Abstract
Transcription factors (TFs) preferentially bind sites contained in regions of computationally predicted high nucleosomal occupancy, suggesting that nucleosomes are gatekeepers of TF binding sites. However, because of their complexity mammalian genomes contain millions of randomly occurring, unbound TF consensus binding sites. We hypothesized that the information controlling nucleosome assembly may coincide with the information that enables TFs to bind cis-regulatory elements while ignoring randomly occurring sites. Hence, nucleosome would selectively mask genomic sites contacted by TFs and thus potentially functional. The hematopoietic TF Pu.1 maintained nucleosome depletion at macrophage-specific enhancers that displayed a broad range of nucleosome occupancy in other cell types and in reconstituted chromatin. We identified a minimal set of DNA sequence and shape features that accurately predicted both Pu.1 binding and nucleosome occupancy genome-wide. These data reveal a basic organizational principle of mammalian cis-regulatory elements whereby TF recruitment and nucleosome deposition are controlled by overlapping DNA sequence features.
INTRODUCTION
The identification of histone marks (notably H3K4me1) and coregulators (such as the histone acetyltransferase p300) associated with functionally validated enhancers and/or with evolutionary conserved, potential cis-regulatory sequences, recently enabled the characterization of the genomic repertoire of candidate enhancers characteristic of distinct cell types (Heintzman et al., 2007). Terminally differentiated cells have a unique repertoire of enhancers (Creyghton et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011; Stergachis et al., 2013; Visel et al., 2009) that is generated by transcription factors (TFs) that control lineage specification (Calo and Wysocka, 2013; Natoli, 2010). Specialized nucleosome-binding TFs (pioneer factors)(Zaret and Carroll, 2011) increase the local accessibility of nucleosomal DNA as determined by DNase I hypersensitivity assays (Thurman et al., 2012) and promote the deposition of enhancer-associated histone marks (Heintzman et al., 2007). Since most other TFs are unable to bind nucleosomal DNA, transcriptional regulation in a given cell type occurs almost exclusively within the accessible fraction of a much broader genomic repertoire of cis-regulatory elements. Therefore, the mutual interplay between nucleosomes, pioneer TFs, and TFs opportunistically binding to accessible DNA controls cell type-specific transcriptional outputs.
Factors that determine nucleosome occupancy can be broadly classified into three groups (Struhl and Segal, 2013): DNA sequence, trans-acting factors (including TFs and the transcriptional machinery), and chromatin remodeling enzymes. The role of DNA sequence in nucleosome occupancy has been the object of a long controversy (Struhl and Segal, 2013) centered on the relative role of nucleotide composition (Segal et al., 2006) vs. DNA-bound barriers (Mavrich et al., 2008) and remodeler-driven nucleosome packing against barriers (Zhang et al., 2011) in determining nucleosome positioning in vivo. It is now clear that each of these mechanisms has a specific role in controlling nucleosomal organization and that sequence-driven nucleosome assembly can be overcome by trans-acting factors in specific instances and at specific locations.
The affinity of DNA sequences for the histone octamer spans several orders of magnitude and computational models that use sequence features to predict nucleosome occupancy have been described (Ioshikhes et al., 2006; Kaplan et al., 2009; Segal et al., 2006; Tillo and Hughes, 2009). In particular, poly(dA:dT) tracts are stiff regions unable to bend around the histone octamer (Nelson et al., 1987; Suter et al., 2000), which accounts for nucleosome depletion at poly(dA:dT) sequences of > 5 bp in length in S. cerevisiae gene promoters. In human cells, container sites (sequences able to generate positioned nucleosomes in in vitro assembly experiments)(Valouev et al., 2011) are demarcated by nucleosome-repelling poly(dA:dT) tracts flanking moderately (dG:dC)-rich, high-affinity regions for nucleosomes.
Both in yeast (Charoensawan et al., 2012) and in mammals (Gaffney et al., 2012; Lidor Nili et al., 2010) some TFs have been shown to contact genomic sequences encoding high nucleosome occupancy. This is consistent with the notion that cis-regulatory elements, albeit nucleosome-depleted in those cells in which they are bound by TFs, have an intrinsic propensity to be incorporated into nucleosomes (Tillo et al., 2010). These observations suggest that nucleosomes actively mask TF consensus sites but can be displaced by cooperatively binding TFs. However, although a trend linking sequence-encoded nucleosome occupancy and TF recruitment is detectable when analyzing genomic data (Tillo et al., 2010), this simple conceptual scheme does not explain the much higher complexity of the relationship between TF binding and sequence determinants controlling nucleosome occupancy.
The notion that nucleosomes restrict the access of TFs to the underlying regulatory DNA leads to a simple yet critical inference: In complex mammalian genomes high nucleosome occupancy must be selectively encoded by sequences containing engaged TF binding sites but not by the millions of randomly occurring sequences that are apparently identical to TF consensus binding sites but are not engaged by TFs (Pan et al., 2010). Clearly, while TF binding to a genomic site will not always and necessarily cause functional effects, consensus sequences that are not engaged are not likely to contribute to transcriptional control.
In this study, we have addressed the hypothesis that in mammalian genomes the information controlling the ability of TFs to recognize cognate sites in cis-regulatory elements (while ignoring randomly occurring consensus binding sites, heretofore indicated as non-functional sites) may at least partially coincide with the information that controls incorporation of the same sequence into nucleosomes. An attractive and biologically meaningful implication of this hypothesis is that conservation of nucleosome occupancy and TF binding sites would be subjected to the same evolutionary forces.
We used primary mouse macrophages in which the ETS family TF Pu.1, the master regulator of the myeloid lineage (Nerlov and Graf, 1998; Rosenbauer and Tenen, 2007; Scott et al., 1994), binds virtually the entire repertoire of H3K4me1-positive regions and a large fraction of transcription start sites (TSSs) (Ghisletti et al., 2010; Heinz et al., 2010). Pu.1 expression in fibroblasts (Ghisletti et al., 2010) or in Pu.1-negative myeloid precursors (Heinz et al., 2010) is sufficient to drive the deposition of H3K4me1 and to locally increase DNA accessibility. This suggests that Pu.1, together with other TFs expressed at different phases of myeloid differentiation (Lichtinger et al., 2012), may act as a pioneer factor to create the macrophage-specific repertoire of accessible cis-regulatory elements.
We found that Pu.1-bound consensus sites, but not those sites that are not bound in any of the Pu.1-expressing cell types, were shielded by nucleosomes in cells that do not express Pu.1. However, nucleosomes covering Pu.1 sites displayed a broad spectrum of occupancy and positioning, thus unveiling a complexity in the interplay between TF binding and nucleosome occupancy that was overlooked by previous analyses. Both at distal and TSS-proximal Pu.1 sites, nucleosome occupancy and positioning were encoded in the DNA sequence and could be recapitulated in vitro. We identified a minimal set of DNA features, including three-dimensional DNA shape, which discriminated bound from unbound Pu.1 consensus sites at genome scale with unprecedented accuracy for a mammalian TF. Critically, the same set of features predicted nucleosome occupancy of the same DNA elements with improved or similar performances compared to computational models specifically designed for this aim, thus suggesting that overlapping DNA sequence features control both nucleosome deposition and binding competence of Pu.1 consensus sites.
RESULTS
DNA sequence features correlate with distinct nucleosome profiles in vivo
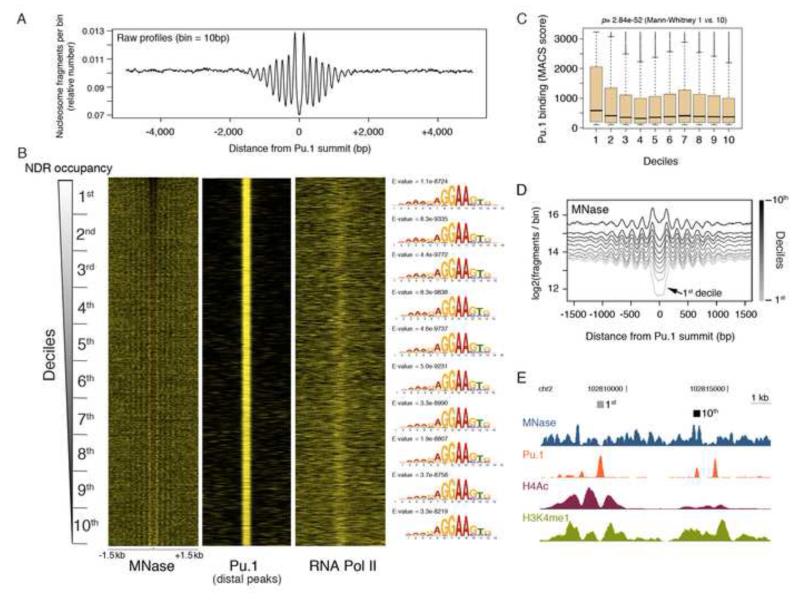
Mononucleosome-sized DNA fragments from limited micrococcal nuclease (MNase) digestion of mouse macrophage nuclei were subjected to paired-end sequencing. Each of the four biological replicates used was sequenced to generate ca. 200 million uniquely aligned, filtered and properly paired sequence reads. By pooling these four replicates we obtained 825 million sequencing reads that allowed us to obtain a high-resolution view of nucleosome arrays. Sequencing reads centered on annotated TSSs generated a canonical asymmetric pattern with a nucleosome-depleted region (NDR) bracketed by the −1 and a more prominent +1 nucleosome (Fig. S1A). Conversely, when TSS-distal Pu.1 peaks were used as central anchors, we detected regular arrays of nucleosomes (Fig. 1A). Nucleosome depletion surrounding Pu.1-bound sites was independently observed in ChIP-seq experiments based on sonicated chromatin (Ghisletti et al., 2010; Heinz et al., 2010), thus suggesting that it was not due to digestion of labile nucleosomes with high sensitivity to MNase.
Fig. 1. Regular arrays of nucleosomes centered at Pu.1-bound enhancers in macrophages.
A) Cumulative distribution of midpoints of nucleosomal sequencing fragments centered on the summit of TSS-distal Pu.1 sites in macrophages. The number of fragments in each 10-bp bin was normalized by the total number of fragments in the area. The same information is shown in B) as heatmap (first from the left), ordered from top to bottom based on decreasing occupancy of the NDR and divided in deciles. Heatmaps of Pu.1 and Pol_II are also shown on the right of the MNase data. The counts exceeding the 95th percentile of the overall distribution were set to its value. Considering MNase data, these counts were then normalized in the range 0-1 separately for each region. The same procedure was applied to ChIP-seq data except that the 0-1 normalization was applied to the entire dataset. Sequence logos on the right show the Pu.1 binding motifs identified de novo in individual deciles and their E-values. C) ChIP-seq scores (MACS) of the Pu.1 peaks in in the different deciles. D) Cumulative distributions of the midpoints of the nucleosomal fragments at Pu.1 bound enhancers (divided in deciles according to panel B). E) A representative snapshot showing two NDRs of the 1st and the 10th decile. See also Fig. S1.
Pu.1 summit-centered nucleosome maps were ordered based on the decreasing occupancy of the central NDR and divided in deciles (Fig. 1B). De novo motif discovery on the sequences from each decile returned as first hit the known Pu.1 binding site with very similar statistical significance (Fig. 1B, right). The median distance of the best motif match to the Pu.1 peak center was very similar in all deciles and comprised between 7 and 10 nt. Pu.1 binding scores were significantly higher in the 1st decile but similar across all the others (Fig. 1C). Taken together, these results indicate that different Pu.1 binding affinities do not contribute to a different NDR occupancy. Considering a + 1.5 kbp central region centered on the Pu.1 peaks, the 1st decile showed a lower overall nucleosome occupancy than the 10th one (Fig. 1D), indicating that differences in nucleosome organization extend beyond the central regulatory region. The two NDR-flanking nucleosomes were prominent in the 10th decile and almost absent in the 1st, thus contributing to the lower occupancy and to the apparently broader width of the NDR in this group. Therefore, qualitatively different classes of NDRs surrounding Pu.1 peaks could be identified, and these classes did not correlate with differences in Pu.1 occupancy. A representative snapshot is shown in Fig. 1E.
Since RNA Polymerase II (Pol_II) is associated with a subset of enhancers (De Santa et al., 2010; Kim et al., 2010; Koch et al., 2011) we analyzed its density in the deciles. Pol_II reads showed higher density in the NDRs of higher deciles (Fig. 1B). While this result suggests that Pol_II did not contribute to the maintenance of the low-occupancy and broad NDR characteristic of the lower deciles, it may point to a role of Pol_II in determining the occupancy and positioning properties of the higher deciles. However, depletion of the large Pol_II subunit Rpb1 by a 4h alpha-amanitin treatment did not significantly alter nucleosome occupancy (MS, IB and GN, unpublished observations). At TSS-proximal Pu.1 sites, the relationship between NDR occupancy and Pol_II was opposite than at enhancers, with higher Pol_II loading in less occupied regions (Fig. S1B, C).
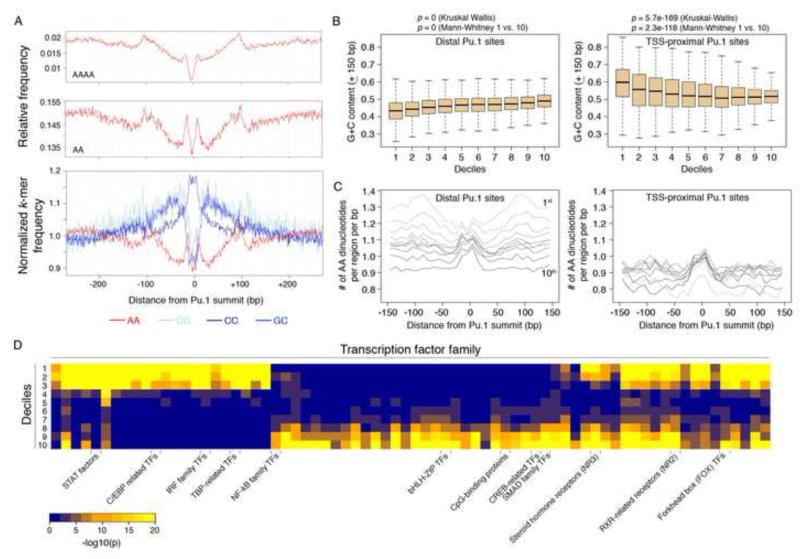
We next analyzed the sequence features of the DNA associated with the distal Pu.1 binding sites. When considering the ensemble of all distal Pu.1-bound regions in macrophages, we detected features characteristic of nucleosome container sites (Valouev et al., 2011): an increase in the relative frequency of nucleosome-repelling AA dinucleotides and AAAA polynucleotides peaking at the −100 and +100 bp, with a central core of G+C rich sequences that promote nucleosome occupancy (Tillo and Hughes, 2009) (Fig. 2A). Next, we analyzed individual deciles separately. Consistent with the progressive increase in nucleosome occupancy, the G+C content increased from the 1st to the 10th decile (p < 1e-15; Kruskal-Wallis test)(Fig. 2B, left). Conversely, AA dinucleotides were more represented in the 1st decile and peaked at +100nt (Fig. 2C, left). In a reciprocal manner, the 1st decile showed a relative depletion of GC and CC dinucleotides in the flanks (Fig. S2). Therefore, a signature of container sites was selectively found in the lower deciles.
Fig. 2. Sequence features discriminate among enhancers with different nucleosome occupancy and positioning.
A) Cumulative distribution of AAAA tetranucleotides (top panel), AA dinucleotides (middle panel) and G/C containing dinucleotides (bottom panel) are shown relative to the summit of TSS-distal Pu.1 peaks (the strong enrichment of CC/GG dinucleotides at the anchor point is enhanced by the central invariant nucleotides of the Pu.1 site, 5′-AGAGGAAGTG-3′). G+C content (B) and distribution of AA dinucleotides (C) in deciles at Pu.1-bound distal (left) and TSS-proximal (right) sites. See also Fig. S2. D) Statistical over-representation of binding sites for TF families at Pu.1 bound distal sites divided in deciles (according to Fig. 1B).
Compared to distal sites, sequence composition at TSS-proximal bound sites showed some fundamental differences. The G+C content at Pu.1-bound TSSs was much higher than the one at the distal sites (Fig. 2B, right). At TSSs, the lower deciles (namely those with the lowest NDR occupancy, Fig. S1C) showed the highest G+C content (Fig. 2B, right). This is consistent with the notion that a very high G+C content (such as the one found at CpG islands) disfavors nucleosome assembly (Fenouil et al., 2012; Ramirez-Carrozzi et al., 2009). In fact, at TSSs the correlation between G+C content and deciles was inverted, with a progressive reduction from the 1st to the 10th decile. Therefore the relationship between G+C content and nucleosome occupancy was bimodal, nucleosome occupancy being anti-correlated with both a very low G+C content (such as the one found at enhancers in the lower deciles) and a very high G+C content (such as the one found at promoters in the lower deciles). The AA frequency of the Pu.1-bound TSS-proximal regions was much lower than that observed at distal Pu.1 sites (Fig. 2C, right). Moreover, TSS-proximal sites did not display the AA-rich flanks that delimit container sites and that could instead be detected in the lower deciles of the distal Pu.1-bound regions (Fig. 2C, left).
Finally, we analyzed the co-occurrence of binding sites for other TFs at distal Pu.1 sites. Binding sites for some TF families (e.g. STAT and IRF) were over-represented (relative to the distal Pu.1-bound sequences in their entirety) in the lower deciles, and some others (e.g. NF-kB and CREB) in the higher ones (Fig. 2D).
Overall, these data indicate qualitative differences in sequence composition of the deciles that may directly impact both nucleosome assembly and cooperation with other TFs.
Selective masking of engaged Pu.1 sites by nucleosomes
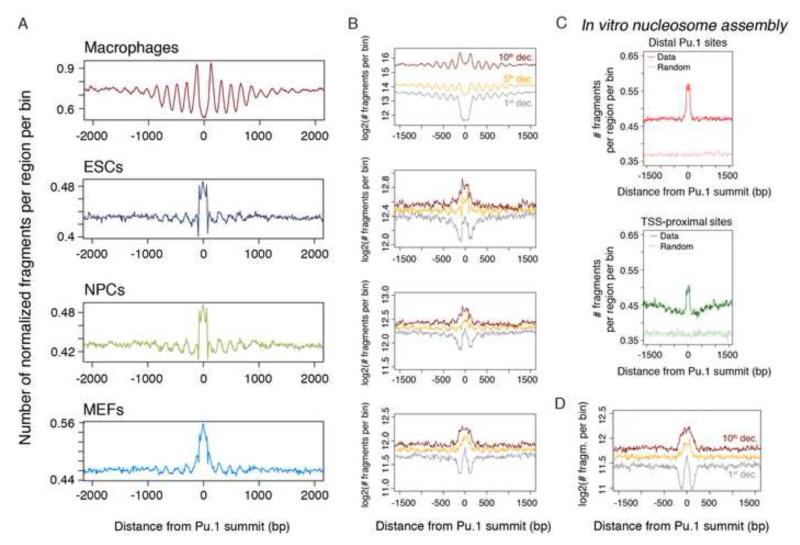
To determine the impact of DNA sequence on nucleosomes at enhancers, we analyzed nucleosome occupancy in cell types that do not express Pu.1 (Fig. 3A, B) and in in vitro reconstituted chromatin generated from recombinant histones (Fig. 3C, D). Nucleosomal sequences from embryonic stem cells (ESCs), neural precursors (NPCs) and mouse embryonic fibroblasts (MEFs)(Teif et al., 2012) were aligned to the summit of Pu.1 peaks. In all the three cases, high nucleosome occupancy overlapping the macrophage Pu.1-bound nucleosome-depleted regions was detected (Fig. 3A). Considering the deciles shown in Fig. 1, the central NDR observed in the 1st decile in macrophages, showed instead a focused nucleosomal signal bracketed by two narrow areas of nucleosome depletion in all other cells (Fig. 3B). This result suggests a strongly positioned nucleosome controlled by container sites demarcated by AA-rich flanks (Fig. 2C). The 10th decile was instead characterized by central nucleosomes with higher occupancy but weaker positioning.
Fig. 3. Pu.1-bound, nucleosome-depleted macrophage enhancers are covered by nucleosomes in unrelated cell types and in vitro.
(A) Cumulative distributions of the midpoints of the nucleosomal fragments centered on distal Pu.1 sites in macrophages and in unrelated cells that do not express Pu.1 (ESCs, NPCs and MEFs). The number of midpoints in each 10-bp bin was scaled according to the total number of regions and sequencing depth. In (B) data were split in deciles (only the 1st, 5th and 10th deciles are shown). C) Midpoint distributions from in vitro assembled nucleosomes. Data for distal and TSS-proximal sites are shown. See also Fig. S3. D) MNase-seq data from in vitro nucleosomes are shown divided in deciles.
Although nucleosome occupancy is affected by several factors, these data suggest that DNA sequence features contribute to promote nucleosome assembly at genomic regions that in macrophages are nucleosome-depleted due to Pu.1 binding. To directly define the role of DNA sequence in controlling the nucleosomal landscape at Pu.1 sites, we assembled nucleosomes in vitro. Assembly conditions in which DNA was not limiting were used to focus on the effects of the primary sequence on nucleosome assembly (Luger et al., 1999; Valouev et al., 2011). Sequencing data recapitulated previously reported features of in vitro assembled nucleosomes, such as the nucleosome depletion at CpG islands (Fenouil et al., 2012) that increased with CpG content but was less marked than that observed in vivo (Valouev et al., 2011) (Fig. S3). The cumulative distribution of nucleosome fragments (Fig. 3C) indicates that genomic sequence features are sufficient to generate a focused increase in nucleosomal density at both TSS-distal and proximal sites bound by Pu.1 in macrophages. Consistent with the notion that formation of nucleosome arrays requires the activity of ATP-dependent remodelers (Zhang et al., 2011), we did not detect arrays in these conditions. When data were split according to the deciles shown in Fig. 1, in vitro generated nucleosomes recapitulated the behaviors observed in cells that do not express Pu.1 (Fig. 3D).
These data indicate that the DNA sequence encodes the deposition of nucleosomes at Pu.1-bound genomic sites. In cells that do not express Pu.1, a range of behaviors was found. At one end (1st decile), the regulatory region was covered by a strongly positioned nucleosome within an area of low nucleosome occupancy. In macrophages, the removal of this centrally positioned nucleosome resulted in the broad NDR characteristic of the 1st decile. At the opposite end (10th decile), the sequence encoded higher occupancy but weak positioning. In macrophages, Pu.1 binding correlated with the partial displacement and repositioning of nucleosomes, thus resulting in a narrower NDR.
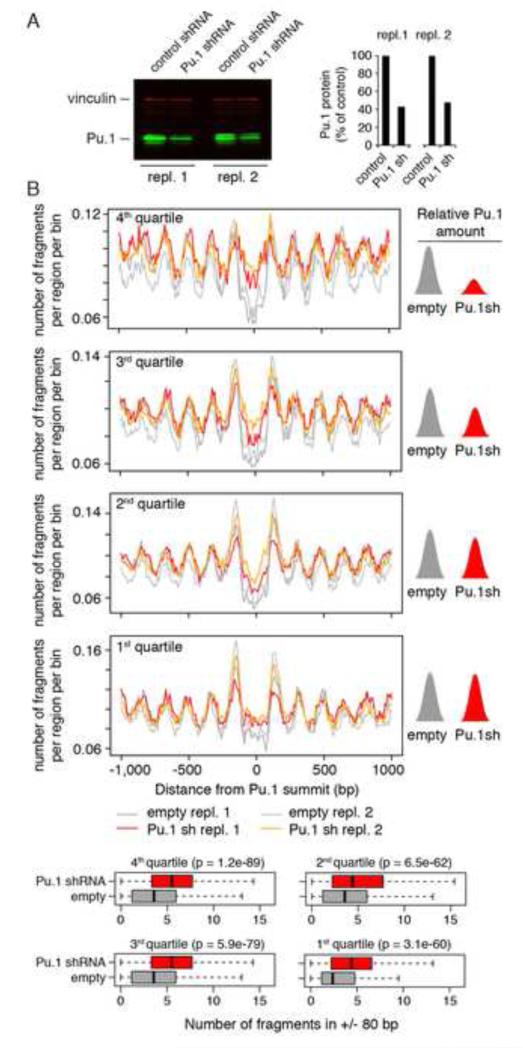
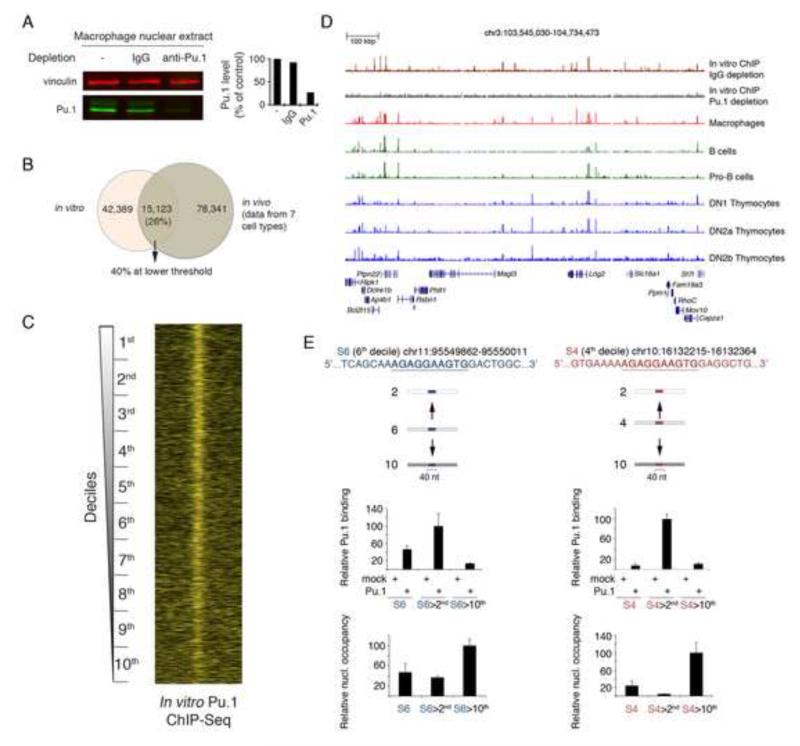
To address the role of Pu.1 in counteracting DNA sequence-driven nucleosome occupancy, we used an inducible retroviral vector to deplete Pu.1 from macrophages. 48h after shRNA induction a 60% depletion of Pu.1 was obtained in two independent experiments (repl. 1 and 2 in Fig. 4A; notably, a complete depletion of Pu.1 would not be compatible with macrophage survival). We next carried out a Pu.1 ChIP-seq experiment to classify regulatory regions based on the level of reduction of Pu.1 binding and we simultaneously analyzed nucleosome profiles. TSS-distal Pu.1 peaks identified by ChIP-seq were divided in quartiles based on the Pu.1 signal ratio in Pu.1-depleted vs. control cells, the fourth quartile corresponding to the stronger reduction in Pu.1 binding. A strong and statistically significant (p << 0.01, Wilcoxon signed-rank test) increase in nucleosomal reads at Pu.1-bound enhancers was detected, particularly in the fourth quartile (Fig. 4B, upper panel). Qualitatively similar results were obtained when considering the entire repertoire of Pu.1 peaks (Fig. S4). Overall, these data indicate that genomic sites vacated by Pu.1 upon depletion tend to be reincorporated into nucleosomes.
Fig. 4. Effects of Pu.1 depletion on nucleosome occupancy.
A) Acute depletion of Pu.1 in terminally differentiated macrophages using a retrovirus-encoded Tet-regulated shRNA. Data from two biological replicates are shown. Vinculin was used as loading control. B) Nucleosomal occupancy in Pu.1-depleted macrophages. Pu.1 peaks were divided in quartiles based on the degree of signal reduction in Pu.1-depleted vs. control cells. The 4th quartile corresponds to Pu.1 peaks with the higher reduction in binding in Pu.1-depleted cells. Quartile-specific distributions of nucleosome fragments midpoints centered on the summit of Pu.1 peaks are shown. Midpoints found within + 80 bp from the Pu.1 summit are also summarized in the box plots on the right. For each quartile, the statistical significance of the difference is expressed by the p-value of a Wilcoxon signed-rank test. See also Fig. S4.
To determine the ability of Pu.1 to bind different types of nucleosomal sites, we incubated in vitro-assembled nucleosomes with macrophage nuclear extracts in order to allow the formation of protein-DNA complexes. Pu.1-bound nucleosomes were immunoprecipitated and sequenced. A Pu.1-immunodepleted nuclear extract was used as a reference (Fig. 5A). Depending on the stringency, between 26% and 40% of the Pu.1 binding events observed in vivo were recapitulated in the in vitro assay (Fig. 5B). When Pu.1 binding to in vitro-assembled chromatin was analyzed considering the deciles shown in Fig. 1, it became clear that while Pu.1 was able to strongly interact with sites in the 1st decile, it was less efficient at binding sites in the 10th decile (p = 1.84e-278, Kruskal-Wallis test; Fig. 5C), which is consistent with the in vivo binding data (Fig. 1C). A representative in vitro ChIP-seq snapshot is shown in Fig. 5D. Supplemental Fig. S5 shows a genomic snapshot of in vivo and in vitro assembled nucleosomes.
Fig. 5. In vitro analysis of Pu.1 binding to nucleosomal DNA.
A) Pu.1 ChIP-seq on in vitro assembled nucleosomes. Macrophage nuclear lysates (used as a source of Pu.1) were incubated with in vitro assembled chromatin. Prior to incubation with nucleosomes, nuclear lysates were reacted twice either with control rabbit IgG or anti-Pu.1 antibody coupled to paramagnetic beads. Immunodepletion with anti-Pu.1 antibodies resulted in an almost complete loss of Pu.1 from lysates. Vinculin: loading control. B) Venn diagram showing the overlap between in vitro and in vivo Pu.1 binding. C) Heatmap of in vitro Pu.1 binding, showing the relative density of nucleosome midpoints. In vitro ChIP signals were sorted according to nucleosome occupancy in macrophages (Fig. 1B). See also Fig. S5. D) A representative ChIP-seq snapshot is shown. E) Pu.1 sites and flanks (40nt) from the 6th or 4th decile were transferred to sequences of higher (10th) or lower (2nd) deciles and used for nucleosome assembly and ChIP-qPCR. Mock transfected extracts and extracts from HEK-293 cells transfected with a Pu.1 expression vector were used as indicated.
Since the Pu.1 sites in different deciles are virtually identical (Fig. 1B, right), these data and the in vivo data (Fig. 1B, C) suggest that a high level of nucleosome occupancy has a detrimental impact on Pu.1 binding. To further address this issue, we analyzed the impact of transferring a 10 nt Pu.1 site (with a 15 nt extension on both sides) from intermediate deciles into the sequence context of lower or higher deciles. Nucleosomes were then assembled in vitro onto these chimeric sequences and Pu.1 binding was measured by ChIP. As shown in Fig. 5E, upon transferring the binding site from its original context (4th or 6th decile) to the one of a higher (10th) decile, an increase in nucleosome occupancy was paralleled by a reduction in Pu.1 binding; the opposite was observed when the same sequences were moved into the context of a lower decile. Therefore, sequences that intrinsically favor nucleosome occupancy correlate with weaker Pu.1 binding in vivo and interfere with the association of Pu.1 with its binding site in vitro.
Usage of Pu.1 binding sites correlates with nucleosome occupancy
Data shown above demonstrate that the DNA sequence promotes nucleosome assembly at regions containing Pu.1 consensus sites that are bound in vivo. However, randomly occurring nucleotide combinations in mammalian genomes lead to the casual generation of non-functional TF consensus sites. Randomly occurring sites outnumber TF consensus sites contained in functional cis-regulatory elements and bound by their cognate TF in vivo. We reasoned that the information that discriminates between these two groups of sites might be coupled with the information relevant for nucleosome assembly.
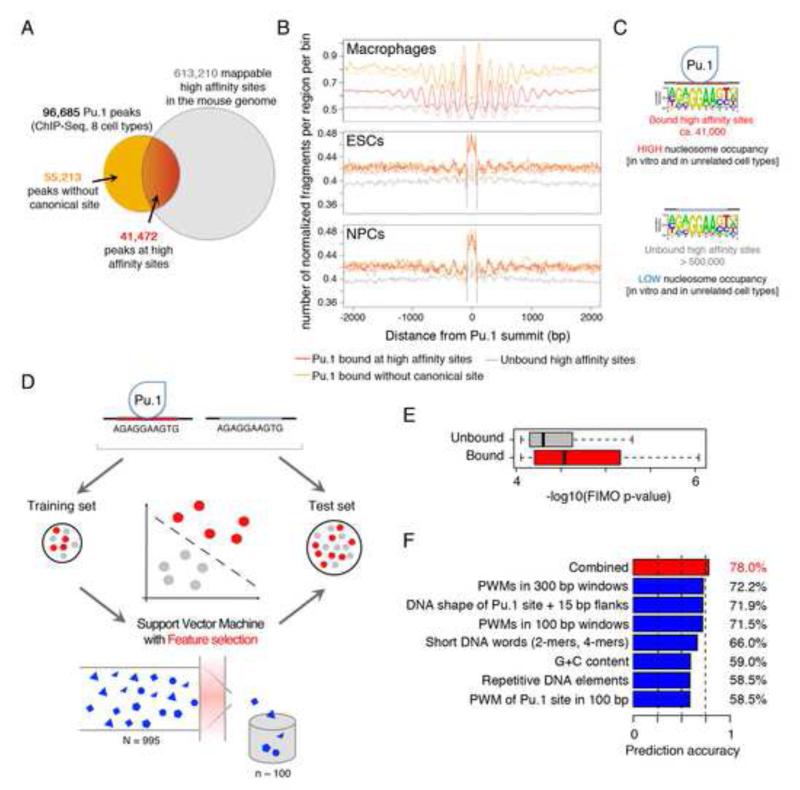
Pu.1 is expressed only in the hematopoietic system and specifically in myeloid cells, B and early T lymphocytes. We collected eight high-quality ChIP-seq datasets from Pu.1 expressing cells (Heinz et al., 2010; Mullen et al., 2011; Ostuni et al., 2013; Zhang et al., 2012) (Table S2). Collectively, Pu.1 peaks from these datasets (n=96,685) virtually represent the entire repertoire of genomic regulatory elements that can be bound by Pu.1 in mouse cells (Fig. 6A). 41,472 of these peaks (42.9%) contain a canonical high-affinity Pu.1 binding site, which differs from those bound by other ETS proteins (Wei et al., 2010). The reference mouse genome contains additional 571,738 Pu.1 sites with a computationally predicted high-affinity (for a total of 613,210 Pu.1 sites): even assuming that a fraction of them may be bound in conditions not recapitulated in the datasets we collected, it is clear that the vast majority of them are not bound in vivo.
Fig. 6. Pu.1 binding site usage correlates with nucleosome occupancy.
A) Venn diagram showing the overlap between Pu.1 peaks identified in ChIP-seq experiments from multiple cell types and computationally identified genomic Pu.1 sites. B) Cumulative distributions of nucleosome midpoints in macrophages, ESCs and NPCs at Pu.1-bound high affinity consensus sites (red), Pu.1-bound non-canonical sites (orange) and computationally identified consensus sites that are not bound in vivo (grey). C) Schematic representation of the relationship between Pu.1 binding and nucleosome occupancy. D) Schematic of the SVM approach used to predict in vivo binding competence of Pu.1 sites and to identify their distinctive DNA sequence and shape features. E) Computationally measured affinity of Pu.1 towards bound and unbound genomic sites. F) Bar plots showing the prediction accuracies of the most predictive features selected by the SVM, divided in categories (blue) or all in combination (red). See also Fig. S6.
We next re-analyzed MNase-seq data separately for different groups of Pu.1 binding sites. In macrophages, nucleosome arrays were very similar at Pu.1 peaks with or without the presence of a canonical binding site, while unbound sites were not associated with detectable nucleosome arrays (Fig. 6B, upper panel). Pu.1-bound elements showed instead an increase in nucleosome occupancy over the binding site in ESCs and NPCs, irrespective of the presence or absence of a canonical binding site. Conversely, canonical sites that were not bound by Pu.1 in any of the cell types analyzed did not display any clear increase in occupancy over the flanking regions (Fig. 6B). Therefore, the ability of a cis-regulatory region containing a Pu.1 site to bind Pu.1 in vivo correlated with its affinity for nucleosomes (Fig. 6C).
Identification of DNA sequence and shape determinants of Pu.1 binding site occupancy
These data suggest that nucleosomes may selectively mask Pu.1 sites contained in cis-regulatory elements. Therefore we set out to identify DNA features that discriminate bound from unbound Pu.1 sites and to test whether the same determinants predict nucleosome occupancy in cells that do not express Pu.1.
We considered the whole set of Pu.1 binding events at high affinity sites (41,472) and randomly extracted the same number of regions from the unbound sites as negative set. We used Support Vector Machines (Cortes and Vapnik, 1995) to evaluate to what extent the local genomic sequence is informative for Pu.1 binding in vivo. 995 DNA features were assessed in 300 bp windows aligned to the summit of the ChIP-seq peaks in the case of bound regions, and to the invariant GGAA core of the Pu.1 binding site in the case of the unbound ones. Features tested included: i) Position Weight Matrices (PWMs) describing known binding preferences for TFs; ii) nucleotide words of length 2 or 4 (k-mers); iii) G+C content; iv) the average theoretical nucleosome occupancy of the region calculated with a published algorithm (Kaplan et al., 2009); v) the overlap with classes of repetitive elements, and vi) three-dimensional DNA shape features predicted for the 10 bp in the ETS core motif and for additional 15 bp on each side of the core. DNA shape was shown to affect protein-DNA recognition (Rohs et al., 2009) and to improve the prediction of binding specificities for bHLH TFs in yeast (Gordan et al., 2013) and human (Yang et al., 2014), and homeodomain TFs in mouse and Drosophila (Dror et al., 2014). Given this large amount of features, we devised a selection procedure (Guyon et al., 2003) to identify the smallest set with the highest predictive power (Fig. 6D).
We first assessed the prediction accuracy of the Pu.1 binding preferences as determined only by in vitro protein binding microarrays (Wei et al., 2010). Bound sites showed better FIMO p-values (Fig. 6E), indicating a higher median affinity of the target sites in these regions compared to the unbound sites (p = 2.31e-294, Mann-Whitney test). Nevertheless, Pu.1 sequence preferences alone were poor predictors of binding, resulting in an average accuracy of 58.5% (Fig. 6F). Instead, starting with the entire set of 995 features and through feature selection, we achieved an average accuracy of 78% (Fig. 6F).
We then analyzed the contribution of individual groups of features to the prediction accuracy (Fig. 6F). Theoretical nucleosome occupancy and G+C content had similar performances (accuracy of 59-60%), consistent with the notion that G+C content is a proxy for nucleosome occupancy (Tillo and Hughes, 2009). The role of cooperativity in Pu.1 binding to genomic sites is demonstrated by the high prediction accuracy of PWMs for partner TFs (72.2%). Remarkably, a small number of DNA shape features (Zhou et al., 2013) alone achieved an average prediction accuracy of 71.9%. In the combined model, DNA shape features of the ETS core boundaries and the −2, −1 and +1 flanking nucleotides were systematically selected (Table S3). Of the four DNA shape features used in this study (minor groove width, roll, propeller twist, and helix twist), minor groove width and roll were the predominant structural determinants of Pu.1 binding (Table S3).
To directly assess the impact of DNA shape features on Pu.1 binding, we carried out competitive electrophoretic mobility shift assays (EMSA). We used two labeled Pu.1 sites (10 nt flanked by 7 nt on both sides) and a panel of unlabeled competitors corresponding to high affinity mouse genomic Pu.1 sites (Table S4). These sites were either unchanged or mutated in the two nucleotides upstream or/and downstream the 10 nt core Pu.1 site. Mutations were designed to cause effects on DNA shape that would be detrimental for Pu.1 binding. While mutations either upstream or downstream of the Pu.1 site had a negative impact on the competition efficiency (being downstream mutations collectively more efficient than the upstream ones), their combination showed the most negative effect (Fig. S6A, B).
Besides ETS family motifs, motifs for TFs that are known to cooperatively bind at Pu.1 sites (such as Fos/AP-1 and IRF family TFs)(Ghisletti et al., 2010) were systematically selected. Moreover, we found the recurrent inclusion of G+C content/theoretical nucleosome occupancy as well as CG/GC/CC and AT/TA dinucleotides, which correlates with our previous observation that bound and unbound sites show a different potential to assemble nucleosomes when Pu.1 is not expressed.
Prediction of nucleosome occupancy using features that predict Pu.1 site occupancy
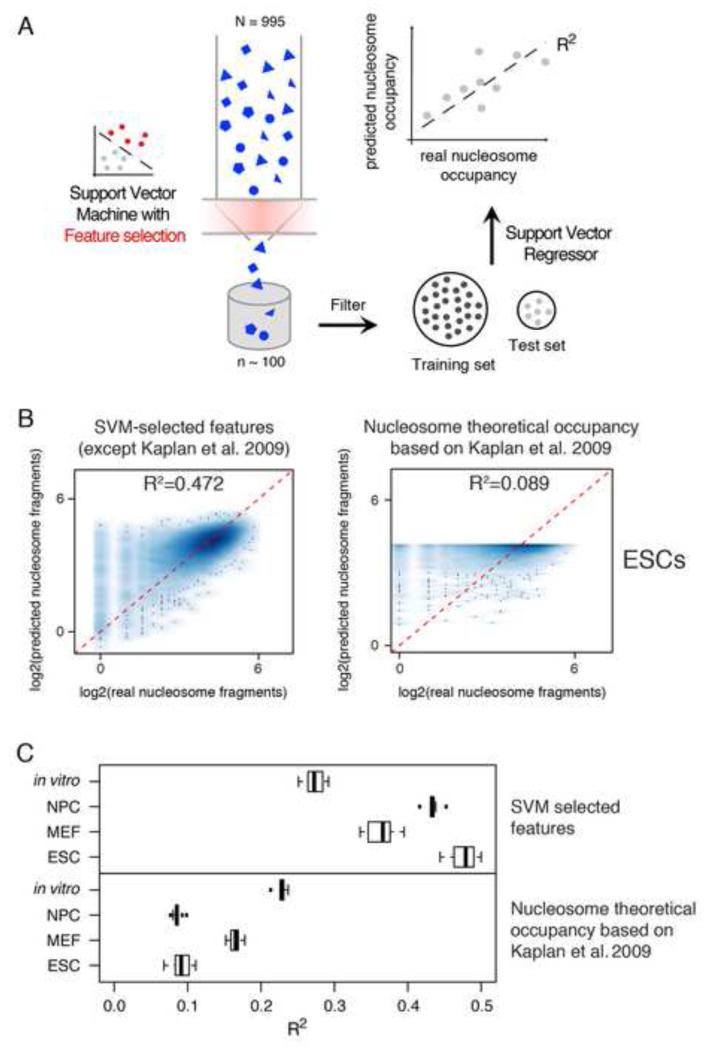
We next asked whether the set of features that predict Pu.1 binding also predict nucleosomal patterns in cells that do not express Pu.1. The local nucleosome occupancy of regions containing Pu.1-bound sites was extracted from ESCs, NPCs, MEFs and in vitro patterns. The number of nucleosome fragments spanning the center of each region was counted and the log2-transformed value used as a proxy for occupancy. The information for all the features except the theoretical nucleosome occupancy (Kaplan et al., 2009) was used to feed a Support Vector Regressor (SVR)(Drucker et al., 1997), a variant of SVM for regression (Fig. 7A). The set of bound and unbound sites was split into 90% training and 10% test data. The training dataset was used to fit the experimentally determined nucleosome counts based on the sequence features. The model obtained was then used to predict the nucleosome counts over the test dataset. Performance was evaluated through the coefficient of determination (R2), calculated as the squared Pearson Correlation Coefficient among the predicted and the observed counts.
Fig. 7. Nucleosome positioning prediction using DNA sequence and shape features associated with engaged TF binding sites.
A) Schematic of the SVR approach used to predict nucleosome occupancy from the DNA sequence and shape features predictive for Pu.1 binding. B) Smoothed scatterplots of the predicted values against the observed log2-transformed values of nucleosome occupancy in ESCs over Pu.1 sites (using the set of features selected for one of the training-test randomizations that served as input for the SVM). The scatterplot on the right shows the results on the test dataset using only theoretical nucleosome occupancy. The one on the left shows the results using all the features selected except for it. C) Box plots showing the distribution of the R2 for the sets of features from the ten training-test randomizations of the SVM.
Results for a representative set of features are shown in Fig. 7C as smoothed scatterplots of the predicted values in function of the observed values. The features discriminating Pu.1-bound from unbound sites explained 45% of the variability in the nucleosome occupancy pattern at these sites in ESC. Conversely, an SVR trained and tested using only the theoretical nucleosomes occupancy (Kaplan et al., 2009) explained less than 10% of the variability in the same data, which is in agreement with previous analyses (Tillo et al., 2010). These results were robust when slightly different sets of features (corresponding to multiple re-initializations of the SVM-based procedure used to predict Pu.1 binding) and different cell types were considered (Fig. 7C). The results we obtained indicated improved or similar performance compared to previous models specifically developed to predict nucleosome occupancy from the genomic sequence (Kaplan et al., 2009; Tillo and Hughes, 2009; van der Heijden et al., 2012).
Therefore, sequence determinants of Pu.1 binding could also encode part of the information for nucleosome affinity. Nevertheless, different DNA features are predicted to bring about quantitatively different effects on DNA binding and nucleosome occupancy. For instance, the DNA shape changes in the vicinity of the Pu.1 site should greatly impair Pu.1 binding without causing major effects on nucleosome assembly. Consistently, using the in vitro nucleosome assembly and Pu.1 ChIP assay described above, we found that mutations affecting DNA shape had a small but measurable detrimental effect on nucleosome assembly and a much higher impact on Pu.1 binding (Fig. S6C).
DISCUSSION
The interplay between TF binding and nucleosome-mediated occlusion of the regulatory DNA sequences that TFs recognize is at the hearth of regulated gene expression. However, understanding the determinants of this relationship has been hampered by some objective difficulties. First, engaged TF consensus sites are outnumbered by sites that, while characterized by an apparently identical affinity, represent non-functional random nucleotide arrangements. Moreover, signals controlling nucleosome occupancy and positioning are degenerate and loose, and in vivo they can be overcome by DNA-bound barriers, thus complicating identification and dissection of cis- and trans-acting components in studies carried out in a single cell type. Finally, the huge number of nucleosomes associated with complex mammalian genomes imposes the requirement of a high sequencing depth (that until recently was unavailable or exceedingly expensive) to achieve the resolution required to faithfully measure their occupancy and positioning. The strategy used in this study allowed us to overcome some of these limitations and to reach a more advanced understanding of the basic properties of this essential regulatory relationship.
An important aspect of our strategy is that we anchored our analysis to a single TF, Pu.1, which pervasively marks enhancers in macrophages and imposes nucleosome depletion at these regulatory elements. Since Pu.1 is expressed exclusively in hematopoietic cells and since its genomic distribution in Pu.1-expressing cell types has already been determined, it is possible to discriminate those randomly occurring Pu.1 sites that do not have binding competence from those that are contacted in vivo and are therefore potentially involved in transcriptional control. The classification of Pu.1 target sites based on their ability to bind Pu.1 in vivo allowed us to identify several molecular determinants of binding competence, to characterize different properties of binding sites in terms of their ability to drive nucleosome occupancy and positioning, and finally to determine whether and to what extent features controlling binding competence also affect nucleosome occupancy.
Collectively, Pu.1-bound cis-regulatory elements differed from unbound high-affinity Pu.1 sites in that, consistent with previous sequence based predictions (Kaplan et al., 2009; Tillo and Hughes, 2009), they were preferentially associated with nucleosomes both in vitro and in vivo in unrelated cell types that do not express Pu.1. However, previous analyses missed the complexity of the relationship between nucleosomal occupancy and TF recruitment. In fact, when our MNase-seq data were deconvolved based on the occupancy of the central Pu.1-bound NDR, it became clear that the nucleosome-assembly ability of the genomic sequences bound by Pu.1 was not homogeneous. At one end of the spectrum we found nucleosome container sequences (Valouev et al., 2011) able to drive the formation of a single, strongly positioned nucleosome within regions of overall lower nucleosome occupancy. At the other end we observed sequences determining a broad higher occupancy context that extended on both sides of a centrally located, prominently but less strongly positioned nucleosome. Importantly, when analyzing Pu.1 recruitment to in vitro assembled chromatin, only the second configuration inhibited Pu.1 binding, thus suggesting that in spite of this broad nucleosome-mediated enforcement of Pu.1 sites, chromatin remodelers may be selectively required for full Pu.1 binding only to sequences characterized by an extended high nucleosomal occupancy.
An important issue relates to the functional impact of the different levels of intrinsic nucleosome occupancy and positioning in the deciles. In addition to the possibility that chromatin remodelers may be differentially required depending on the affinity of the underlying sequence for nucleosomes, lower and higher deciles were specifically enriched for binding sites of distinct TF families. For instance, binding sites for NF-κB, which controls the rapid induction of hundreds of inflammatory genes, showed their maximal relative enrichment in the higher deciles. Since NF-κB binding is greatly impaired by nucleosomes (Lone et al., 2013), the preferential inclusion of its binding sites within sequences that promote nucleosomal occupancy may provide the basis for a tight enforcement of its recruitment to regulatory sequences.
Another important aspect is that TSS-proximal and distal cis-regulatory elements bound by Pu.1 displayed fundamental differences in their sequence composition that resulted in distinct effects on nucleosome assembly. For instance, container sites were exclusively found at distal, but not at TSS-proximal Pu.1 sites. Moreover, differently from distal sites, nucleosome depletion at TSS-proximal sequences with the lowest occupancy of the NDR was dependent on their high G+C content and not on Pu.1 occupancy.
Our data also demonstrate that the DNA sequence of cis-regulatory elements contains information that controls both binding competence of TF consensus sequences and nucleosome assembly. This co-occurrence explains the ability of the same DNA sequence features that discriminate bound from unbound Pu.1 sites to predict nucleosome occupancy in cells that do not express Pu.1. Whether such co-occurrence underlies direct causal relationships between DNA features that control TF recruitment (such as DNA shape characteristics) (Rohs et al., 2009) and nucleosome assembly, remains to be determined. Moreover, the general relevance of this model outside of this specific set of regulatory sites will have to be assessed.
The notion that overlapping DNA sequence and shape features control both the ability of a genomic DNA sequence to recruit transcription factors and its propensity to be incorporated into nucleosomes might have fundamental implications for both genomic biology and transcriptional control. First of all, it explains at the molecular level and at a genomic scale how regulatory elements can be selectively maintained under the gatekeeper activity of nucleosomes. Second, it implies that the same evolutionary forces that act to maintain the functionality of TF binding sites jointly control nucleosome deposition, thus preserving the gatekeeper function of nucleosomes during the evolution of regulatory DNA.
EXPERIMENTAL PROCEDURES
Nucleosome mapping
A limited MNase digestion was carried out on intact macrophage nuclei to generate a mixture of mono- and poly-nucleosomes. Mono-nucleosomal DNA was isolated from agarose gels and used for library construction. A detailed description of the computational analyses is provided in the Supplemental Experimental Procedures.
ChIP sequencing
ChIP was carried out starting from 5-8 × 106 cells, using a previously described protocol (Ghisletti et al., 2010).
In vitro nucleosome assembly and in vitro ChIP
Naked genomic DNA purified from mouse macrophages was sonicated to obtain fragments ranging from 600 to 2000 bp. DNA was combined with recombinant histones (EpiMark™ Nucleosome Assembly Kit, NEB E5350) to generate nucleosomes by salt dialysis (Luger et al., 1999). In vitro assembled nucleosomes were digested with MNase and then incubated with macrophage-derived nuclear extracts to generate TF-nucleosome complexes. The in vitro ChIP-seq was carried out as described in the Supplemental Experimental Procedures.
Computational methods
MNase-seq paired-end reads were mapped to the mouse genome using Bowtie (Langmead et al., 2009). Wiggle tracks at single bp resolution were generated with BedTools (Quinlan and Hall, 2010). PeakSplitter (Salmon-Divon et al., 2010) was used to extract nucleosomal positions from this population-averaged profile. Paired-end fragments for ESCs, NPCs and MEFs were retrieved from the literature (Teif et al., 2012).
ChIP-seq reads were aligned to the mouse genome using Bowtie and Peak calling was performed using MACS (Zhang et al., 2008). Peaks were annotated over Ensembl genes (Flicek et al., 2012). Pu.1-bound regions were sorted according to the NDR width. The number of midpoints of the nucleosomal fragments falling into the central 300 bp of each region was calculated and used as a proxy for the overall occupancy of the area. The genome-wide map of canonical Pu.1-binding sites (De Santa et al., 2010) was generated using FIMO (Creyghton et al., 2011).
Support vector machines (SVMs) (Cortes and Vapnik, 1995) were used to classify Pu.1-bound and unbound sites. Given a set of examples an SVM training algorithm builds a model that can be used to categorize new examples. The LibSVM implementation (Chang and Lin, 2011) was used to train and test two-class SVMs. Given the large amount of features used, a selection procedure (Guyon et al., 2003) to identify the smallest set with the highest predictive power was devised. Support Vector Regressors (SVRs)(Drucker et al., 1997) were applied to assess the fraction of variability in the nucleosomal occupancy patterns at Pu.1-bound and unbound sites that can be explained by the features selected by the SVM. DNA shape features were derived from a high-throughput data mining approach of all-atom Monte-Carlo predictions (Zhou et al., 2013). A detailed description of the computational methods and feature groups is provided in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Nucleosomes that mask TF binding sites show a spectrum of occupancy and positioning
Nucleosomes selectively mask TF bound sites but not random, unbound target sites
Coinciding DNA sequence and shape features control TF binding and nucleosome assembly
ACKNOWLEDGEMENTS
This work was supported by the European Research Council (ERC grant NORM to G.N.) and in part the National Institutes of Health (grants R01GM106056 and U01GM103804 to R.R.). R.R. is an Alfred P. Sloan Research Fellow. We thank B. Amati (IEO/IIT, Milan), JC. Andrau (CIML, Marseille) and A. Agresti (HSR, Milan) for comments on the manuscript; M. Pelizzola, L. Riva, M. Morelli (IIT, Milan) and L. Fornasari (IFOM, Milan) for suggestions; L. Ferrarini (IFOM) for help with machine learning; N. Habib, A. Weiner and N. Friedman (HUJI, Jerusalem) for initial help with the SVM and the analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers. Raw data sets are available for download at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds/) under the accession number GSE50762.
Supplemental information includes Supplemental Experimental Procedures, 6 figures and 3 tables.
REFERENCES
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Molecular cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- Charoensawan V, Janga S, Bulyk M, Babu M, Teichmann S. DNA sequence preferences of transcriptional activators correlate more strongly than repressors with nucleosomes. Molecular cell. 2012;47:183–192. doi: 10.1016/j.molcel.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2011;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I, Zhou T, Mandel-Gutfreund Y, Rohs R. Covariation between homeodomain transcription factors and the shape of their DNA binding sites. Nucleic Acids Research. 2014;42:430–441. doi: 10.1093/nar/gkt862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker H, Burges CJ, Kaufman L, Smola A, Vapnik V. Support vector regression machines. Advances in neural information processing systems. 1997:155–161. [Google Scholar]
- Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza J, Innocenti C.n., Ferrier P, Spicuglia S, Gut M, Gut I, et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome research. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic acids research. 2012;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney D, McVicker G, Pai A, Fondufe-Mittendorf Y, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard J. Controls of nucleosome positioning in the human genome. PLoS genetics. 2012:8. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gordân R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell reports. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Elisseeff An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. Andr, #233. [Google Scholar]
- Heintzman N, Stuart R, Hon G, Fu Y, Ching C, Hawkins R, Barrera L, Van Calcar S, Qu C, Ching K, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes I, Albert I, Zanton S, Pugh B. Nucleosome positions predicted through comparative genomics. Nature Genetics. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett A, Tillo D, Field Y, LeProust E, Hughes T, Lieb J, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Müller D, Clarke D, Assi S, Lie-A-Ling M, Noailles L, Vijayabaskar M, Wu M, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. The EMBO journal. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidor Nili E, Field Y, Lubling Y, Widom J, Oren M, Segal E. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome research. 2010;20:1361–1368. doi: 10.1101/gr.103945.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone IN, Shukla MS, Charles Richard JL, Peshev ZY, Dimitrov S, Angelov D. Binding of NF-kappaB to nucleosomes: effect of translational positioning, nucleosome remodeling and linker histone H1. PLoS genetics. 2013;9:e1003830. doi: 10.1371/journal.pgen.1003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Mavrich T, Ioshikhes I, Venters B, Jiang C, Tomsho L, Qi J, Schuster S, Albert I, Pugh B. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome research. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Nelson H, Finch J, Luisi B, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pan Y, Tsai C-J, Ma B, Nussinov R. Mechanisms of transcription factor selectivity. Trends in genetics: TIG. 2010;26:75–83. doi: 10.1016/j.tig.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West S, Sosinsky A, Liu P, Mann R, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen D. Transcription factors in myeloid development: balancing differentiation with transformation. Nature reviews Immunology. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore I, Wang J-PZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Segal E. Determinants of nucleosome positioning. Nature structural & molecular biology. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Schnappauf G, Thoma F. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic acids research. 2000;28:4083–4089. doi: 10.1093/nar/28.21.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif V, Vainshtein Y, Caudron-Herger MØ, Mallm J-P, Marth C, Höfer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nature structural & molecular biology. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes T. G+C content dominates intrinsic nucleosome occupancy. BMC bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett A, Field Y, Lieb J, Widom J, Segal E, Hughes T. High nucleosome occupancy is encoded at human regulatory sequences. PloS one. 2010:5. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson S, Boyd S, Smith C, Fire A, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden T, van Vugt JJ, Logie C, van Noort J. Sequence-based prediction of single nucleosome positioning and genome-wide nucleosome occupancy. Proceedings of the National Academy of Sciences. 2012;109:E2514–E2522. doi: 10.1073/pnas.1205659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhou T, Dror I, Mathelier A, Wasserman WW, Gordân R, Rohs R. TFBSshape: a motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Res. 2014;42:D148–155. doi: 10.1093/nar/gkt1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K, Carroll J. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mortazavi A, Williams B, Wold B, Rothenberg E. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo C, Wal M, Ward E, Korber P, Pugh B. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science (New York, NY) 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic acids research. 2013;41:W56–62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.