that are characterized by a massive accumulation of autophagosomes. However, it is not clear whether the myophaty is caused by autophagosome accumulation, leading to myofibrillar disorganization, or, vice versa, whether it is due to lysosomal impairement: In this case, autophagosome accumulation would be an attempt to maintain homeostasis (10).
To summarize, it is currently believed that a correct balance between activation and inhibition of autophagy is critical for muscle homeostasis. Too much autophagy causes an excessive removal of crucial cellular components, which leads to muscle atrophy. On the other hand, insufficient autophagy leads to the accumulation of dysfunctional organelles, thus impairing myofiber homeostasis (161).
Autophagy in exercise-mediated plasticity
Increasing evidence suggests that exercise triggers autophagy in skeletal muscle and that autophagy mediates some beneficial effects due to exercise. Grumati et al. have revealed an important connection between autophagy and exercise physiology. They have shown that physical training stimulates autophagy in mice skeletal muscles, and that autophagy was able to prevent the accumulation of damaged organelles and to maintain myofiber homeostasis. By contrast, exercise is detrimental for collagen VI-deficient muscles, in which, however, basal autophagy is extremely beneficial (58, 59). Interestingly, He et al. have found that, in the skeletal muscle of autophagy-deficient mice, exercise is not able to induce beneficial effects to the same extent as in wild-type mice (68). The authors have used Bcl2AAA mutant mice and Beclin1 heterozygous mice that are deficient in exercise-induced but not in basal autophagy. Bcl2AAA mice contain mutations in some Bcl2 phosphorylation sites, which hinder Bcl2 phosphorylation. This prevents exercise-induced dissociation of the Bcl2-Beclin1 complex and autophagy activation. These mutant mice, in which exercise-induced autophagy is defective, display decreased endurance although they have normal muscle strength and fiber size. Redistribution of the GLUT4 to the sarcolemma, increase of glucose uptake, and increase of insulin sensitivity, usually triggered by exercise, are reduced in these mice and this might explain their impaired muscle performance (57). Therefore, unaffected autophagy seems to be required for glucose homeostasis and metabolism in the skeletal muscle during exercise. In addition, in autophagy-deficient mice, phosphorylation and activation of AMPK (which plays a role in the redistribution of GLUT4 to the plasma membrane in the skeletal muscle on exercise) are reduced (65, 161).
Other researchers have reported a connection betweeen autophagy and exercise: It has been revealed that Vps34 is activated by an acute bout of resistance exercise and that autophagy is involved in protein catabolism after exercise (53, 111). By contrast, a study performed on humans after an acute bout of resistance exercise indicates a decrease of LC3 lipidation, thus suggesting a potential down-regulation of autophagy in muscles (46). Moreover, endurance exercise protects skeletal muscle against excessive activation of autophagy triggered by doxorubicin (175). Conversely, increased Atg7, Beclin1, and LC3 protein levels have been reported to occur in rat soleus in response to moderate endurance exercise (40). Accordingly, moderate aerobic exercise increases autophagy (Atg7 and Beclin1) in old mice and triggers autophagy (LC3 and Atg12) in skeletal muscles of old obese women (82, 194). It has also been reported that 9 weeks of resistance exercise training, along with preventing the loss of muscle mass and strength, also increase autophagy and reduce apoptosis in skeletal muscles of aged rats (108): In particular, reduced p62 protein levels; increased levels of Beclin1, Atg5-Atg12, and Atg7; and reduced LC3-II/LC3-I ratio have been documented. The specific autophagy pathway CASA seems to be induced by mechanical signals that are typically triggered by exercise. Mechanical tension is an essential stimulus for the development as well as for the homeostasis of the locomotory system, and CASA plays a role in muscle mechano-transduction (164).
The induction of autophagy has also been detected in humans after ultra-endurance exercise (training lasting more than 6 h) by Jamart et al. In this study, vastus lateralis samples were acquired from subjects who had performed a 200-km running race, and an increased expression of autophagy proteins (Atg4, LC3, and Atg12) was found. In addition, levels of autophagy proteins were found to be higher after an ultra-marathon race (76, 77). Induction of lysosomal enzyme activity and an increased number of autophagic vacuoles after strenuous endurance exercise had already been reported in rodents (158). However, while physical exercise exerts a beneficial effect on health, strenuous ultra-endurance exercise is extreme and high energy demanding, thus also causing a mechanical damage to muscle and a metabolic stress impairing mitochondria. In this context, autophagy might be triggered in order to remove damaged proteins and organelles and to provide amino acids that can be utilized in case of energetic stress (161). The role of autophagy in the skeletal muscle in response to exercise needs to be clarified while taking into account that it depends, among the other things, on the type of muscles studied and on the mode of exercise performed.
Interplay between autophagy and the other signaling involved in muscle plasticity
The involvement of autophagy in exercise-induced remodeling might be related to the two most important functions of autophagy: providing new sources of energy and removing dysfunctional organelles. During exercise, more energy is needed; the requirement of energy generally induces autophagy and it is possible that, as stated earlier, the increase of glucose uptake triggered by exercise depends on autophagy. Moreover, autophagy is the main mechanism for the removal of damaged mitochondria that is necessary to protect myfibers from atrophy. Damaged mitochondria removal is especially needed during exercise when oxidative metabolism and turnover of mitochondria increase. Interestingly, many of the sensors and pathways triggered by exercise in skeletal muscle are involved in the modulation of autophagy (Fig. 8).
The main intracellular pathways regulating autophagy involve mTOR, which, as discussed earlier, is crucial for the modulation of protein synthesis and degradation. In a favorable energetic status, nutrients are sensed by mTORC1, which becomes activated via a cascade involving the PI3K/Akt pathway (90, 115). Active mTORC1 inhibits autophagy by blocking the formation of the Atg1 (unc-51-like kinase1 [ULK1]) complex, which acts at the autophagy initiation stage. Conversely, nutrient starvation and/or hypoxia cause mTORC1 signaling inhibition and, consequently, autophagy activation. mTORC1 is inhibited by rapamycin, a potent inducer of autophagy, even under nutrient-rich conditions. The phosphatase and tensin homologue (PTEN) is a major negative regulator of the PI3K/Akt signaling pathway, thus enabling autophagy initiation (3). mTORC2 regulates both protein degradation and autophagy (160). In fact, the PI3K/AKT/mTORC2 pathway phosphorylates and inhibits the FoxO transcription factors, which, when active, up-regulate genes encoding both ubiquitin-proteasome proteins (atrogin-1 and MuRF-1) and autophagy-lysosome proteins such as Beclin1, LC3, Atg16, and Atg5 (Fig. 8) (114). FoxO transcription factors have been found to also induce PGC-1α transcription, and PGC-1α induction has been associated with increased autophagy (14, 99, 132, 151, 177). However, little data are available on this issue and, by contrast, some of them also suggest that during catabolic conditions high PGC-1α levels prevent the excessive activation of proteolytic systems by inhibiting the transcriptional activity of FoxO3 (18).
Several studies have suggested that AMPK is required for autophagy (Fig. 8). Interestingly, AMPK integrates stress stimuli with autophagy initiation. Nutrient deprivation, hypoxia, and oxidative stress increase the intracellular AMP/ATP ratio, thus activating the AMPK sensor, which, in turn, promotes autophagy. It has been suggested that AMPK induces autophagy by inhibiting mTORC1 through phosphorylation of the tuberosus sclerosis complex 2 (TSC2) (which regulates the activity of the mTOR) (69, 90, 137, 154, 200). Autophagy seems to be modulated by a mechanism involving AMPK, mTOR, and ULK1. ULK1 forms a complex with AMPK, and AMPK activation results in ULK1 phosphorylation, which regulates the localization of a critical component of the phagophore, Atg9 (110, 196). AMPK also leads to the activation of FoxOs and to the expression of the ATG LC3 and Beclin1 (159).
The induction of autophagy has also been related to p38 MAPK. Although this connection is still unclear and controversial, it has been suggested that p38MAPK might regulate the expression of ATG via FoxO transcription factors (34, 109, 120, 121, 129, 172).
In addition to mTOR and AMPK, protein (de)acetylation by sirtuins can regulate autophagy under nutrient depletion (97, 136) (Fig. 8). More specifically, SIRT1 can deacetylate essential components of the autophagy machinery such as Atg5, Atg7, and LC3. Conversely, p300 acetyltransferase acetylates Atg5, Atg7, Atg8, and Atg12, thus inhibiting starvation-induced autophagy (98). Although controversial, nuclear SIRT1 seems to induce the expression of autophagy genes through the activation of FoxOs (66). HDAC1 and HDAC2 have also been found to regulate muscle autophagy by controlling the expression of autophagy genes (128).
As discussed earlier, an important role in muscle adaptation to exercise is played by ROS. Very interestingly, ROS can also regulate autophagy. The mechanism through which ROS induce autophagy has not been fully elucidated, but it is believed that ROS could increase Beclin1 expression and/or regulate the activity of Atg4 (27, 28, 165). Both ROS and autophagy might have either beneficial or detrimental effects, depending on their balance.
Although there are many links suggesting an involvement of autophagy in muscle remodeling due to exercise, the underlying signalings are still obscure, sometimes contradictory and need to be further investigated.
Metabolic Modulating Agents
As reported earlier, some pharmacological compounds defined as “exercise mimetics” can produce metabolic effects similar to those induced by exercise. However, as suggested by Booth and Laye (11), recapitulating the complexity of all the molecular effects at the level of various organs with a single “exercise pill” is unlikely to be achieved. Such drugs might be used to reproduce only some of the multiple effects due to exercise. Nevertheless, considering such limits, pharmaco-therapy that replicates some exercise-induced effects could be extremely useful in potentiating the adaptive response to exercise or under conditions where there is a physiologically low response of muscle to exercise, such as in some insulin-resistent individuals or in old people whose response to exercise in terms of muscle mass increase is low (33, 92).
We have already discussed how metabolic remodeling agents such as GW1516 and AICAR recapitulate some effects of exercise that are aimed at achieving the best metabolic energy efficiency. Other drugs might modulate metabolism through different mechanisms. For example, the category of “metabolic modulators” (including trimetazidine and perhexiline) optimizes metabolism by shifting ATP production from FFA β-oxidation toward glucose oxidation. This increases the energy production efficiency, as ATP synthesis during FFA β-oxidation requires more oxygen than during glucose oxidation. Metabolic modulators would likely trigger metabolic sensors as in the case of GW1516 and AICAR and might be beneficial during exercise where there is a high oxygen expenditure. The choice of glucose as a substrate triggered by these drugs induces a more efficient utilization of the oxygen available; this could increase skeletal muscle metabolism efficiency and contractile performance, as already observed for cardiac muscle function under transitory hypoxia (106). It has already been demonstrated that trimetazidine improves exercise capability in patients suffering from chronic stable angina (187), and that it has a strong hypertrophic effect on cultured myotubes (42). Moreover, as in the case of exercise, metabolic modulators increase glucose uptake, thereby possibly enhancing insuline sensitivity, although they reduce the FFA catabolism, which is strongly enhanced during exercise.
Conclusions
The effects of exercise on skeletal muscle are various and involve several molecular and metabolic players, which are activated to a larger or lesser extent depending on the modalities of training. The recent findings about the involvement of autophagy and reactive species, with their ambivalent effects, in adaptation to exercise demonstrate how this phenomenon is extremely complicated. Exercise's physiological and therapeutic relevance to health justifies the efforts of the scientific community that are aimed at unraveling the complex network of signaling and metabolic pathways triggered by physical training on skeletal muscle. This will enable an optimization of exercise prescription in the case of chronic diseases along with deeper knowledge of the several effects that drugs such as antioxidants and metabolic remodeling agents might exert on adaptation to exercise.
Abbreviations Used
- 4E-BP1
eukaryotic translation initiation factor 4E-binding protein-1
- ACC
acetyl-CoA carboxylase
- AICAR
5-aminoimidazole-4-carboxamide-1-β-4-ribofuranoside
- AK
adenylate kinase
- AMPK
AMP-activated protein kinase
- ATF
activating transcription factor
- ATG
autophagy-related genes
- Bcl2
B-cell lymphoma 2
- CaMKs
Ca2+/calmodulin-dependent kinases
- CASA
chaperone-assisted selective autophagy
- CK
creatine kinases
- CPT
carnitine palmitoyltransferase
- Cr
creatine
- CREB
cAMP-response element-binding protein
- CSA
cross-sectional area
- Cyt c
cytochrome c
- DMD
Duchenne muscular dystrophy
- EPG5
ectopic P-granules autophagy-protein-5
- ERK
extracellular-signal-regulated kinase
- ERR-α
estrogen-related receptor-α
- FAK
focal adhesion kinase
- FAT/CD36
fatty acyl translocase
- FFAs
free fatty acids
- FoxO
forkhead box
- GCN5
general control of amino-acid synthesis
- Glu
glucose
- Glu-6-P
glucose-6-phosphate
- GLUT4
glucose transporter 4
- GPX
glutatione peroxidase
- H2O2
hydrogen peroxide
- HDAC
histone deacetylase
- HIF-1
hypoxia-inducible factor-1
- HK
hexokinase
- IGF
insulin growth factor
- IM
inner membrane
- JNK
c-Jun N-terminal kinase
- LC3
microtubule-associated protein 1-light chain 3
- LDH
lactate dehydrogenase
- LKB1
liver kinase B1
- MAFbx
muscle atrophy F-box
- MEF2
myocyte enhancer factor 2
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- MuRF-1
muscle ring finger protein 1
- MyHC
myosin heavy chain
- myoD
myogenic differentiation factor
- NAC
N-acetyl-l-cysteine
- Naf1
nutrient deprivation autophagy factor-1
- NAMPT
nicotinamide phosphoribosyltransferase
- NF-κB
nuclear factor-kappa B
- NFAT
nuclear factor of activated T-cells
- NO•
nitric oxide
- NOS
nitric oxide synthases
- NRF-1
nuclear respiratory factor-1
- O2•−
superoxide anion
- OH•
hydroxyl radical
- OM
outer membrane
- ONOO−
peroxynitrite
- p38 MAPK
p38 mitogen-activated protein kinase
- p70S6K
p70 ribosomal protein S6 kinase
- PCr
phosphocreatine
- PDH
pyruvate dehydrogenase
- PDK
pyruvate dehydrogenase kinase
- PE
phosphatidyl-ethanolamine
- PGC-1α
peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α
- PHD
prolyl hydroxylase enzymes
- PI3K
phosphoinositide 3-kinase
- PLA2
phospholipase A2
- PO2
partial pressure of oxygen
- PPARγ
peroxisome proliferator-activated receptor γ
- PTEN
phosphatase and tensin homologue
- Pyr
pyruvate
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SERCAs
sarcoplasmic reticulum Ca2+ ATPases
- SDH
succinate dehydrogenase
- SIRT1
sirtuin 1
- SOD
superoxide dismutase
- Sox9
sex determining region Y-box 9
- TCA
tricarboxylic acid
- Tfam
transcription factor A mitochondrial
- TSC
tuberosus sclerosis complex
- Ub
ubiquinon
- ULK1
unc-51-like kinase1
- Vps
vacuolar protein sorting
Acknowledgments
The authors are grateful to Martin Bennet for the valuable editorial work. This work was supported by the Italian Ministry of Health (Ricerca finalizzata; RF-2010-2318508) and Studies Investigating Co-morbidities Aggravating Heart Failure (SICA-HF) (Grant Agreement no. 241558, Collaborative Project, Large Scale EU FP7 Health).
References
- 1.Abruzzo PM, Esposito F, Marchionni C, di Tullio S, Belia S, Fulle S, Veicsteinas A, and Marini M. Moderate exercise training induces ROS-related adaptations to skeletal muscles. Int J Sports Med 34: 676–687, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, and Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem 280: 19587–19593, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, and Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 276: 35243–35246, 2001 [DOI] [PubMed] [Google Scholar]
- 4.Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, and Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J 19: 786–788, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, and Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 16: 1879–1886, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Barany M. ATPase activity of myosin correlated with speed of muscle shortening. J Gen Physiol 50Suppl: 197–218, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, and Ruegg MA. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab 8: 411–424, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, and Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab 281: E1340–E1346, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, and Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Bonaldo P. and Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 6: 25–39, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth FW. and Laye MJ. Lack of adequate appreciation of physical exercise's complexities can pre-empt appropriate design and interpretation in scientific discovery. J Physiol 587: 5527–5539, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booth FW. and Roberts CK. Linking performance and chronic disease risk: indices of physical performance are surrogates for health. Br J Sports Med 42: 950–952, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Booth FW. and Thomason DB. Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol Rev 71: 541–585, 1991 [DOI] [PubMed] [Google Scholar]
- 14.Borniquel S, Garcia-Quintans N, Valle I, Olmos Y, Wild B, Martinez-Granero F, Soria E, Lamas S, and Monsalve M. Inactivation of Foxo3a and subsequent downregulation of PGC-1 alpha mediate nitric oxide-induced endothelial cell migration. Mol Cell Biol 30: 4035–4044, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borst SE. Interventions for sarcopenia and muscle weakness in older people. Age Ageing 33: 548–555, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Bottinelli R. Functional heterogeneity of mammalian single muscle fibres: do myosin isoforms tell the whole story? Pflugers Arch 443: 6–17, 2001 [DOI] [PubMed] [Google Scholar]
- 17.Branvold DJ, Allred DR, Beckstead DJ, Kim HJ, Fillmore N, Condon BM, Brown JD, Sudweeks SN, Thomson DM, and Winder WW. Thyroid hormone effects on LKB1, MO25, phospho-AMPK, phospho-CREB, and PGC-1alpha in rat muscle. J Appl Physiol (1985) 105: 1218–1227, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Brault JJ, Jespersen JG, and Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem 285: 19460–19471, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, and Rangwala SM. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol 104: 1304–1312, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, and Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, and Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11: 213–219, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, and Collins S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol 24: 3057–3067, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carling D. and Hardie DG. The substrate and sequence specificity of the AMP-activated protein kinase. Phosphorylation of glycogen synthase and phosphorylase kinase. Biochim Biophys Acta 1012: 81–86, 1989 [DOI] [PubMed] [Google Scholar]
- 24.Carlstrom M, Larsen FJ, Nystrom T, Hezel M, Borniquel S, Weitzberg E, and Lundberg JO. Dietary inorganic nitrate reverses features of metabolic syndrome in endothelial nitric oxide synthase-deficient mice. Proc Natl Acad Sci U S A 107: 17716–17720, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers MA, Moylan JS, Smith JD, Goodyear LJ, and Reid MB. Stretch-stimulated glucose uptake in skeletal muscle is mediated by reactive oxygen species and p38 MAP-kinase. J Physiol 587: 3363–3373, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen GQ, Mou CY, Yang YQ, Wang S, and Zhao ZW. Exercise training has beneficial anti-atrophy effects by inhibiting oxidative stress-induced MuRF1 upregulation in rats with diabetes. Life Sci 89: 44–49, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, McMillan-Ward E, Kong J, Israels SJ, and Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci 120: 4155–4166, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, McMillan-Ward E, Kong J, Israels SJ, and Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ 15: 171–182, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Chin ER. Intracellular Ca2+ signaling in skeletal muscle: decoding a complex message. Exerc Sport Sci Rev 38: 76–85, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, and Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev 12: 2499–2509, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colberg SR, Albright AL, Blissmer BJ, Braun B, Chasan-Taber L, Fernhall B, Regensteiner JG, Rubin RR, and Sigal RJ. Exercise and type 2 diabetes: American College of Sports Medicine and the American Diabetes Association: joint position statement. Exercise and type 2 diabetes. Med Sci Sports Exerc 42: 2282–2303, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, Simpson MA, Yau S, Bertini E, McClelland V, Al-Owain M, Koelker S, Koerner C, Hoffmann GF, Wijburg FA, ten Hoedt AE, Rogers RC, Manchester D, Miyata R, Hayashi M, Said E, Soler D, Kroisel PM, Windpassinger C, Filloux FM, Al-Kaabi S, Hertecant J, Del Campo M, Buk S, Bodi I, Goebel HH, Sewry CA, Abbs S, Mohammed S, Josifova D, Gautel M, and Jungbluth H. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 45: 83–87, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Filippis E, Alvarez G, Berria R, Cusi K, Everman S, Meyer C, and Mandarino LJ. Insulin-resistant muscle is exercise resistant: evidence for reduced response of nuclear-encoded mitochondrial genes to exercise. Am J Physiol Endocrinol Metab 294: E607–E614, 2008 [DOI] [PubMed] [Google Scholar]
- 34.de la Cruz-Morcillo MA, Valero ML, Callejas-Valera JL, Arias-Gonzalez L, Melgar-Rojas P, Galan-Moya EM, Garcia-Gil E, Garcia-Cano J, and Sanchez-Prieto R. P38MAPK is a major determinant of the balance between apoptosis and autophagy triggered by 5-fluorouracil: implication in resistance. Oncogene 31: 1073–1085, 2012 [DOI] [PubMed] [Google Scholar]
- 35.De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, Rovere-Querini P, Moggio M, Ripolone M, Francolini M, Sandri M, and Clementi E. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis 3: e418, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derbre F, Ferrando B, Gomez-Cabrera MC, Sanchis-Gomar F, Martinez-Bello VE, Olaso-Gonzalez G, Diaz A, Gratas-Delamarche A, Cerda M, and Vina J. Inhibition of xanthine oxidase by allopurinol prevents skeletal muscle atrophy: role of p38 MAPKinase and E3 ubiquitin ligases. PLoS One 7: e46668, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolmetsch RE, Lewis RS, Goodnow CC, and Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386: 855–858, 1997 [DOI] [PubMed] [Google Scholar]
- 38.Egan B, Carson BP, Garcia-Roves PM, Chibalin AV, Sarsfield FM, Barron N, McCaffrey N, Moyna NM, Zierath JR, and O'Gorman DJ. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J Physiol 588: 1779–1790, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan B. and Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17: 162–184, 2013 [DOI] [PubMed] [Google Scholar]
- 40.Feng Z, Zou X, Jia H, Li X, Zhu Z, Liu X, Bucheli P, Ballevre O, Hou Y, Zhang W, Wang J, Chen Y, and Liu J. Maternal docosahexaenoic acid feeding protects against impairment of learning and memory and oxidative stress in prenatally stressed rats: possible role of neuronal mitochondria metabolism. Antioxid Redox Signal 16: 275–289, 2012 [DOI] [PubMed] [Google Scholar]
- 41.Ferraro E. and Cecconi F. Autophagic and apoptotic response to stress signals in mammalian cells. Arch Biochem Biophys 462: 210–219, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Ferraro E, Giammarioli AM, Caldarola S, Lista P, Feraco A, Tinari A, Salvatore AM, Malorni W, Berghella L, and Rosano G. The metabolic modulator trimetazidine triggers autophagy and counteracts stress-induced atrophy in skeletal muscle myotubes. FEBS J 280: 5094–5108, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Figueira TR, Barros MH, Camargo AA, Castilho RF, Ferreira JC, Kowaltowski AJ, Sluse FE, Souza-Pinto NC, and Vercesi AE. Mitochondria as a source of reactive oxygen and nitrogen species: from molecular mechanisms to human health. Antioxid Redox Signal 18: 2029–2074, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, and Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Formenti F, Constantin-Teodosiu D, Emmanuel Y, Cheeseman J, Dorrington KL, Edwards LM, Humphreys SM, Lappin TR, McMullin MF, McNamara CJ, Mills W, Murphy JA, O'Connor DF, Percy MJ, Ratcliffe PJ, Smith TG, Treacy M, Frayn KN, Greenhaff PL, Karpe F, Clarke K, and Robbins PA. Regulation of human metabolism by hypoxia-inducible factor. Proc Natl Acad Sci U S A 107: 12722–12727, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Volpi E, and Rasmussen BB. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci 68: 599–607, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, and Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 14: 661–673, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaitanos GC, Williams C, Boobis LH, and Brooks S. Human muscle metabolism during intermittent maximal exercise. J Appl Physiol 75: 712–719, 1993 [DOI] [PubMed] [Google Scholar]
- 49.Geng T, Li P, Okutsu M, Yin X, Kwek J, Zhang M, and Yan Z. PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Am J Physiol Cell Physiol 298: C572–C579, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, and Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J 26: 1913–1923, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gielen S, Sandri M, Kozarez I, Kratzsch J, Teupser D, Thiery J, Erbs S, Mangner N, Lenk K, Hambrecht R, Schuler G, and Adams V. Exercise training attenuates MuRF-1 expression in the skeletal muscle of patients with chronic heart failure independent of age: the randomized Leipzig Exercise Intervention in Chronic Heart Failure and Aging catabolism study. Circulation 125: 2716–2727, 2012 [DOI] [PubMed] [Google Scholar]
- 52.Glass DJ. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. Curr Top Microbiol Immunol 346: 267–278, 2010 [DOI] [PubMed] [Google Scholar]
- 53.Glynn EL, Fry CS, Drummond MJ, Dreyer HC, Dhanani S, Volpi E, and Rasmussen BB. Muscle protein breakdown has a minor role in the protein anabolic response to essential amino acid and carbohydrate intake following resistance exercise. Am J Physiol Regul Integr Comp Physiol 299: R533–R540, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gohil K. and Brooks GA. Exercise tames the wild side of the Myc network: a hypothesis. Am J Physiol Endocrinol Metab 303: E18–E30, 2012 [DOI] [PubMed] [Google Scholar]
- 55.Gomez-Cabrera MC, Domenech E, Romagnoli M, Arduini A, Borras C, Pallardo FV, Sastre J, and Vina J. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am J Clin Nutr 87: 142–149, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Gong MC, Arbogast S, Guo Z, Mathenia J, Su W, and Reid MB. Calcium-independent phospholipase A2 modulates cytosolic oxidant activity and contractile function in murine skeletal muscle cells. J Appl Physiol 100: 399–405, 2006 [DOI] [PubMed] [Google Scholar]
- 57.Goodyear LJ. and Kahn BB. Exercise, glucose transport, and insulin sensitivity. Annu Rev Med 49: 235–261, 1998 [DOI] [PubMed] [Google Scholar]
- 58.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, Blaauw B, Urciuolo A, Tiepolo T, Merlini L, Maraldi NM, Bernardi P, Sandri M, and Bonaldo P. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med 16: 1313–1320, 2010 [DOI] [PubMed] [Google Scholar]
- 59.Grumati P, Coletto L, Schiavinato A, Castagnaro S, Bertaggia E, Sandri M, and Bonaldo P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy 7: 1415–1423, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gundersen K. Excitation-transcription coupling in skeletal muscle: the molecular pathways of exercise. Biol Rev Camb Philos Soc 86: 564–600, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutteridge JM. and Halliwell B. Antioxidants: molecules, medicines, and myths. Biochem Biophys Res Commun 393: 561–564, 2010 [DOI] [PubMed] [Google Scholar]
- 62.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, and Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem 282: 30014–30021, 2007 [DOI] [PubMed] [Google Scholar]
- 63.Handschin C, Rhee J, Lin J, Tarr PT, and Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A 100: 7111–7116, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Handschin C. and Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454: 463–469, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 25: 1895–1908, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, and Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res 107: 1470–1482, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haskell WL, Lee IM, Pate RR, Powell KE, Blair SN, Franklin BA, Macera CA, Heath GW, Thompson PD, and Bauman A. Physical activity and public health: updated recommendation for adults from the American College of Sports Medicine and the American Heart Association. Med Sci Sports Exerc 39: 1423–1434, 2007 [DOI] [PubMed] [Google Scholar]
- 68.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, and Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481: 511–515, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inoki K, Kim J, and Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 52: 381–400, 2012 [DOI] [PubMed] [Google Scholar]
- 70.Irrcher I, Adhihetty PJ, Sheehan T, Joseph AM, and Hood DA. PPARgamma coactivator-1alpha expression during thyroid hormone- and contractile activity-induced mitochondrial adaptations. Am J Physiol Cell Physiol 284: C1669–C1677, 2003 [DOI] [PubMed] [Google Scholar]
- 71.Irrcher I, Ljubicic V, and Hood DA. Interactions between ROS and AMP kinase activity in the regulation of PGC-1alpha transcription in skeletal muscle cells. Am J Physiol Cell Physiol 296: C116–C123, 2009 [DOI] [PubMed] [Google Scholar]
- 72.Irrcher I, Ljubicic V, Kirwan AF, and Hood DA. AMP-activated protein kinase-regulated activation of the PGC-1alpha promoter in skeletal muscle cells. PLoS One 3: e3614, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, and Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472, 2001 [DOI] [PubMed] [Google Scholar]
- 74.Jager S, Handschin C, St-Pierre J, and Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A 104: 12017–12022, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jahnke VE, Van Der Meulen JH, Johnston HK, Ghimbovschi S, Partridge T, Hoffman EP, and Nagaraju K. Metabolic remodeling agents show beneficial effects in the dystrophin-deficient mdx mouse model. Skelet Muscle 2: 16, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jamart C, Benoit N, Raymackers JM, Kim HJ, Kim CK, and Francaux M. Autophagy-related and autophagy-regulatory genes are induced in human muscle after ultraendurance exercise. Eur J Appl Physiol 112: 3173–3177, 2012 [DOI] [PubMed] [Google Scholar]
- 77.Jamart C, Francaux M, Millet GY, Deldicque L, Frere D, and Feasson L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J Appl Physiol 112: 1529–1537, 2012 [DOI] [PubMed] [Google Scholar]
- 78.Jeong HW, Cho SY, Kim S, Shin ES, Kim JM, Song MJ, Park PJ, Sohn JH, Park H, Seo DB, Kim WG, and Lee SJ. Chitooligosaccharide induces mitochondrial biogenesis and increases exercise endurance through the activation of Sirt1 and AMPK in rats. PLoS One 7: e40073, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang LQ, Garcia-Roves PM, de Castro Barbosa T, and Zierath JR. Constitutively active calcineurin in skeletal muscle increases endurance performance and mitochondrial respiratory capacity. Am J Physiol Endocrinol Metab 298: E8–E16, 2010 [DOI] [PubMed] [Google Scholar]
- 80.Kahn BB, Alquier T, Carling D, and Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25, 2005 [DOI] [PubMed] [Google Scholar]
- 81.Kang C, O'Moore KM, Dickman JR, and Ji LL. Exercise activation of muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha signaling is redox sensitive. Free Radic Biol Med 47: 1394–1400, 2009 [DOI] [PubMed] [Google Scholar]
- 82.Kim YA, Kim YS, Oh SL, Kim HJ, and Song W. Autophagic response to exercise training in skeletal muscle with age. J Physiol Biochem 69: 697–705, 2013 [DOI] [PubMed] [Google Scholar]
- 83.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937, 2007 [DOI] [PubMed] [Google Scholar]
- 84.Klossner S, Durieux AC, Freyssenet D, and Flueck M. Mechano-transduction to muscle protein synthesis is modulated by FAK. Eur J Appl Physiol 106: 389–398, 2009 [DOI] [PubMed] [Google Scholar]
- 85.Koh TJ. and Tidball JG. Nitric oxide synthase inhibitors reduce sarcomere addition in rat skeletal muscle. J Physiol 519Pt 1: 189–196, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kopp R, Koblitz L, Egg M, and Pelster B. HIF signaling and overall gene expression changes during hypoxia and prolonged exercise differ considerably. Physiol Genomics 43: 506–516, 2011 [DOI] [PubMed] [Google Scholar]
- 87.Kou R, Greif D, and Michel T. Dephosphorylation of endothelial nitric-oxide synthase by vascular endothelial growth factor. Implications for the vascular responses to cyclosporin A. J Biol Chem 277: 29669–29673, 2002 [DOI] [PubMed] [Google Scholar]
- 88.Kraft CS, LeMoine CM, Lyons CN, Michaud D, Mueller CR, and Moyes CD. Control of mitochondrial biogenesis during myogenesis. Am J Physiol Cell Physiol 290: C1119–C1127, 2006 [DOI] [PubMed] [Google Scholar]
- 89.Kramer DK, Al-Khalili L, Guigas B, Leng Y, Garcia-Roves PM, and Krook A. Role of AMP kinase and PPARdelta in the regulation of lipid and glucose metabolism in human skeletal muscle. J Biol Chem 282: 19313–19320, 2007 [DOI] [PubMed] [Google Scholar]
- 90.Kroemer G, Marino G, and Levine B. Autophagy and the integrated stress response. Mol Cell 40: 280–293, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuma A. and Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol 21: 683–690, 2010 [DOI] [PubMed] [Google Scholar]
- 92.Kumar V, Selby A, Rankin D, Patel R, Atherton P, Hildebrandt W, Williams J, Smith K, Seynnes O, Hiscock N, and Rennie MJ. Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. J Physiol 587: 211–217, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kusuhara K, Madsen K, Jensen L, Hellsten Y, and Pilegaard H. Calcium signalling in the regulation of PGC-1alpha, PDK4 and HKII mRNA expression. Biol Chem 388: 481–488, 2007 [DOI] [PubMed] [Google Scholar]
- 94.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, and Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127: 1109–1122, 2006 [DOI] [PubMed] [Google Scholar]
- 95.Lamb GD. and Westerblad H. Acute effects of reactive oxygen and nitrogen species on the contractile function of skeletal muscle. J Physiol 589: 2119–2127, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leblanc PJ, Howarth KR, Gibala MJ, and Heigenhauser GJ. Effects of 7 wk of endurance training on human skeletal muscle metabolism during submaximal exercise. J Appl Physiol 97: 2148–2153, 2004 [DOI] [PubMed] [Google Scholar]
- 97.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, and Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 105: 3374–3379, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee IH. and Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 284: 6322–6328, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lempiainen J, Finckenberg P, Mervaala EE, Sankari S, Levijoki J, and Mervaala EM. Caloric restriction ameliorates kidney ischaemia/reperfusion injury through PGC-1alpha-eNOS pathway and enhanced autophagy. Acta Physiol (Oxf) 208: 410–421, 2013 [DOI] [PubMed] [Google Scholar]
- 100.Levine B, Sinha S, and Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 4: 600–606, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin J, Handschin C, and Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370, 2005 [DOI] [PubMed] [Google Scholar]
- 102.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, and Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418: 797–801, 2002 [DOI] [PubMed] [Google Scholar]
- 103.Lira VA, Benton CR, Yan Z, and Bonen A. PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. Am J Physiol Endocrinol Metab 299: E145–E161, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y, Randall WR, and Schneider MF. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J Cell Biol 168: 887–897, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Long YC, Barnes BR, Mahlapuu M, Steiler TL, Martinsson S, Leng Y, Wallberg-Henriksson H, Andersson L, and Zierath JR. Role of AMP-activated protein kinase in the coordinated expression of genes controlling glucose and lipid metabolism in mouse white skeletal muscle. Diabetologia 48: 2354–2364, 2005 [DOI] [PubMed] [Google Scholar]
- 106.Lopaschuk GD, Barr R, Thomas PD, and Dyck JR. Beneficial effects of trimetazidine in ex vivo working ischemic hearts are due to a stimulation of glucose oxidation secondary to inhibition of long-chain 3-ketoacyl coenzyme a thiolase. Circ Res 93: e33–e37, 2003 [DOI] [PubMed] [Google Scholar]
- 107.Lundby C, Calbet JA, and Robach P. The response of human skeletal muscle tissue to hypoxia. Cell Mol Life Sci 66: 3615–3623, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luo L, Lu AM, Wang Y, Hong A, Chen Y, Hu J, Li X, and Qin ZH. Chronic resistance training activates autophagy and reduces apoptosis of muscle cells by modulating IGF-1 and its receptors, Akt/mTOR and Akt/FOXO3a signaling in aged rats. Exp Gerontol 48: 427–436, 2013 [DOI] [PubMed] [Google Scholar]
- 109.Lv XC. and Zhou HY. Resveratrol protects H9c2 embryonic rat heart derived cells from oxidative stress by inducing autophagy: role of p38 mitogen-activated protein kinase. Can J Physiol Pharmacol 90: 655–662, 2012 [DOI] [PubMed] [Google Scholar]
- 110.Mack HI, Zheng B, Asara JM, and Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 8: 1197–1214, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mackenzie MG, Hamilton DL, Murray JT, and Baar K. mVps34 is activated by an acute bout of resistance exercise. Biochem Soc Trans 35: 1314–1316, 2007 [DOI] [PubMed] [Google Scholar]
- 112.Madaro L, Marrocco V, Carnio S, Sandri M, and Bouche M. Intracellular signaling in ER stress-induced autophagy in skeletal muscle cells. FASEB J 27: 1990–2000, 2013 [DOI] [PubMed] [Google Scholar]
- 113.Malinska D, Kudin AP, Bejtka M, and Kunz WS. Changes in mitochondrial reactive oxygen species synthesis during differentiation of skeletal muscle cells. Mitochondrion 12: 144–148, 2012 [DOI] [PubMed] [Google Scholar]
- 114.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, and Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6: 458–471, 2007 [DOI] [PubMed] [Google Scholar]
- 115.Martelli AM, Evangelisti C, Chiarini F, Grimaldi C, Cappellini A, Ognibene A, and McCubrey JA. The emerging role of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling network in normal myelopoiesis and leukemogenesis. Biochim Biophys Acta 1803: 991–1002, 2010 [DOI] [PubMed] [Google Scholar]
- 116.Martins KJ, St-Louis M, Murdoch GK, MacLean IM, McDonald P, Dixon WT, Putman CT, and Michel RN. Nitric oxide synthase inhibition prevents activity-induced calcineurin-NFATc1 signalling and fast-to-slow skeletal muscle fibre type conversions. J Physiol 590: 1427–1442, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mason SD, Howlett RA, Kim MJ, Olfert IM, Hogan MC, McNulty W, Hickey RP, Wagner PD, Kahn CR, Giordano FJ, and Johnson RS. Loss of skeletal muscle HIF-1alpha results in altered exercise endurance. PLoS Biol 2: e288, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mathai AS, Bonen A, Benton CR, Robinson DL, and Graham TE. Rapid exercise-induced changes in PGC-1alpha mRNA and protein in human skeletal muscle. J Appl Physiol 105: 1098–1105, 2008 [DOI] [PubMed] [Google Scholar]
- 119.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, and Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11: 385–396, 2009 [DOI] [PubMed] [Google Scholar]
- 120.Matsuzawa T, Kim BH, Shenoy AR, Kamitani S, Miyake M, and Macmicking JD. IFN-gamma elicits macrophage autophagy via the p38 MAPK signaling pathway. J Immunol 189: 813–818, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McClung JM, Judge AR, Powers SK, and Yan Z. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol 298: C542–C549, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McConell GK, Ng GP, Phillips M, Ruan Z, Macaulay SL, and Wadley GD. Central role of nitric oxide synthase in AICAR and caffeine-induced mitochondrial biogenesis in L6 myocytes. J Appl Physiol 108: 589–595, 2010 [DOI] [PubMed] [Google Scholar]
- 123.McGee SL, Fairlie E, Garnham AP, and Hargreaves M. Exercise-induced histone modifications in human skeletal muscle. J Physiol 587: 5951–5958, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Michael LF, Wu Z, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, Kelly DP, and Spiegelman BM. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A 98: 3820–3825, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mikines KJ, Sonne B, Farrell PA, Tronier B, and Galbo H. Effect of physical exercise on sensitivity and responsiveness to insulin in humans. Am J Physiol 254: E248–E259, 1988 [DOI] [PubMed] [Google Scholar]
- 126.Mizushima N. and Komatsu M. Autophagy: renovation of cells and tissues. Cell 147: 728–741, 2011 [DOI] [PubMed] [Google Scholar]
- 127.Momken I, Stevens L, Bergouignan A, Desplanches D, Rudwill F, Chery I, Zahariev A, Zahn S, Stein TP, Sebedio JL, Pujos-Guillot E, Falempin M, Simon C, Coxam V, Andrianjafiniony T, Gauquelin-Koch G, Picquet F, and Blanc S. Resveratrol prevents the wasting disorders of mechanical unloading by acting as a physical exercise mimetic in the rat. FASEB J 25: 3646–3660, 2011 [DOI] [PubMed] [Google Scholar]
- 128.Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, and Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143: 35–45, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moruno-Manchon JF, Perez-Jimenez E, and Knecht E. Glucose induces autophagy under starvation conditions by a p38 MAPK-dependent pathway. Biochem J 449: 497–506, 2013 [DOI] [PubMed] [Google Scholar]
- 130.Murray J. and Huss JM. Estrogen-related receptor alpha regulates skeletal myocyte differentiation via modulation of the ERK MAP kinase pathway. Am J Physiol Cell Physiol 301: C630–C645, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nader GA. and Esser KA. Intracellular signaling specificity in skeletal muscle in response to different modes of exercise. J Appl Physiol 90: 1936–1942, 2001 [DOI] [PubMed] [Google Scholar]
- 132.Nakae J, Cao Y, Oki M, Orba Y, Sawa H, Kiyonari H, Iskandar K, Suga K, Lombes M, and Hayashi Y. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes 57: 563–576, 2008 [DOI] [PubMed] [Google Scholar]
- 133.Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, and Evans RM. AMPK and PPARdelta agonists are exercise mimetics. Cell 134: 405–415, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, and Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem 275: 4545–4548, 2000 [DOI] [PubMed] [Google Scholar]
- 135.Nemazanyy I, Blaauw B, Paolini C, Caillaud C, Protasi F, Mueller A, Proikas-Cezanne T, Russell RC, Guan KL, Nishino I, Sandri M, Pende M, and Panasyuk G. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med 5: 870–890, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ng F. and Tang BL. Sirtuins' modulation of autophagy. J Cell Physiol 228: 2262–2270, 2013 [DOI] [PubMed] [Google Scholar]
- 137.Ng S, Wu YT, Chen B, Zhou J, and Shen HM. Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy 7: 1173–1186, 2011 [DOI] [PubMed] [Google Scholar]
- 138.Nisoli E, Falcone S, Tonello C, Cozzi V, Palomba L, Fiorani M, Pisconti A, Brunelli S, Cardile A, Francolini M, Cantoni O, Carruba MO, Moncada S, and Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A 101: 16507–16512, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ortenblad N, Westerblad H, and Nielsen J. Muscle glycogen stores and fatigue. J Physiol 591Pt 18: 4405–4413, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pattwell DM, McArdle A, Morgan JE, Patridge TA, and Jackson MJ. Release of reactive oxygen and nitrogen species from contracting skeletal muscle cells. Free Radic Biol Med 37: 1064–1072, 2004 [DOI] [PubMed] [Google Scholar]
- 141.Pedersen BK. and Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol 8: 457–465, 2012 [DOI] [PubMed] [Google Scholar]
- 142.Periasamy M. and Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve 35: 430–442, 2007 [DOI] [PubMed] [Google Scholar]
- 143.Pette D. and Staron RS. Transitions of muscle fiber phenotypic profiles. Histochem Cell Biol 115: 359–372, 2001 [DOI] [PubMed] [Google Scholar]
- 144.Phillips SM, Green HJ, Tarnopolsky MA, Heigenhauser GJ, and Grant SM. Progressive effect of endurance training on metabolic adaptations in working skeletal muscle. Am J Physiol 270: E265–E272, 1996 [DOI] [PubMed] [Google Scholar]
- 145.Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, Baar K, and Schenk S. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J Biol Chem 286: 30561–30570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pilegaard H, Saltin B, and Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J Physiol 546: 851–858, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pogozelski AR, Geng T, Li P, Yin X, Lira VA, Zhang M, Chi JT, and Yan Z. p38gamma mitogen-activated protein kinase is a key regulator in skeletal muscle metabolic adaptation in mice. PLoS One 4: e7934, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Powers SK, Duarte J, Kavazis AN, and Talbert EE. Reactive oxygen species are signalling molecules for skeletal muscle adaptation. Exp Physiol 95: 1–9, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Powers SK, Kavazis AN, and McClung JM. Oxidative stress and disuse muscle atrophy. J Appl Physiol (1985) 102: 2389–2397, 2007 [DOI] [PubMed] [Google Scholar]
- 150.Powers SK, Nelson WB, and Hudson MB. Exercise-induced oxidative stress in humans: cause and consequences. Free Radic Biol Med 51: 942–950, 2011 [DOI] [PubMed] [Google Scholar]
- 151.Puigserver P. and Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 24: 78–90, 2003 [DOI] [PubMed] [Google Scholar]
- 152.Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839, 1998 [DOI] [PubMed] [Google Scholar]
- 153.Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J, Feramisco JR, Karin M, and Wang JY. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev 14: 574–584, 2000 [PMC free article] [PubMed] [Google Scholar]
- 154.Qin L, Wang Z, Tao L, and Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6: 239–247, 2010 [DOI] [PubMed] [Google Scholar]
- 155.Radak Z, Zhao Z, Koltai E, Ohno H, and Atalay M. Oxygen consumption and usage during physical exercise: the balance between oxidative stress and ROS-dependent adaptive signaling. Antioxid Redox Signal 18: 1208–1246, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ristow M, Zarse K, Oberbach A, Kloting N, Birringer M, Kiehntopf M, Stumvoll M, Kahn CR, and Bluher M. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Natl Acad Sci U S A 106: 8665–8670, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ryall JG. Metabolic reprogramming as a novel regulator of skeletal muscle development and regeneration. FEBS J 280: 4004–4013, 2013 [DOI] [PubMed] [Google Scholar]
- 158.Salminen A. Lysosomal changes in skeletal muscles during the repair of exercise injuries in muscle fibers. Acta Physiol Scand Suppl 539: 1–31, 1985 [PubMed] [Google Scholar]
- 159.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, and Candau R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem 113: 695–710, 2012 [DOI] [PubMed] [Google Scholar]
- 160.Sandri M. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am J Physiol Cell Physiol 298: C1291–C1297, 2010 [DOI] [PubMed] [Google Scholar]
- 161.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol 45: 2121–2129, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, and Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A 103: 16260–16265, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sandstrom ME, Zhang SJ, Bruton J, Silva JP, Reid MB, Westerblad H, and Katz A. Role of reactive oxygen species in contraction-mediated glucose transport in mouse skeletal muscle. J Physiol 575: 251–262, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, McDonald K, Stajich JM, Mahjneh I, Vihola A, Raheem O, Penttila S, Lehtinen S, Huovinen S, Palmio J, Tasca G, Ricci E, Hackman P, Hauser M, Katsanis N, and Udd B. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet 44: 450–455, S1–S2, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, and Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schiaffino S. Fibre types in skeletal muscle: a personal account. Acta Physiol (Oxf) 199: 451–463, 2010 [DOI] [PubMed] [Google Scholar]
- 167.Schiaffino S. and Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 76: 371–423, 1996 [DOI] [PubMed] [Google Scholar]
- 168.Schwer B. and Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab 7: 104–112, 2008 [DOI] [PubMed] [Google Scholar]
- 169.Sellman JE, DeRuisseau KC, Betters JL, Lira VA, Soltow QA, Selsby JT, and Criswell DS. In vivo inhibition of nitric oxide synthase impairs upregulation of contractile protein mRNA in overloaded plantaris muscle. J Appl Physiol 100: 258–265, 2006 [DOI] [PubMed] [Google Scholar]
- 170.Semenza GL. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 24: 97–106, 2009 [DOI] [PubMed] [Google Scholar]
- 171.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, and Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci U S A 98: 13108–13113, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shimada Y, Kobayashi H, Kawagoe S, Aoki K, Kaneshiro E, Shimizu H, Eto Y, Ida H, and Ohashi T. Endoplasmic reticulum stress induces autophagy through activation of p38 MAPK in fibroblasts from Pompe disease patients carrying c.546G>T mutation. Mol Genet Metab 104: 566–573, 2011 [DOI] [PubMed] [Google Scholar]
- 173.Silveira LR, Pereira-Da-Silva L, Juel C, and Hellsten Y. Formation of hydrogen peroxide and nitric oxide in rat skeletal muscle cells during contractions. Free Radic Biol Med 35: 455–464, 2003 [DOI] [PubMed] [Google Scholar]
- 174.Silveira LR, Pilegaard H, Kusuhara K, Curi R, and Hellsten Y. The contraction induced increase in gene expression of peroxisome proliferator-activated receptor (PPAR)-gamma coactivator 1alpha (PGC-1alpha), mitochondrial uncoupling protein 3 (UCP3) and hexokinase II (HKII) in primary rat skeletal muscle cells is dependent on reactive oxygen species. Biochim Biophys Acta 1763: 969–976, 2006 [DOI] [PubMed] [Google Scholar]
- 175.Smuder AJ, Kavazis AN, Min K, and Powers SK. Exercise protects against doxorubicin-induced markers of autophagy signaling in skeletal muscle. J Appl Physiol (1985) 111: 1190–1198, 2011 [DOI] [PubMed] [Google Scholar]
- 176.Suwa M, Nakano H, and Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol 95: 960–968, 2003 [DOI] [PubMed] [Google Scholar]
- 177.Takikita S, Schreiner C, Baum R, Xie T, Ralston E, Plotz PH, and Raben N. Fiber type conversion by PGC-1alpha activates lysosomal and autophagosomal biogenesis in both unaffected and Pompe skeletal muscle. PLoS One 5: e15239, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Talbert EE, Smuder AJ, Min K, Kwon OS, Szeto HH, and Powers SK. Immobilization-induced activation of key proteolytic systems in skeletal muscles is prevented by a mitochondria-targeted antioxidant. J Appl Physiol (1985) 115: 529–538, 2013 [DOI] [PubMed] [Google Scholar]
- 179.Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, Watanabe Y, Uchiyama Y, Sumi K, Iguchi H, Ito S, Doi T, Hamakubo T, Naito M, Auwerx J, Yanagisawa M, Kodama T, and Sakai J. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A 100: 15924–15929, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, and Tabata I. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun 296: 350–354, 2002 [DOI] [PubMed] [Google Scholar]
- 181.Terada S, Kawanaka K, Goto M, Shimokawa T, and Tabata I. Effects of high-intensity intermittent swimming on PGC-1alpha protein expression in rat skeletal muscle. Acta Physiol Scand 184: 59–65, 2005 [DOI] [PubMed] [Google Scholar]
- 182.Thomson DM, Porter BB, Tall JH, Kim HJ, Barrow JR, and Winder WW. Skeletal muscle and heart LKB1 deficiency causes decreased voluntary running and reduced muscle mitochondrial marker enzyme expression in mice. Am J Physiol Endocrinol Metab 292: E196–E202, 2007 [DOI] [PubMed] [Google Scholar]
- 183.Tureckova J, Wilson EM, Cappalonga JL, and Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J Biol Chem 276: 39264–39270, 2001 [DOI] [PubMed] [Google Scholar]
- 184.Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, Vakeel P, Stadel D, Haas A, Saftig P, Behrends C, Furst DO, Volkmer R, Hoffmann B, Kolanus W, and Hohfeld J. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol 23: 430–435, 2013 [DOI] [PubMed] [Google Scholar]
- 185.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, and Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 28: 2244–2258, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Vina J, Gomez-Cabrera MC, Lloret A, Marquez R, Minana JB, Pallardo FV, and Sastre J. Free radicals in exhaustive physical exercise: mechanism of production, and protection by antioxidants. IUBMB Life 50: 271–277, 2000 [DOI] [PubMed] [Google Scholar]
- 187.Vitale C, Marazzi G, Pelliccia F, Volterrani M, Cerquetani E, Spoletini I, Mercuro G, Bonassi S, Dall'Armi V, Fini M, and Rosano GM. Trimetazidine improves exercise performance in patients with peripheral arterial disease. Pharmacol Res 63: 278–283, 2011 [DOI] [PubMed] [Google Scholar]
- 188.Wadley GD, Choate J, and McConell GK. NOS isoform-specific regulation of basal but not exercise-induced mitochondrial biogenesis in mouse skeletal muscle. J Physiol 585: 253–262, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wadley GD, Nicolas MA, Hiam DS, and McConell GK. Xanthine oxidase inhibition attenuates skeletal muscle signaling following acute exercise but does not impair mitochondrial adaptations to endurance training. Am J Physiol Endocrinol Metab 304: E853–E862, 2013 [DOI] [PubMed] [Google Scholar]
- 190.Wende AR, Schaeffer PJ, Parker GJ, Zechner C, Han DH, Chen MM, Hancock CR, Lehman JJ, Huss JM, McClain DA, Holloszy JO, and Kelly DP. A role for the transcriptional coactivator PGC-1alpha in muscle refueling. J Biol Chem 282: 36642–36651, 2007 [DOI] [PubMed] [Google Scholar]
- 191.Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, and Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A 106: 20405–20410, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 192.Winder WW. and Hardie DG. Inactivation of acetyl-CoA carboxylase and activation of AMP-activated protein kinase in muscle during exercise. Am J Physiol 270: E299–E304, 1996 [DOI] [PubMed] [Google Scholar]
- 193.Winder WW, Holmes BF, Rubink DS, Jensen EB, Chen M, and Holloszy JO. Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol 88: 2219–2226, 2000 [DOI] [PubMed] [Google Scholar]
- 194.Wohlgemuth SE, Lees HA, Marzetti E, Manini TM, Aranda JM, Daniels MJ, Pahor M, Perri MG, Leeuwenburgh C, and Anton SD. An exploratory analysis of the effects of a weight loss plus exercise program on cellular quality control mechanisms in older overweight women. Rejuvenation Res 14: 315–324, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wojtaszewski JF, Nielsen P, Hansen BF, Richter EA, and Kiens B. Isoform-specific and exercise intensity-dependent activation of 5′-AMP-activated protein kinase in human skeletal muscle. J Physiol 528Pt 1: 221–226, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wong PM, Puente C, Ganley IG, and Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy 9: 124–137, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, and Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. J Biol Chem 282: 194–199, 2007 [DOI] [PubMed] [Google Scholar]
- 198.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, and Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296: 349–352, 2002 [DOI] [PubMed] [Google Scholar]
- 199.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, and Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124, 1999 [DOI] [PubMed] [Google Scholar]
- 200.Yoshida S, Hong S, Suzuki T, Nada S, Mannan AM, Wang J, Okada M, Guan KL, and Inoki K. Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J Biol Chem 286: 32651–32660, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]