Abstract
Despite the presence of the blood-brain barrier (BBB) that restricts the entry of immune cells and mediators into the central nervous system (CNS), a small number of peripheral leukocytes can traverse BBB and infiltrate into the CNS. Cerebrospinal fluid (CSF) is one of the major routes through which trafficking leukocytes migrate into the CNS. Therefore, the number of leukocytes and their phenotypic compositions in CSF may represent important sources to investigate immune-to-brain interaction, or diagnose and monitor neurodegenerative diseases. Due to the paucity of trafficking leucocytes in CSF, a technology capable of efficient isolation, enumeration, and molecular typing of these cells in the clinical settings has not been achieved. In this study, we report on a biofunctionalized silicon nanowire array chip for highly efficient capture and multiplexed phenotyping of rare trafficking leukocytes in small quantities (50 microliters) of clinical CSF specimens collected from neurodegenerative disease patients. The antibody-coated 3D nanostructured materials exhibited vastly improved rare cell capture efficiency due to high-affinity binding and enhanced cell-substrate interactions. Moreover, our platform creates multiple cell capture interfaces, each of which can selectively isolate specific leukocyte phenotype. Comparison with the traditional immunophenotyping using flow cytometry demonstrated that our novel silicon nanowire-based rare cell analysis platform can perform rapid detection and simultaneous molecular characterization of heterogeneous immune cells. Multiplexed molecular typing of rare leukocytes in CSF samples collected from Alzheimer’s disease patients revealed the elevation of white blood cell counts and significant alterations in the distribution of major leukocyte phenotypes. Our technology represents a practical tool potentially for diagnosing and monitoring the pathogenesis of neurodegenerative diseases by allowing an effective hematological analysis of CSF from patients.
Introduction
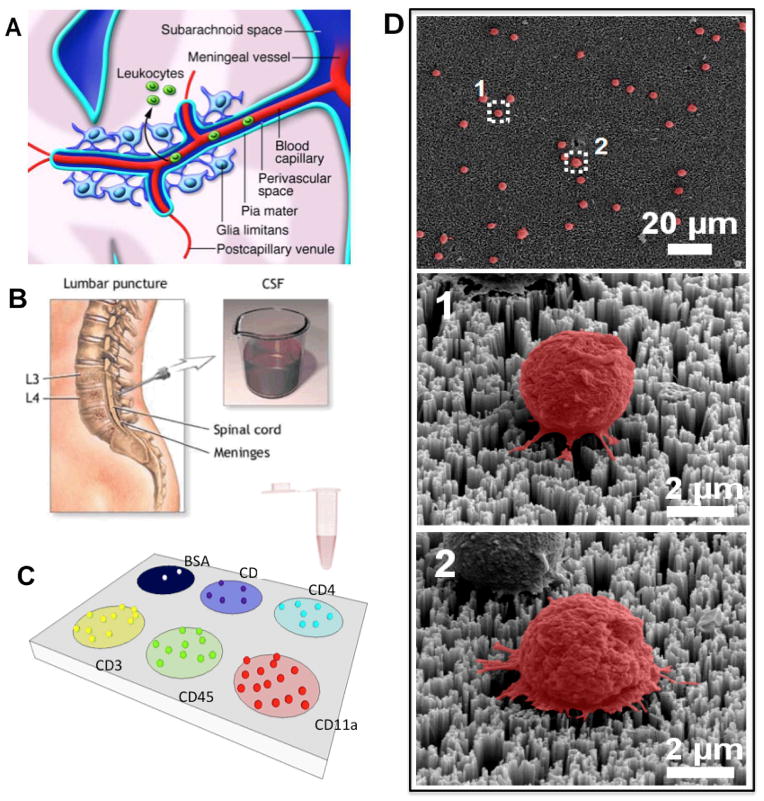
Although Alzheimer’s disease (AD) is one of the most common neurodegenerative diseases1,2 and its prevalence is expected to double over next 30 years, there is no currently accepted early diagnosis for AD3. Development of sensitive biological markers for neurodegenerative diseases will allow for early diagnosis and longitudinal monitoring of AD in a routine clinical setting, resulting in more effective therapeutic intervention1. Increasing evidence has suggested a substantial contribution of leukocyte trafficking and inflammation in the pathogenesis of major neurological disorders including AD1,4–7. Despite the blood-brain barrier, a wide variety of peripherally derived leukocytes would traverse the BBB and infiltrate into the CNS during numerous neuropathological circumstances8,5,9. The cerebrospinal fluid (CSF) has been suggested as the major hub for leukocytes trafficking into the CNS9,10 (Figure 1A). While healthy CSF typically contains a very small number of leukocytes11–14, neurological infection or inflammation in the CNS trigger marked elevation of leukocyte extravasation and accumulation into the CSF20,21. While abnormal CSF leukocyte count and variations in distributions of phenotypically different leukocyte subsets are typical observations among patients with CNS inflammatory neurological diseases [reference], little information regarding phenotypically diverse trafficking leukocytes in the CSF of AD patients is currently available6. Therefore, multiplexed detection of phenotypically distinct leukocyte subsets, and determination of phenotypic distribution may introduce a promising approach for clinical diagnosis and monitoring of neurological disorders6,19 (Figure 1B).
Figure 1.
Overall experiment design. A) The graphical representation of trafficking leukocyte migration into the central nervous system. In normal physiological conditions, immune cells enter the CNS for immune-surveillance. During pathological conditions, leukocytes increasingly traverse the blood-brain barrier and accumulate, leading to inflammation. One of the leukocyte migration pathways follows the formation of cerebrospinal fluid (CSF). B) The cartoon shows lumbar puncture, a conventional procedure to acquire CSF. Counting and immunophenotyping of trafficking leukocytes in CSF can yield valuable diagnostic information and help monitoring the status of the central nervous system. C) Conceptual illustration of highly efficient capture and multiplexed immunophenotyping of rare immune cells in CSF using a biochemically functionalized silicon nanowire platform. Integration of a PDMS construct with multiple loading chambers allows for functionalization of multiple antibodies at distinct regions. The platform contains multiple cell capture regions, each of which is functionalized with a specific antibody. Each region captures only the cells expressing specific surface antigens against a coated antibody. D) The SEM images of the CSF trafficking leukocytes captured on the silicon nanowire substrate. The images show that the cells are immobilized via a strong cell-nanostructure interaction.
To better evaluate immunogenicity in patients, simultaneous measurement of multiple phenotypic parameters of immune cells using a single analytical platform is increasingly needed to quantify complex cellular immune responses 20,21. Flow cytometry detection of distinct T cell surface markers is considered to be the gold standard of cellular immunophenotyping22–26. Flow cytometric immunophenotyping has been used to evaluate biological specimen such as blood or tissue to identify phenotypically abnormal cells with aberrant antigen expressions and characterize hematologic neoplasms and neurological disorders based on distinct immunophenotypes27,28. However, recovery of viable CSF leukocytes and characterization of their surface marker phenotypes have been technically limited due to extremely low abundance of trafficking leukocytes in cerebrospinal fluid combined with the rapidly decreasing CSF leukocyte viability following the centrifugation steps29. While sorting and characterization of heterogeneous cell population using flow cytometry requires between 1–10 million cells for optimal performance, cerebrospinal fluid of a normal adult may contain only about 1000 leukocytes per mL9,12, indicating the need of 100mL of CSF in order to perform flow cytometric analysis of trafficking leukocytes, which is impractical for clinical testing. Moreover, because the detection of leukocyte subsets with distinct surface markers is based upon distinct optical signals, the number of different cell phenotypes that can be simultaneously analyzed on the same sample is limited by spectral overlap30. Although the degree of multiplexing can be improved by the recent development of mass cytometry, it remains impractical in a clinical setting to perform rare cell immunophenotyping due to the high cost and technical expertise required for advanced flow cytometry and the requirement of large quantities of cells22,23.
Recent studies have reported antibody microarray-based T cell detection and sorting technologies, which can potentially increase the degree of multiplexing as cells are sorted and characterized based on their locations instead of colors30,31. However, those antibody microarray platforms were not used to capture rare cells and were developed based on traditional 2D materials. Many 3D nanostructured cell-capture substrates have been developed for highly efficient capture of cells of extremely low abundance32–37. Recent studies have shown 3D nanostructures coated with capture agents are advantageous over traditional 2D surface materials for high-quality rare cell capture as 3D nanostructures exhibit vastly improved cell capture efficiency32,34,37–40. The 3D nanotopography influences diverse cellular behaviors such as cell adhesion and motility, and enhances local topographic interactions between substrates and nanoscale cellular surface components (microvilli or filopodia)33–37,41–43. Moreover, the surface structures with size and shape-matched nanotopography can be readily fabricated from a range of materials to enhance interactions between the substrate and target cells32,38,44–47. Despite a significant improvement in cell capture efficiency, these nanostructure-based cell capture devices have not been demonstrated for multiplexed detection and analysis of phenotypically different immune cell subtypes in a single platform. Simultaneous detection and phenotyping of multiple cellular subsets with a single nanostructure has a number of advantages48,29,49. It can significantly reduce fabrication time for nanostructures, required reagent volumes, and cost of the assay. It also reduces consumption of clinical specimen, making the use of nano-enabled platforms in a clinical setting more practical.
We report on the development of a biofunctionalized silicon nanowire (SiNW) array for quantitative and multiplexed molecular typing of rare trafficking leukocytes from CSF specimens from neurodegenerative patients. Previous reports of 3D nanostructure-based rare cell capture platforms have inspired us to evolve the nanowire-based platform for rare cell capture into a novel platform for multiplexed detection and molecular phenotyping of rare and heterogeneous trafficking leukocytes populations. This platform combines nanostructure-enabled high-quality rare cell capture and “panning” of specific leukocyte subsets on the different locations of a single microchip to perform capture and quantificaiton of various lymphocyte phenotypes based upon their surface antigens (Figure 1C, Supplementary Figure S1). Additionally, integration of laser scanning cytometry method, a powerful technology for high-content, quantitative characterization of cellular functions, enables the large-area, automated detection and enumeration of captured leukocytes, and rapid on-chip determination of multiple leukocyte phenotypes at a single-cell level35. The validation of the platform using immune cell lines and primary T lymphocytes showed that multiplexed phenotyping using functionalized nanowire arrays was in a good agreement with a traditional flow cytometric immunophenotyping. Our study also demonstrated that the nanowire-enabled platform can achieve not only high capture efficiency, but also the ability to quantitatively measure leukocyte subsets of specific phenotypes from a mixture of primary immune cells. It was then successfully applied to the cellular phenotyping of rare trafficking leukocytes from AD patients. Based on multiplexed phenotyping using our platform, this study demonstrated a number of abnormalities and variations in the distribution pattern of diverse trafficking leukocyte subsets. Our results indicate that the total CSF white blood cell count and T lymphocytes of Alzheimer’s disease patients was elevated from the normal value, and the distribution patterns of major T lymphocyte subsets were significantly altered and varies from patient to patient. Our research represents the potential of biofunctionalized nanostructure as diagnostic or analytical tools based on rapid, highly multiplexed, and clinically practical molecular phenotyping of rare and heterogeneous immune cell populations.
Results and Discussion
Development of biofunctionalized silicon nanowire arrays for multiplexed detection and phenotyping of immune cells
We constructed the rare cell capture/analysis system using a SiNW array covalently linked with streptavidin for rapid biofunctionalization with protein capture agents such as antibodies as described in the experimental method section37,35. In brief, SiNW arrays were fabricated by a silver-catalyzed wet etching process of p-type Si (100) wafers with a resistivity of 1–10 Ω cm. assisted by nanosphere lithography (Materials and Method)50. To accurately count and analyze ultra-rare trafficking leukocytes in CSF samples with good statistical power, large-area SiNW substrate (>1 in.x1 in.) was fabricated for the device. Next, the SiNW substrate was treated with oxygen plasma that generated high density of silano group, and then sequentially applied with (3-aminopropyl)-triethoxysilane (APTES) and glutaraldehyde (GA), which provide chemical linker to functionalize nanowires with streptavidin in the next step (Supplementary Fig. S1). The cell-capture reservoirs with multiple circular loading chambers was fabricated using PDMS elastomer, and carefully aligned onto the SiNW substrate so that all of loading chambers were directly above the substrate region containing silicon nanowire. While loading chambers were made as large as possible (diameter ~8mm) so that the interaction between the nano-substrate and the cells can be maximized, the size of the chamber is adjustable depending on the area of a nanostructure and the number of phenotypes of interest. After streptavidin (STR) functionalization, 100 μL of biotin-antibody solution against specific leukocyte antigen was introduced into the streptavidin-immobilized region. Each distinct region was immuno-functionalized with one of different antibodies against distinct leukocyte phenotypic markers together with a negative control of bovine serum albumin (BSA). CD45 is a common leukocyte marker, which is expressed by most leukocytes. CD11a is a structural subunit of LFA-1, which plays a vital role in leukocyte trafficking and is present in all monocytic leukocytes. CD3 is a biomarker for T lymphocytes, while CD4 and CD8 are surface antigens expressed in helper T cells and cytotoxic T cells, respectively. The resultant platform (Figure 1C) contained multiple distinct substrate regions coated with leukocyte-specific antibodies that simultaneously detection of multiple leukocyte subsets of specific surface phenotypes to achieve multiplexed immunophenotyping. To use this microchip, the sample would be loaded into each loading chamber and incubated on ice for 30 minutes, during which the whole platform is gently agitated on the electronic shaker to facilitate the interaction between the substrate and target cells. The loading volume per each chamber can vary, ranging from 50μL to 200μL. Following multiple washing steps with PBS to remove unbound or non-specific cells, the standard immunostaining procedure that includes fixing (~20 min), permeabilizing (~10 min), and antibody staining (~45 min) was followed to generate detectable fluorescent signals from captured leukocytes. Conventional hematocymeter method would not be efficient or reliable enough to perform enumeration and characterization of all captured cells over such a large area at the single-cell resolution. We integrated a biofunctionalized nanowire arrays with laser scanning cytometry (LSC) for rapid, high-throughput, and automated phenotypic assessment of all trafficking leukocytes separated and captured on a single platform35. The assay typically takes no more than 2 hours from the sample loading to the LSC imaging (Materials and Method). To quantitatively characterize the cellular morphologies of the CSF trafficking leukocytes captured on STR- and antibody-functionalized SiNW arrays, scanning electron microscopy (SEM) analysis was performed using a cell freezing technique41. SEM images of representative leukocytes from cerebrospinal fluid captured on the fabricated silicon nanowire array (Figure 1D) showed that the captured leukocytes are well bound onto the substrate without significant morphological alterations and there are distinctive local topographical interactions between nanostructures and cellular surface components. The SEM images showed the captured leukocytes with smaller diameters (~5μm) than the typical diameter of leukocyte (8μm). It has been known that a sample prepared through conventional fixation method typically retains 72% of its original size43,51.
Determination of distinct immunophenotypes of well-characterized immune cell lines using nanowire-enabled multiplexed molecular phenotyping
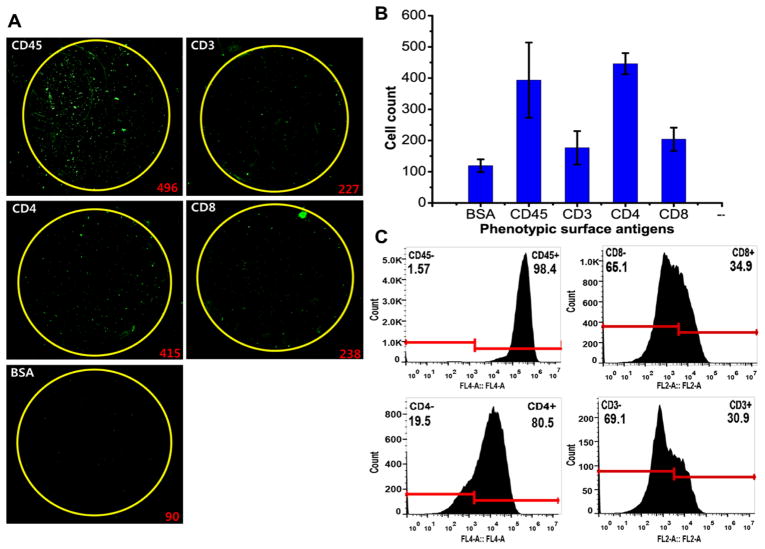
Before we applied this platform to clinical specimens from Alzheimer’s disease patients, we first tested the ability of our cell capture-analysis integrated platform to perform multiplexed phenotyping and quantitative determination of phenotypic profiles using immune cell lines. We used CCRF-CEM, which was derived from human acute lymphoblastic leukemia. Cells were prepared in single cell suspension at the cell density of 400 cells per loading chamber (~100μL). This density range is 1 order of magnitude higher than the leukocyte density in normal CSF, but is necessary in order to perfume head-to-head comparative validation using the gold standard technology such as flow cytometry. The captured cells were immuno-stained with phycoerythrin (PE)-labeled CD45 antibody to enable detection by fluorescence imaging and identify specific phenotypes of captured cells. The fluorescent image of the captured cells on the substrate was acquired by scanning the entire chip surface with a laser microarray scanner36,35,52. Figure 2A show the fluorescence micrographs of CCRF-CEM cells captured on different sample wells of a single chip. Quantification of the cells captured on different capture regions showed that CCRF-CEM cells express multiple surface antigens at the varying levels and exhibits heterogeneous expression patterns (Figure 2B), which agrees with previous studies20,53.
Figure 2.
Validation of multiplexed detection and immunophenotyping analysis using CCRF-CEM cells. (A) Fluorescence images of CCRF-CEM cells captured on different capture regions of a silicon nanowire platform functionalized with antibody against the specific leukocyte surface molecule. To detect and identify the captured trafficking leukocytes, the captured cells on the substrate were stained with PE-conjugated antibody against universal leukocyte surface antigen (CD11a). Excitation wavelength: 532 nm. The number in red on the bottom right corner of each image represents the number of cells captured and enumerated. (B) The enumeration of CCRF-CEM cells captured on different nanowire capture interfaces coated with phenotypic surface markers. 500 CCRF-CEM cells were loaded into each loading chamber. Captured cells which were stained positive for CD45 were counted. N=3. (C) Flow cytometric immunophenotyping result of CCRF-CEM cell populations. The cell suspension at the cell density of >106 cells/mL was reacted with fluorophore-conjugated detection antibodies against cell surface antigens (APC-CD11a, APC-CD4, PE-CD8, and PE-CD3). The number on the top left corner in each plot indicates the proportion of cells that were negative for the specific antigen expression. The number on the top right corner in each plot indicates the proportion of cells that were positive for the specific antigen expression. The locations of the gates were determined based on isotype control samples (Supplementary Figure S3).
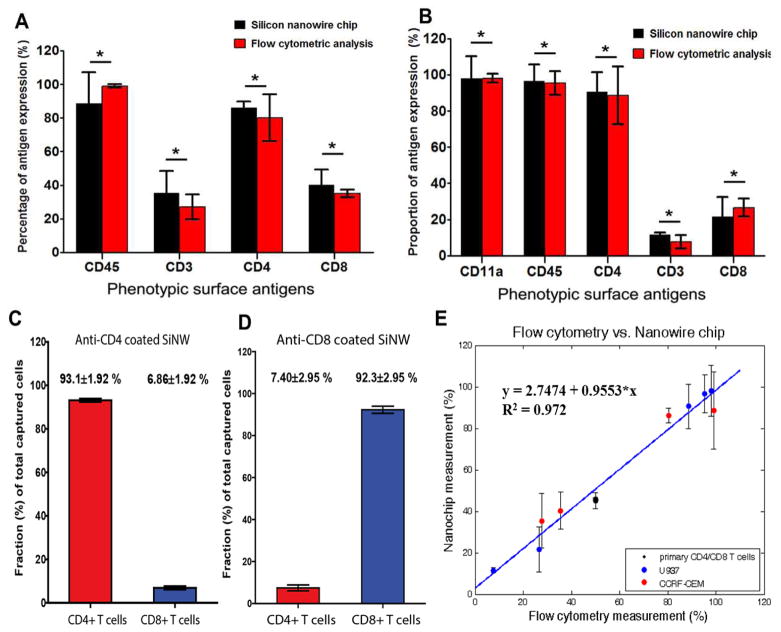
To quantitatively validate our platform, we performed dual-colored flow cytometric analysis for head-to-head comparison using the same cell samples (Figure 2C). To distinguish non-specific background signals, the gates were located to the right of the negative peaks of the negative control samples labeled with isotype control antibodies conjugated with corresponding fluorophores so that nearly 100% events were negative (Supplementary Figure S2). The side-by-side comparison of two independent technologies illustrates that the phenotypic antigen expression pattern of CCRF-CEM cells obtained by the nanowire-based multiplexed molecular typing approach is consistent with phenotypic expression profiles assessed by flow cytometry (Figure 3A). The percentage of the cells captured on a specific antibody-coated region is proportional to the frequency of the cells expressing the cognate antigen determined by flow cytometry. High capture yields for CD4 (~86.2%) and CD45 (~88.6%) antibody-coated wells correlated with immunophenotyping by flow cytometry that determined CD4 and CD45 expression on −80.4% and −99.8% of the total CCRF-CEM population, respectively. This observation also corroborates the notion that these acute lymphoblastic leukemia cells exhibit the high expression level of CD4 surface antigen20. Furthermore, the lower percentages of the cells captured on anti-CD3 (~35.3%) and –CD8 (~40.2%) coated interfaces correspond with FACS analysis that revealed antigen expression on 25–40% of the population. The differences between two immunophenotyping results were not statistically significant (p-value greater than 0.1) for all five surface antigens tested. As a further validation of the technology, we also performed multiplexed immunophenotyping of U937, which is derived from human monocytes, and observed that U937 cells exhibit a unique molecular phenotype by differentially expressing multiple surface phenotypic markers (Supplementary Figure S3A–B, Supplementary Table S2). The side-by-side comparison with immunophenotyping by flow cytometry reveals that the phenotypic antigen expression profiles of U937 obtained using silicon nanowire-based phenotyping platform are also in good agreement with immunophenotype determined by flow cytometry (Figure 3B).
Figure 3.
Comparison of multiplexed molecular phenotyping of two different immune cell populations with flow cytometric immunophenotyping, and capture specificities on primary T lymphocyte mixture on the SiNW-based platform. (A) Side-by-side graphical illustration of multiplexed immunophenotyping results of CCRF-CEM cells by both our silicon nanowire rare cell analysis platform and conventional flow cytometry approach. For each surface antigen expression level, the proportions from flow cytometry and our silicon nanowire-enabled platform were statistically compared using unpaired two-tail t test. * denotes the differences of proportion determined by flow cytometry phenotyping and nanowire-enabled rare cell analysis platform are not statistically significant (p-value>0.05). (B) Side-by-side graphical comparison of immunophenotyping results by flow cytometric analysis and our novel rare cell analysis chip. The percent proportion of each cellular phenotype was determined as the number of cells detected versus total number of U937 cells loaded in each well. For each surface antigen expression level, the proportions from flow cytometry and our silicon nanowire-enabled platform were statistically compared using unpaired two-tail t test. * indicates the differences in percentages of leukocyte expressing specific phenotypic antigens determined by two methods are not statistically significant (p>0.05). (C) The capture purity (capture specificity) of CD4+ and CD8+ T cells on the substrates coated with CD4 antibody. The red bar indicates the average proportion of target CD4+ T cells among all captured cells, while the blue bar represents the average proportion of non-target CD8+ T cells among all captured cells. (n=4) (D) The capture purity (capture specificity) of CD4+ and CD8+ T cells on the substrates coated with CD8 antibody. The red bar indicates the average proportion of non-target CD4+ T cells among all captured cells, while the blue bar represents the average proportion of target CD8+ T cells among all captured cells. (n=4) (E) Correlation of the proportions of leukocyte subsets expressing different surface antigens determined by the bio-functionalized silicon nanowire arrays as a function of the proportions measured by flow cytometric immunophenotyping for three different cell types (black-primary CD4 and CD8 T cell mixture, blue-U937, and red-CCRF-CEM). The solid blue line represents a linear regression, indicating a strong linear correlation (R2=0.972). The linear regression illustrates that the agreement between immunophenotyping based on flow cytometry and our silicon nanowire substrate is statistically significant with the slope is close to 1 (0.955). Each point shows the mean proportion of the specific leukocyte phenotype and the standard deviation with the sample size (n) greater than or equal to 3.
Furthermore, we evaluated the platform’s capability for phenotyping of primary immune cells using the mixture of human CD4+ and CD8+ T cells (Astarte Biologics, Redmond, VA, USA). Varying numbers of CD4 and CD8 T lymphocytes were mixed in a ratio of 1:1, and the mixture was loaded into a silicon nanowire array coated either with anti-CD4 or anti-CD8. The captured cells were treated with PE-anti-CD8 and APC-anti-CD4 so that CD4+ and CD8+ T cells could be detected with red and green, respectively (Supplementary Fig. S4A). As shown in Fig. S4B, the SiNW substrates demonstrated excellent capture yields for both CD4+ and CD8+ T cells (90.1±7.74% and 91.6±1.30%, respectively). In anti-CD4 coated surfaces, the specific capture efficiency for target cells (CD4+ T cells) was significantly higher than that for non-target cells (CD8+ T cells). In anti-CD8 coated substrates, the capture efficiency for CD8+ T cells was higher than that of CD4+ T cells (Supplementary Fig. S4B). The numbers of captured CD4+ and CD8+ T cells were nearly equal at varying cell loading numbers, consistent with the fact that the mixture contained 50% CD4 and 50% CD8 T cells (Supplementary Fig. S4C). We could also evaluate the cell-capture specificity. As shown in Fig. S4A, the cells captured on the SiNW array surfaces coated with anti-CD4 or anti-CD8 were predominantly T cells expressing the surface antigens that react with the substrate-functionalizing antibody. The capture purity was defined as (# of captured target cells) divided by (total # of all captured cells). The capture purities of both CD4 antibody-coated and CD8 antibody-coated substrates were high, (93.1±1.92%) and (93.9±1.10%), respectively (Figure 3C–D). While previously developed nanostructured-based cell capture platforms have reported excellent separation efficiency for primary T cells of single phenotype, the platform described in our current study, to our knowledge, is the first one to achieve highly efficient and multiplexed capture of different immune cell phenotypes and exhibit excellent capture specificity for specific target leukocyte phenotypes when multiple immune cell types are mixed. This observation of excellent capture purity demonstrates that our platform is capable of capturing target cells of specific phenotypes with high selectivity with insignificant level of non-specific capture.
To quantitatively evaluate how strong the correlation between immunophenotyping results from two platforms is, we performed linear regression for the proportions of different leukocyte phenotypes from all three immune cell populations, CCRF-CEM, U937, and primary CD4 and CD8 T cell mixture, determined by two platforms. The linear regression results showed that there is a statistically significant correlation between a nanowire array and flow cytometry as the slope of linear regression equation is ~0.955 (Figure 3E). The slope of linear regression that is close to 1 indicates that immunophenotypes determined by flow cytometry and our nanowire-based platform agree very well. R2 value is also close to 1 (0.972), indicating a strong linear correlation (Figure 3E). These observations indicate that our technology can achieve highly efficient and specific capture of multiple leukocyte phenotypes, quantitative molecular typing, and all together give rise to the immunophenotypes defined by surface antigen expression profiles with good accuracy and sensitivity while requiring only as few as hundreds of cells.
Efficient isolation and multiplexed phenotyping of rare trafficking leukocytes in the cerebrospinal fluid of Alzheimer’s disease patients
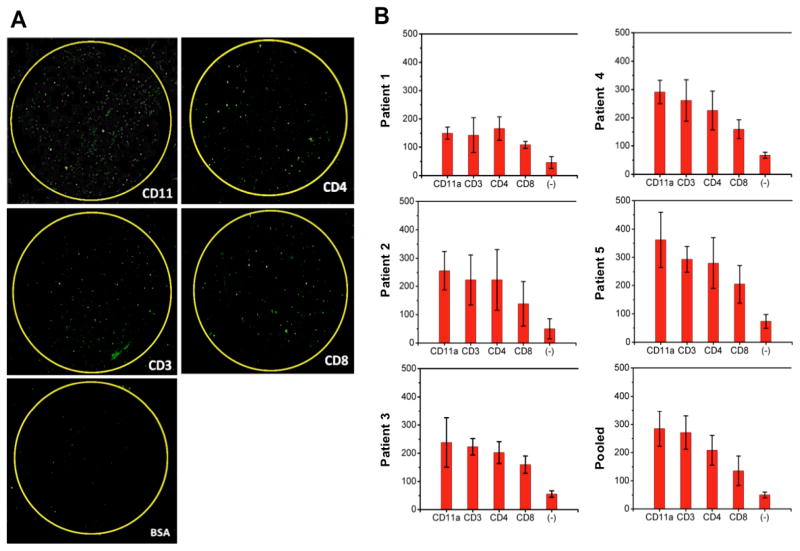
Upon the validation of our platform, it was applied for multiplexed molecular typing of rare leukocytes from human cerebrospinal fluid collected from Alzheimer’s disease patients. We directly measured the as-collected cerebrospinal fluid samples from 5 CSF samples from individual AD patients and a pooled CSF sample from a large cohort of patients (Figure 1B). For each patient’s CSF sample, the experiment was repeated at least 3 times in multiple independent tests. In each test, we used a microchip that has five streptavidin-conjugated capture miniwells, four of which were further functionalized with biotinylated capture antibodies specific to different phenotypic markers and the last miniwell was left blank without adding any antibody that served as a negative control. The captured cells were fixed and stained with DRAQ and anti-CD45-PE and quantified using a laser scanning cytometry method36,54,55. Due to extreme cellular paucity in cerebrospinal fluid, the significantly fewer number of leukocytes were captured and detected from patient CSF samples (Figure 4A) than from the validation experiment using cell lines.
Figure 4.
Capture and multiplexed quantification of leukocytes in five CSF samples collected from individual Alzheimer’s disease (AD) patients and a pooled CSF aliquot. A) Fluorescence images of trafficking leukocytes in 50 μL CSF captured on biochemically functionalized silicon nanowire substrate. The fluorescent image of a whole chip was acquired through microarray laser scanner. Biotinylated antibodies against leukocyte surface antigens such as CD11a, CD4, CD3, and CD8 were conjugated with streptavidin-coated silicon nanowire arrays. The captured leukocytes illuminate with green fluorescence as they are tagged with PE-conjugated antibody against a common leukocyte antigen, CD45. The regions highlighted with yellow circles represent the antibody functionalized capture interface. Very few cells were detected in the region, which is not functionalized with antibody and serves as negative control. B) Quantification of trafficking leukocytes in CSF captured on each region functionalized with different antibodies. Counting of the captured leukocytes was fully automated via our image analysis scheme (described in ref. 10) that distinguishes the cells based on their physical characteristics. For each CSF sample from one patient, the experiment was repeated at least three times, and the average number of the cells captured at different regions was plotted in the bar graph.
Cerebrospinal fluid abnormalities, including increased leukocyte numbers and alternation in leukocyte subset distribution, have been observed in neuropathological conditions, affecting the CNS56–58. The absolute count of white blood cell (WBC) and percentages of leukocyte subsets in cerebrospinal fluid are routine and important assessment parameters for the diagnosis and examination of many neurological disorders29. Although immunophenotyping of CSF leukocytes has been recognized as a potentially informative tool for diagnosing and monitoring pathogenesis of inflammatory conditions in the CNS59, to our knowledge, it remains challenging to perform multiplex molecular typing of leukocytes from the AD patients’ CSF to determine the absolute counts of phenotypically distinct leukocyte subsets and establish the representative immunophenotype profile for Alzheimer’s disease CSF. We quantified the absolute number of white blood cells and the relative proportion of different leukocyte phenotypes in cerebrospinal fluid in order to evaluate their relative prevalence for diagnosing brain inflammation and understanding the immune responses during neurodegenerative diseases (Figure 4, Supplementary Table 3). The highest proportion of cells from CSF was consistently captured in anti-CD11a leukocyte function-associated antigen-1 (LFA-1) coated region in all six AD CSF samples (Figure 4A–B). This observation is consistent with the fact that trafficking leukocytes migrating via CSF are known to express high levels of LFA-1 molecules because it has been proposed that infiltrating T cells are recruited into CSF through the increased LFA-1 expression60. CD11a is the structural components of LFA-1 which plays a critical role in the adhesion and migration of trafficking leukocytes at the sites of inflammation, and it has been shown to be present in the leukocytes in the brain of AD patients1. Our observation corroborates the previous reports that showed the up-regulated LFA-1 expression on infiltrating leukocytes in AD brain and the significant increase in the number of trafficking leukocytes expressing LFA-1 in Parkinson’s disease patients61. Also, a significant number of leukocytes were captured on anti-CD4 functionalized substrates than on anti-CD8 coated regions. This is supported by the previous reports that leukocytes contained in normal adult CSF are mainly CD4+ memory T cells9,11. We observed a small number of leukocytes captured in the substrate surface serving as a negative control, which is consistent with previous studies that biofunctionalized nanostructured substrates can capture rare cells more efficiently regardless of cells types33 (Supplementary Figure S5).
We assessed the utility of the SiNW-based platform to determine proportions of specific leukocyte subsets in AD CSF and detect significant abnormalities in AD CSF leukocyte phenotype. First, it is known that the total white blood cell count in CSF is correlated to the development of a variety of neurological pathologies62. In our platform, the total trafficking leukocytes count could be estimated from detecting and enumerating the cells captured on anti-CD11a grafted region. Since LFA-1 plays a critical role in leukocyte trafficking and is present on all leukocytes, we could estimate the CSF trafficking leukocyte counts by enumerating the cells captured on the capture surface coated with antibody for CD11a, the structural component of LFA-163. The total number of trafficking leukocytes in the CSF of AD patients ranges from 1.28±0.062 to 3.03±0.82 (Figure 5A), which is slightly higher than the average white blood cell count in normal CSF (mean: 1.12 cells/uL [5th and 95th percentile: 0.40 and 3.17]) reported in previous studies62,14,59,64. The results demonstrated that our platform could capture trafficking leukocytes from 50 μL of CSF and estimate the total white blood cell density to examine disease states. Our observation that the number of migrating leukocytes expressing LFA-1 increased in the CSF of AD patients can be explicated by the recent hypothesis that Aβ deposition during AD pathogenesis promotes leukocyte adhesion and transmigration into the CNS by inducing up-regulation of adhesion molecules including LFA-11,65.
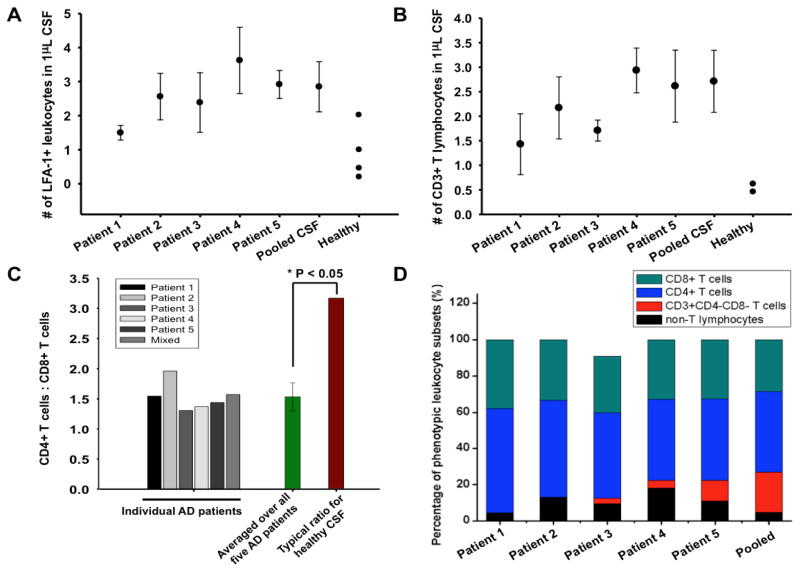
Figure 5.
Molecular phenotyping analysis and interpretation of trafficking leukocytes in CSF of Alzheimer’s disease patients. A) The scatter plot illustrating the cell density of trafficking leukocyte population expressing LFA-1 in CSF of five AD patients and in one pooled CSF sample. For each phenotyping analysis, the number of all trafficking leukocytes contained in 50 μL CSF was first determined, and it was normalized to determine the cell density (absolute count/μL). The values of trafficking leukocyte density in normal adult’s CSF were derived from previous studies. B) The scatter plot illustrating the cell density of CD3-positive T lymphocytes in CSF of AD brains. The cells captured on the region coated with anti-CD3 and stained positive for CD45 were enumerated. For each patient CSF sample, the phenotyping analysis was repeated four times. C) The ratio of CD4+ helper T cells to CD8+ cytotoxic T cells based on immunophenotyping of AD patient’s CSF. The average CD4 helper T cells:CD8 cytotoxic T cells in Alzheimer’s disease CSF was significantly lower than the ratio observed in CSF of healthy subjects. The population average of CD4:CD8 ratio in normal adult CSF was used for comparison. One sample student’s t-test was employed to calculate statistical significance of the difference between Alzheimer’s disease and healthy CSF. *P-value<0.05 (1.2×10−5); n=6. D) Graphical illustration of CSF trafficking leukocyte distribution of five Alzheimer’s disease patients and one pooled mixture. The analysis shows that trafficking leukocytes in AD patient’s CSF are predominantly T cells (CD3+), and there are a small number of T cells that express neither CD4 nor CD8 antigen, indicating the presence of T cells that are not fully developed.
By detecting and enumerating the number of the cells captured in the region functionalized with anti-CD3, a T lymphocyte-specific marker, the platform could determine the total number of T lymphocytes in CSF of AD patients. Multiplexed phenotyping of CSF from AD patients showed that the leukocytes found in the CSF of AD patients were predominantly constituted of CD3 positive T lymphocytes (86.3±9.20%) (Figure 5D). The number of invading CD3+ T lymphocytes in the AD brain is lower than other inflammatory diseases in the CNS such as multiple sclerosis as AD lacks prominent infiltrates of peripheral leukocytes observed in those prototypical autoimmune neurological diseases66. However, multiplexed phenotyping showed that the number of trafficking T lymphocytes in the AD patient CSF detected by the nanowire-enabled rare cell analysis system was significantly greater than the typical number of T cells in normal CSF (mean: 0.46 and 0.62 cells/μL [5th and 95th percentile: 0.16 and 1.88]) (Figure 5B)31,67. Our observation agrees with previous studies that the number of infiltrating CD3 T cells typically increased in the AD brain compared to the control groups without neurological complications4. This result supports the notion that peripheral T lymphocytes may traffic and accumulate in the CNS more frequently in response to increased expression of leukocyte adhesion molecules and inflammatory signals during AD pathogenesis66.
Measurement of the CSF leukocyte phenotypic distribution can be valuable in evaluating pathological conditions and immunological responses in the CNS during AD pathogenesis as the cellular composition of CSF leukocytes significantly changes as peripheral immune cells are migrated to the site of inflammation11. We calculated CD4+/CD8+ T cell ratio in AD patient’s CSF by simultaneously detecting and enumerating CD4+ helper T cells and CD8+ helper T cells within a single platform. We calculated CD4+/CD8+ T cell ratio in AD patient’s CSF by simultaneously detecting and enumerating CD4+ T lymphocytes and CD8+ T lymphocytes within a single nanowire array. Multi-colored fluorescence immunostaining was performed to visualize and identify molecular phenotypes of the leukocytes captured on different capture regions (Figure S6A–B). Enumeration of the captured cells showed increased frequency of both CD4+ and CD8+ T cells (Table 1). Our observation again agrees with previous findings showing increases of CD4+ T cells in CSF of inflammatory diseases in the CNS71,72. Increases of CD4+ T cells in CSF, which can assume a helper–I T cell phenotype and secrete pro-inflammatory cytokines, may indicate brain inflammation and related neuronal damage73. It has been reported that normal CSF exhibits the predominance of CD4+ T lymphocytes, low number of CD8+ cytotoxic T lymphocytes (CTLs), and high CD4/CD8 ratio. Multiplexed phenotyping of T lymphocytes showed that the major (>50%) T lymphocyte phenotype in AD CSF was indeed CD4+ T cells as in normal CSF (Figure 5C–D). Although the patient sample number was small (n=6), our data showed that CD4/CD8 ratio in the CSF of AD patients was consistently reduced as the average CD4/CD8 ratio in all AD patients’ CSF is ~1.53 compared to the ratio (~3.1) observed in healthy CSF (Figure 5B)62,69. Several inflammatory diseases in the CNS are associated with increased extravasation of T lymphocytes including CD8+ CTLs, and their deleterious functions can be crucial for neuronal death and tissue destruction leading to neurodegenerative diseases74. The increased percentage of CD8+ cytotoxic T lymphocytes and the significantly reduced CD4/CD8 ratio suggests that Alzheimer’s disease patients may have developed chronic CNS inflammation that elicits increased CD8+ CTL recruitment and clonal expansion75. Excessive infiltration of cytotoxic T cells might have deleterious effects as CTLs mediate pathologic immune activities leading to inflammation and neuronal destruction in neurodegenerative lesions75,76. Our results using multiplexed molecular typing of trafficking leukocytes in CSF reveal the existence of diverse leukocyte subsets expressing distinct surface antigens and the CSF leukocytes from each patient have a unique spectrum of molecular phenotypes (Figure 5D). Our results also show that the CSF leukocytes are predominantly composed of T lymphocytes with significantly smaller portion (<10%) non- T-lymphocytes (Figure 5D).
Table 1.
Comparison between reference values (absolute count) of leukocyte subsets in normal CSF [1] and measured values of leukocyte subsets in Alzheimer’s disease CSF
| Subset | Immunological phenotype | Reference cell densities in normal CSF; n=84a | Measured cell densities in AD CSF; n=6b |
|---|---|---|---|
| Leukocytes | CD45+ | 1.12 (0.40–3.17) | 2.57 ± 0.88 |
| T lymphocytes | CD3+ | 0.62 (0.16–1.88) | 2.31 ± 0.67 |
| CD4 helper | CD4+ | 0.44 (0.08–1.43) | 2.11 ± 0.52 |
| CD8 cytotoxic | CD8+ | 0.13 (0.04–0.40) | 1.46± 0.44 |
Medians (5th–95th percentile) of absolute counts/μL are given based on flow cytometry immunophenotyping of cerebrospinal fluid from 84 individuals without any neurological disorders,65,54.
Means (± standard deviation) of absolute counts/μL of different leukocyte subsets in CSF of Alzheimer’s disease patients (n=6) are given. The cell densities were determined using biofunctionalized nanowire array-based rare cell capture and phenotyping platform.
We further investigated how the molecular phenotyping results may change with varying the CSF input volume (Figure S7 and supplementary information). With increasing the volume of CSF loaded into each microwell ranging from ~50 to ~200 μL, the number of cells captured increased linearly (Figure S7 A–D). Correlation between the CSF loading volumes and the quantitation of different phenotypes shows a strong linear relationship. Furthermore, as the CSF volume progressively decreased from 200 to 50 uL, the total leukocyte count per 100 μL did not change significantly (Figure S7E–F). CD4/CD8 T cell ratio was found to be consistent as well except for the result from 200uL due to the relatively large error of a single data point (CD8 count in Figure S7D). These results illustrated that our platform can perform reproducible and accurate immunophenotyping of rare CSF trafficking leukocytes from AD patients by using 50 μL of CSF per microwell.
Conclusions
In this study, we described the development of a biofunctionalized nanowire array-based platform for efficient capture and multiplexed phenotyping of infiltrating trafficking leukocytes from the cerebrospinal fluid of Alzheimer’s disease patients. Because the readout of the assay by this platform is dependent on both distinct optical signatures and locations of captured cells, we demonstrated the platform’s multiplexing capacity to simultaneously capture and analyze multiple, phenotypically different leukocyte populations while not limited by spectral overlap. Multiplexed detection and analysis of different leukocyte subsets within a single microchip can permit significantly reduced fabrication time, cost, and required reagent volumes. It also minimized the batch-to-batch effect, the major source of variability29,48,49. Because our microchip permits the use of significantly reduced amounts of CSF samples for quantifying phenotypes of trafficking leukocyte, this platform is uniquely suited for clinical evaluation of neurological pathology, given limited availability of CSF in a clinical setting. Comparison with the conventional technology of immunophenotyping using multi-colored flow cytometry confirmed that the new technology can accurately quantify leukocyte subpopulations that express different surface antigens and accurately determine the immunophenotypes using well-characterized immune cell lines while requiring only a small number of cells.
Although the role of trafficking leukocytes in the CNS has been widely appreciated, it remains unclear whether the absolute number and immunophenotype of trafficking leukocytes in the CSF are associated with the disease status. Jesse et al. observed a very small number of neurodegenerative disease patients that showed elevated CSF leukocyte count, but suggested the absolute count of CSF leukocytes can be helpful for specific cases77. Our preliminary study showed that the total trafficking leukocyte counts in AD patients’ CSF modestly increased compared to normal CSF leukocyte counts. An increased frequency of both CD4+ and CD8+ T cells was observed, which agrees with previous findings that there are elevations of both T cell subsets in the brain of neurodegenerative disease71,72. AD patients exhibited significantly altered profiles of leukocyte phenotypes, confirming the recent notion that the increased migration of CD8+ cytotoxic T cells is associated with AD development. More comprehensive quantitative analysis with a large cohort of patients may ultimately establish total trafficking leukocyte count and proportions of phenotypically different leukocyte populations as biomarkers for neurodegenerative diseases.
This study is a significant step towards addressing the challenge in the clinical analysis of trafficking leukocytes due to the paucity of cells, the limit of sample quantity and the requirement of multiplex molecular analysis. It represents a promising tool for early clinical diagnosis and monitoring of neurological diseases. The platform can be further improved by introducing microfluidic channels. The microfluidic techniques can enhance the capture efficiency by optimization of channel dimension, cell distribution, and flow rate32,78–80. The technology will be expanded by fabricating a larger SiNW array substrate containing more capture interfaces. The expanded platform will provide greater multiplexing capacity to capture and quantify more diverse leukocyte phenotypic subsets, and to identify previously unknown abnormalities in cellular compositions of the CSF during neurological disorders. The multiplexed analysis of trafficking leukocytes in the CSF can lead to a better understanding of the molecular mechanisms responsible for increased infiltration of peripheral leukocytes during pathological circumstances affecting the central nervous system and the impacts of trafficking leukocytes in pathogenesis of various neurological diseases. As a conventional flow cytometric analysis generates a database of phenotypic expression profiles that enable diagnosis of blood-born cancer27, this rare cell phenotyping platform has the potential to enable the recognition of distinct immunophenotypes of the CSF trafficking leukocytes from neurological disease patients and establish new molecular criterion for effective and early diagnosis of various neuropathological conditions.
Experimental Methods
Silicon Nanowire Array Fabrication
Silicon nanowire (SiNW) arrays were fabricated by silver (Ag)-assisted chemical etching of p-type Si (100) wafers with a resistivity of 1–10 Ω·cm81. First, a large-scale Si substrate (4 inch in diameter) was cut to a size of 2.5 cm × 2.5 cm and was then cleaned by ultrasonication in acetone, isopropyl alcohol (IPA), and deionized (DI) water for 15 min, respectively. The cleaned samples were immersed in 10 wt% hydrofluoric (HF) acid for 5 min to remove the native oxide layer, and treated in boiling RCA cleaning solution (H2O2 : NH4OH : H2O = 1 : 1 : 5) for 1 h to convert the surface into a hydrophilic surface. An Ag film (~30 nm) was coated onto the cleaned Si substrates by electroless deposition in an aqueous solution containing 10% HF and 5×10−3 M AgNO3 solution at room temperature for 5 min. The Ag-coated Si samples were then immersed in an aqueous solution containing 10% HF and 0.3% H2O2 at room temperature for 30 min. Finally, the Ag metal remaining on the Si substrates was completely removed by aqua regia (HCl : HNO3 = 3 : 1) for 1 h and additional amorphous Si etching for 30 s in buffered oxide etchant (BOE, NH4F : HF = 6 : 1). The SiNWs were typically 60 – 100 nm in diameter and 5 – 10 μm in length, which are strongly dependent on the size of Ag nanoparticles and etching time, respectively82,42.
Chemical functionalization of silicon nanowire arrays
Prior to the bio-functionalization, the silicon nanowire arrays were treated with H2O2:H2SO4 (1:1) for 30 minutes to remove all the organic materials and impurities on the surface. The substrates were washed in acetone, isopropyl alcohol, and distilled water sequentially, and dried with air. The cleaned silicon nanowire (SiNW) surfaces are treated with oxygen plasma for 2–3 minutes to present the hydroxyl groups on the substrate. The surface of the substrate is modified with 5% (volume/volume) 3-mercaptoproyl trimethoxysilane (APTES, Sigma-Aldrich, USA) in ethanol at room temperature for 45 minutes with gentle agitation, immediately followed by rinsing with ethanol. The reaction with APTES presents and functionalizes the substrate surface with amine groups. Next, the SiNWs are treated with 50% (v/v) glutaraldehyde (GA, Sigma-Aldrich, USA) in deionized water at room temperature for 2 hours, followed by gentle rinse with deionized water. The amine groups rendered by APTES are reacted with glutaraldehyde that functionalizes the Si surface with aldehyde groups. The chemical modification is to render stable and efficient deposition of streptavidin (STR) onto the nanostructured substrate.
Integration of a PDMS loading chamber and streptavidin deposition
To enable multiplexed detection and phenotyping, multiple capture regions have to be created. To generate multiple, distinct locations, each of which can be functionalized with different agents, the chemically activated SiNW substrate is coupled with a polydimethylsiloxane (PDMS) elastomer mold. A mixture of GE RTV 615 PDMS prepolymer part A and B (10:1) is thoroughly mixed and degassed for overnight. The PDMS mixture is cured at 80°C for 2 hours and is solidified. The solidified mold is cut in the size of 1×1 inch2, which is also the size of a SiNW substrate, and a number of cylindrical wells (six) are drilled with a stainless steel hole punch with a diameter of 6.5 mm. The PDMS mold and the SiNW substrate are carefully aligned and permanently coupled after they are thermally cured at 80°C for 1 hour. The coupling has created the multiple loading chambers into which 50 μL of 5% streptavidin in PBS is applied and incubated for overnight in an incubator (373°C, 5% CO2), leading to deposition of streptavidin. Previous study by Kim et al (2010) observed that the modification with oxygen plasma, APTES, and GA resulted in successful functionalization of streptavidin on the surface of the SiNW1,4. Now, there are specific regions on the SiNW surface that are functionalized with STR.
Biochemical functionalization with biotinylated capture antibodies
The loading chambers of the platform are carefully washed several times with PBS to remove excess streptavidin without disintegrating the PDMS mold from the SiNW surface. To establish cell analysis system that enables the separation and phenotyping of multiple distinctive leukocyte subsets in a single platform, we used the fact that phenotypically distinct immune cell types tend to express a unique set of surface molecules. For example, helper T cells express CD4 molecules on their surface, but do not express CD8, which cytotoxic T lymphocytes express. The solution of biotinylated antibody against specific leukocyte surface antigen was prepared by dilution with PBS which contains 1% BSA with 1:200 (v:v) ratio. BSA would block the substrate surface to prevent non-specific cell binding. 30μL of the antibody solution against the specific surface biomarker of a distinctive leukocyte subset was loaded into one of the platform’s six cylindrical loading reservoirs, and reacted for 1 hour at room temperature. The SiNW surface functionalized with high density streptavidin allows strong immobilization of biotin-conjugated monoclonal antibodies due to high-affinity binding interaction between STR and biotin. Because of strong chemical affinity between biotin and streptavidin, the substrate would be immobilized with the high density of antibody in 20 min. Each circular capture region on the SiNW surface was functionalized with the monoclonal antibody against the specific surface antigen, serving as the capture antibody that attracts and immobilizes the phenotypically specific leukocyte type.
Multiplexed capture and phenotyping analysis of existing heterogeneous cell lines (U937 and CCRF-CEM)
To validate the platform’s capability for accurate multiplexed immunophenotyping, two cell lines, U937 (monocyte, CRL-1593.2) and CCRF-CEM (immature lymphoblast, CCL-119) were used. They were purchased from Amercian Type Culture Collection (ATCC, USA). Both cell lines were cultured with the medium composed of RPMI-1640, L-glutamin, antibiotic-antimycotic (1x), and 10% fetal bovine serum. Prior to loading into the platform, cell density was manually determined using a conventional hemocytometer (Hausser Scientific Co. USA). The cell suspension was serially diluted in the culture medium so that cell suspension of about 100 μL could be introduced into each loading reservoir with cell numbers in the varying ranges including 400, 600, 800, and 1000 cells/well. After being loaded into the device, the cells were incubated at 37°C and 5% CO2 for 30 minutes, and allowed to interact with the silicon nanowire arrays functionalized with capture antibodies. After incubation, the PDMS wells and SiNW substrate were washed out with 1xPBS with 1% BSA at least three times to remove unbound non-specific cells and cellular debris.
Multiplexed immunophenotyping of existing cell lines (U937 and CCRF-CEM) using conventional flow cytometric phenotype analysis
To validate the accuracy of multiplexed molecular phenotyping and quantification of phenotypically heterogeneous cell populations using a SiNW-based rare cell analysis platform, flow cytometric immunophenotyping was performed with the same two existing cell lines, U937 and CCRF-CEM. The cell suspensions were appropriately diluted in the culture medium to make the cell density of approximately 105 cells/mL. Accuri® C6 flow cytometer (BD Bioscience, USA) was used to perform the analysis. A cell suspension of 1 mL was fixed with 4% PFA at room temperature for 20 minutes, and reacted with florophore-conjugated antibodies against various leukocyte surface molecules at 4°C for 1 hour. As the flow cytometer contains two excitation laser lines for excitation wavelengths of 488 and 640nm, the cells were stained with two detection antibodies each with two distinct emission wavelengths. For instance, the suspension was reacted with FITC (488nm)-anti CD11 and APC (635nm)-anti CD4 to quantify the proportion of CD11+ and CD4+ cells in the heterogeneous cell populations.
Assessment of capture-efficiency and capture-purity using a primary CD4+ and CD8+ T lymphocytes mixture
Additional experiments were conducted to investigate the capture purity and capture efficiency of target and non-target cells when they were mixed together. The SiNW substrates were coated with either CD4 or CD8 antibody solutions diluted in 3% BSA in PBS (1:100 ratio). Fluorescent activated cell sorting (FACS) was used to sort primary CD4+ and CD8+ T cells from human PBMCs. CD4 and CD8 T cells were mixed in 1:1 ratio at the varying cell numbers. We then introduced the mixture composed of approximately 250 CD4+ and CD8+ T cells, 500 CD4+ and CD8+ T cells, or 650 CD4+ and 650 CD8+ T cells into anti-CD4 coated, anti-CD8 coated, or BSA-treated substrate capture regions, followed by incubation for 30 minutes in an incubator. Then, the cell suspension was gently aspirated from the cell-capture reservoirs, which were washed with PBS for 3 times to remove non-specific cells and impurities. 50 μL of PE-CD8 and APC-CD4 antibody solution (1:100 dilution in 3% BSA PBS) was introduced into the loading reservoirs and incubated for 30 minutes in room temperature to fluorescently stain the cells bound on the substrates for detection. The captured CD4+ T cells, which illuminated red fluorescence, could be easily distinguished from CD8+ T cells, which illuminated green fluorescence (Supplementary Fig. S5A).
Multiplexed immunophenotyping and detection of trafficking leukocytes in cerebrospinal fluid (CSF) of human Alzheimer’s disease patients
The CSF specimens were donated from the Alzheimer’s Association. There were five CSF samples collected from five individual Alzheimer’s disease patients, each of which was 10 mL. There was one pooled CSF mixture sample, which was pooled mixture of CSF collected from many individual patients. All CSF samples had been aliquoted in smaller volumes to reduce the number of freeze-thaw cycles, which can preserve the viability of cells contained in CSF. The CSF sample was thawed in the ice immediately before the experiment. 50 μL CSF was loaded into the device’s multiple loading chambers, and incubated on 4°C for 30 minutes on a gentle shaker to maintain cell viability and facilitate the interaction of trafficking leukocytes with capture antibody-coated SiNW surface. During the incubation, trafficking leukocytes interacted with the silicon nanowire arrays functionalized with the capture antibodies, and they were strongly attracted and captured onto the SiNW surface if leukocytes express the specific surface antigens that induced high affinity interaction with the corresponding capture antibodies. After the experiment, the CSF supernatant was carefully removed with a pipette or a microfluidic pump. The capture regions surrounded by loading chambers were rinsed with PBS solution with 1% BSA to remove non-specific cells and other debris from the CSF sample. Because it is generally considered unethical to perform lumbar puncture for acquisition of the CSF without a clinical indication of malignancy, our multiplexed phenotyping analysis of AD patients’ CSF was compared with the reference values of the CSF from healthy subjects67.
Fixation and staining of captured cells and trafficking leukocytes
Captured leukocytes were fixed by applying 50 μL of 4% PFA in PBS into each PDMS loading reservoir and reacting for 20 minutes. The device was subsequently treated with a solution of 50 μL of 0.1% Triton X-100 in PBS for 10 minutes to increase cellular permeability and allow for intracellular staining (DAPI dye agent stains for cell’s nucleus). To identify and confirm the phenotype of the captured cells, 50 μL of fluorophore-conjugated antibody solution (1:100 dilution) against the surface antigens of specific target leukocyte subsets was introduced into the platform and reacted for 1 hour at room temperature. PE-conjugated CD11, APC-conjugated CD4, FITC-conjugated CD8, AlexaFluora 645-conjugated CD4, and PE-conjugated CD45 were used to label the captured cells with fluorescence. After incubating with detection fluorescent antibodies, the chip was carefully rinsed with PBS for three times.
Scanning electron microscopy analysis for characterization of the leukocytes bound on the SiNW substrates
The immobilized trafficking leukocytes on SiNW substrates were fixed with 4% GA in 4°C for 2 hours and post-fixed in 1% osmium tetroxide for 2 h. The cells were dehydrated by successive immersion in 25%, 50%, 75%, 95%, and 100% ethanol for 5 minutes, and slowly dried under vacuum for 24 h. To make samples conductive before the FE-SEM measurements, the surface-bound leukocytes were then sputter coated with a thick layer of platinum (~5–6 nm).
Detection, immunophenotyping, and quantitative analysis of CSF leukocytes using laser scanning cytometry
We employed a rapid laser scanning cytometry (LSC) method to detect and quantify all the captured leukocytes on the entire substrate surface in a rapid and fully automated fashion. An Axon Genepix microarray laser scanner 4000B (Molecular Devices, USA) was used to acquire the fluorescent images of the CSF leukocytes captured by the device. The microarray laser scanner equipped with three laser beams (635, 532, and 488 nm) was used to scan and visualize the substrate with 5 μm resolution. The leukocytes stained with PE-, APC-, and FITC-conjugated antibodies were detected by the 532 nm laser (green channel), 635 nm laser (red channel), and 488 nm laser (blue channel), respectively. The fluorescent images were exported from the Genepix 6.0 and transported into CellProfiler™, the cell image analysis software. We developed our own software algorithm that allowed for rapid identification and enumeration of potential leukocytes. Color of fluorescent emission, fluorescence intensity (brightness), and physical characteristics such as cell size, shape, and circularity were considered in developing algorithm to identify target CSF leukocytes and exclude cell debris and other non-specific objects. Cells that stained positive for CD11 and met the morphological characteristics of typical human leukocytes were identified as trafficking leukocytes. The proportion of antigen-expressing leukocyte subsets could be calculated by enumerating the cells captured on each distinct location functionalized with specific antibody and dividing the count by total number of loaded cells.
Supplementary Material
Acknowledgments
This study was supported by the Alzheimer Association New Investigator Grant (PI: R.F.), the U.S. National Cancer Institute Howard Temin Pathway to Independence Award (NIH 4R00 CA136759-02, PI: R.F.), the grant for the Single Cell Profiling Core supported by the NIH grant U54CA143798 (subaward PI: R.F.), and the Priority Research Centers Program and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A12012685, PI: S.K.L.). We also acknowledge the Yale Institute for Nanoscience and Quantum Engineering (YINQE) and the Yale Nanofabricaiton Center to allow us to use their facilities.
Footnotes
Supplementary material. Additional data are available in the supplementary information, supplementary tables, and supplementary figures, which are available online.
References
- 1.Rossi B, Angiari S, Zenaro E, Budui SL, Constantin G. J Leukoc Biol. 2011;89:539–56. doi: 10.1189/jlb.0710432. [DOI] [PubMed] [Google Scholar]
- 2.Uchihara T, Akiyama H, Kondo H, Ikeda K. Stroke. 1997;28:1948–1950. doi: 10.1161/01.str.28.10.1948. [DOI] [PubMed] [Google Scholar]
- 3.De Leon MJ, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Segal S, Clark C, Kerkman D, DeBernardis J, Li J, Lair L, Reisberg B, Tsui W, Rusinek H. J Intern Med. 2004;256:205–223. doi: 10.1111/j.1365-2796.2004.01381.x. [DOI] [PubMed] [Google Scholar]
- 4.Togo T, Akiyama H, Iseki E, Kondo H, Ikeda K, Kato M, Oda T, Tsuchiya K, Kosaka K. J Neuroimmunol. 2002;124:83–92. doi: 10.1016/s0165-5728(01)00496-9. [DOI] [PubMed] [Google Scholar]
- 5.Neumann H, Medana IM, Bauer J, Lassmann H. Trends Neurosci. 2002;25:313–9. doi: 10.1016/s0166-2236(02)02154-9. [DOI] [PubMed] [Google Scholar]
- 6.Maxeiner HG, Rojewski MT, Schmitt A, Tumani H, Bechter K, Schmitt M. Brain Behav Immun. 2009;23:134–142. doi: 10.1016/j.bbi.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Brochard V, Combadière B, Prigent A, Laouar Y, Perrin A, Beray-berthat V, Bonduelle O, Alvarez-fischer D, Callebert J, Launay J, Duyckaerts C, Flavell RA, Hirsch EC, Hunot S. 2009:119. doi: 10.1172/JCI36470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rezai-Zadeh K, Gate D, Town T. J Neuroimmune Pharmacol. 2009;4:462–75. doi: 10.1007/s11481-009-9166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ransohoff RM, Kivisäkk P, Kidd G. Nat Rev Immunol. 2003;3:569–81. doi: 10.1038/nri1130. [DOI] [PubMed] [Google Scholar]
- 10.Matsui M, Mori KJ, Saida T, Akiguchi I, Kameyama M. Acta Neurol Scand. 1988;77:202–209. doi: 10.1111/j.1600-0404.1988.tb05895.x. [DOI] [PubMed] [Google Scholar]
- 11.Svenningsson A, Hansson GK, Andersen O, Andersson R, Patarroyo M, Stemme S. Ann Neurol. 1993;34:155–161. doi: 10.1002/ana.410340210. [DOI] [PubMed] [Google Scholar]
- 12.Kivisäkk P, Tucky B, Wei T, Campbell JJ, Ransohoff RM. BMC Immunol. 2006;7:14. doi: 10.1186/1471-2172-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelhardt B, Ransohoff RM. Trends Immunol. 2005;26:485–495. doi: 10.1016/j.it.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Svenningsson a, Andersen O, Edsbagge M, Stemme S. J Neuroimmunol. 1995;63:39–46. doi: 10.1016/0165-5728(95)00126-3. [DOI] [PubMed] [Google Scholar]
- 15.Del Maschio a, De Luigi a, Martin-Padura I, Brockhaus M, Bartfai T, Fruscella P, Adorini L, Martino G, Furlan R, De Simoni MG, Dejana E. J Exp Med. 1999;190:1351–6. doi: 10.1084/jem.190.9.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas J, Schopp L, Storch-Hagenlocher B, Fritzsching B, Jacobi C, Milkova L, Fritz B, Schwarz A, Suri-Payer E, Hensel M, Wildemann B. Blood. 2008;111:761–6. doi: 10.1182/blood-2007-08-104877. [DOI] [PubMed] [Google Scholar]
- 17.Weiner HL, Frenkel D. Nat Rev Immunol. 2006;6:404–16. doi: 10.1038/nri1843. [DOI] [PubMed] [Google Scholar]
- 18.Fisher Y, Nemirovsky A, Baron R, Monsonego A. PLoS One. 2010;5:e10830. doi: 10.1371/journal.pone.0010830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleine TO, Benes L. Cytom Part A. 2006;69A:147–151. doi: 10.1002/cyto.a.20225. [DOI] [PubMed] [Google Scholar]
- 20.Belov L. Cancer Res. 2001;61:4483. [PubMed] [Google Scholar]
- 21.Chattopadhyay PK, Price Da, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup Ra, De Rosa SC, Bruchez MP, Roederer M. Nat Med. 2006;12:972–7. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 22.Perfetto SP, Chattopadhyay PK, Roederer M. Nat Rev Immunol. 2004;4:648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 23.De Rosa SC, Herzenberg La, Roederer M. Nat Med. 2001;7:245–8. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 24.Bergeron M, Faucher S, Ding T, Phaneuf S, Mandy F. Cytometry. 2002;50:62–68. doi: 10.1002/cyto.10089. [DOI] [PubMed] [Google Scholar]
- 25.Brando B, Barnett D, Janossy G, Mandy F, Autran B, Rothe G, Scarpati B, D’Avanzo G, D’Hautcourt JL, Lenkei R, Schmitz G, Kunkl A, Chianese R, Papa S, Gratama JW. Cytometry. 2000;42:327–346. doi: 10.1002/1097-0320(20001215)42:6<327::aid-cyto1000>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 26.Storie I, Sawle A, Goodfellow K, Whitby L, Granger V, Ward RY, Peel J, Smart T, Reilly JT, Barnett D. Cytom Part B Clin Cytom. 2004;57B:47–52. doi: 10.1002/cyto.b.10065. [DOI] [PubMed] [Google Scholar]
- 27.Craig FE, Foon Ka. Blood. 2008;111:3941–67. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 28.French CA, Dorfman DM, Shaheen G, Cibas ES. Diagn Cytopathol. 2000;23:369–374. doi: 10.1002/1097-0339(200012)23:6<369::aid-dc1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 29.de Graaf MT, de Jongste AHC, Kraan J, Boonstra JG, Smitt PAES, Gratama JW. Cytom Part B Clin Cytom. 2011;80B:271–281. doi: 10.1002/cyto.b.20603. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Macal M, Jones CN, George MD, Dandekar S, Revzin A. Anal Chim Acta. 2008;608:186–96. doi: 10.1016/j.aca.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 31.Kwong GA, Radu CG, Hwang K, Shu CJ, Ma C, Koya RC, Comin-Anduix B, Hadrup SR, Bailey RC, Witte ON, Schumacher TN, Ribas A, Heath JR. J Am Chem Soc. 2009;131:9695–9703. doi: 10.1021/ja9006707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S. Adv Mater. 2011;23:4376–80. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew Chemie Int Ed. 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng HR. Angew Chemie Int Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M, Han L, Hyung JH, Seol JK, Sander C, Gonzalez A, Li J, Fan R. Nano Lett. 2012;12:2697–704. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DJ, Seol JK, Wu Y, Ji S, Kim GS, Hyung JH, Lee SY, Lim H, Fan R, Lee SK. Nanoscale. 2012;4:2500–7. doi: 10.1039/c2nr11338d. [DOI] [PubMed] [Google Scholar]
- 37.Kim ST, Kim DJ, Kim TJ, Seo DW, Kim TH, Lee SY, Kim K, Lee KM, Lee SK. Nano Lett. 2010;10:2877–83. doi: 10.1021/nl100942p. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Chen L, Liu H, Yang G, Zhang P, Han D, Wang S, Jiang L. NPG Asia Mater. 2013;5:e63. [Google Scholar]
- 39.Kim DJ, Seol JK, Wu Y, Ji S, Kim GS, Hyung JH, Lee SY, Lim H, Fan R, Lee SK. Nanoscale. 2012;4:2500–2507. doi: 10.1039/c2nr11338d. [DOI] [PubMed] [Google Scholar]
- 40.Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M, Han L, Hyung JH, Seol JK, Sander C, Gonzalez A, Li J, Fan R. Nano Lett. 2012;12:2697–704. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SK, Kim DJ, Lee G, Kim GS, Kwak M, Fan R. Biosens Bioelectron. 2014;54:181–188. doi: 10.1016/j.bios.2013.10.048. [DOI] [PubMed] [Google Scholar]
- 42.Kim DJ, Seol JK, Lee G, Kim GS, Lee SK. Nanotechnology. 2012;23:395102. doi: 10.1088/0957-4484/23/39/395102. [DOI] [PubMed] [Google Scholar]
- 43.Kim DJ, Kim GS, Hyung JH, Lee WY, Hong CH, Lee SK. Nanoscale Res Lett. 2013;8:332. doi: 10.1186/1556-276X-8-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang W-H, He R, Zhao X-Z, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng H-R. Angew Chemie Int Ed. 2013;52:3379–3383. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao L, Lu Y-T, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang W-H, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LWK, Rettig M, Tseng H-R, Posadas EM. Adv Mater. 2013;25:2897–2902. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranella A, Barberoglou M, Bakogianni S, Fotakis C, Stratakis E. Acta Biomater. 2010;6:2711–20. doi: 10.1016/j.actbio.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 47.Fischer KE, Alemán BJ, Tao SL, Daniels RH, Li EM, Bünger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano Lett. 2009;9:716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lea P, Keystone E, Mudumba S, Kahama A, Ding SF, Hansen J, Azad A, Wang S, Weber D. Clin Rev Allergy Immunol. 2011;41:20–35. doi: 10.1007/s12016-009-8189-z. [DOI] [PubMed] [Google Scholar]
- 49.Fung E, Esposito L, Todd Ja, Wicker LS. Nat Protoc. 2010;5:357–70. doi: 10.1038/nprot.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Y, Xu F, Qin Q, Fung WY, Lu W. Nano Lett. 2009;9:3934–3939. doi: 10.1021/nl902132w. [DOI] [PubMed] [Google Scholar]
- 51.Kääb MJ, Richards RG, Walther P, Gwynn I, Nötzli HP. 1999;13:61–69. [Google Scholar]
- 52.Zhou J, Wu Y, Lee SK, Fan R. Lab Chip. 2012;12:5025–33. doi: 10.1039/c2lc40309a. [DOI] [PubMed] [Google Scholar]
- 53.Belov L, Huang P, Barber N, Mulligan SP, Christopherson RI. Proteomics. 2003;3:2147–2154. doi: 10.1002/pmic.200300599. [DOI] [PubMed] [Google Scholar]
- 54.Harnett M. Nat Rev Immunol. 2007;7:897–904. doi: 10.1038/nri2188. [DOI] [PubMed] [Google Scholar]
- 55.Edwards BS, Oprea T, Prossnitz ER, Sklar La. Curr Opin Chem Biol. 2004;8:392–8. doi: 10.1016/j.cbpa.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Pranzatelli MR, Travelstead aL, Tate ED, Allison TJ, Moticka EJ, Franz DN, Nigro Ma, Parke JT, Stumpf Da, Verhulst SJ. Neurology. 2004;62:1526–1532. doi: 10.1212/wnl.62.9.1526. [DOI] [PubMed] [Google Scholar]
- 57.Baba Y, Kuroiwa A, Uitti RJ, Wszolek ZK, Yamada T. Parkinsonism Relat Disord. 2005;11:493–498. doi: 10.1016/j.parkreldis.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Cepok S. Brain. 2001;124:2169–2176. doi: 10.1093/brain/124.11.2169. [DOI] [PubMed] [Google Scholar]
- 59.Spudich SS, Nilsson AC, Lollo ND, Liegler TJ, Petropoulos CJ, Deeks SG, Paxinos EE, Price RW. BMC Infect Dis. 2005;5:98. doi: 10.1186/1471-2334-5-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahad DJ, Callahan MK, Trebst C, Tucky B, Wei T, Wu L, Baekkevold ES, Lassmann H, Staugaitis SM, Campbell JJ, Ransohoff RM. 2003;100:4–9. doi: 10.1073/pnas.1433000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frohman EM, Frohman TC, Gupta S, de Fougerolles A, van den Noort S. J Neurol Sci. 1991;106:105–111. doi: 10.1016/0022-510x(91)90202-i. [DOI] [PubMed] [Google Scholar]
- 62.Neuenburg JK. J Acquir Immune Defic Syndr. 2005;39:16. doi: 10.1097/01.qai.0000155036.03004.a0. [DOI] [PubMed] [Google Scholar]
- 63.Andrew DP, Spellberg JP, Takimoto H, Schmits R, Mak TW, Zukowski MM. Eur J Immunol. 1998;28:1959–1969. doi: 10.1002/(SICI)1521-4141(199806)28:06<1959::AID-IMMU1959>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 64.Craig FE, Ohori NP, Gorrill TS, Swerdlow SH. Am J Clin Pathol. 2011;135:22–34. doi: 10.1309/AJCPANA7ER1ABMZI. [DOI] [PubMed] [Google Scholar]
- 65.Yang X, Askarova S, Lee JM. Mol Neurobiol. 2010;41:138–148. doi: 10.1007/s12035-010-8121-9. [DOI] [PubMed] [Google Scholar]
- 66.Giunti D, Borsellino G, Benelli R, Marchese M, Capello E, Valle MT, Pedemonte E, Noonan D, Albini A, Bernardi G, Mancardi GL, Battistini L, Uccelli A. 2003 doi: 10.1189/jlb.1202598. [DOI] [PubMed] [Google Scholar]
- 67.Craig FE, Ohori NP, Gorrill TS, Swerdlow SH. Am J Clin Pathol. 2011;135:22–34. doi: 10.1309/AJCPANA7ER1ABMZI. [DOI] [PubMed] [Google Scholar]
- 68.Craig FE, Foon KA. Blood. 2008;111:3941–3967. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 69.de Graaf MT, Smitt PAES, Luitwieler RL, van Velzen C, van den Broek PDM, Kraan J, Gratama JW. Cytom Part B Clin Cytom. 2011;80B:43–50. doi: 10.1002/cyto.b.20542. [DOI] [PubMed] [Google Scholar]
- 70.Town T, Tan J, Flavell RA, Mullan M. 2005;7:255–264. doi: 10.1385/NMM:7:3:255. [DOI] [PubMed] [Google Scholar]
- 71.Shi N, Kawano Y, Matsuoka T, Mei F, Ishizu T, Ohyagi Y, Kira J. Mult Scler. 2009;15:120–3. doi: 10.1177/1352458508096871. [DOI] [PubMed] [Google Scholar]
- 72.Özenci V, Kouwenhoven M, Huang YM, Kivisäkk P, Link H. Clin Exp Immunol. 2000;120:147–153. doi: 10.1046/j.1365-2249.2000.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selmaj K, Raine CS, Cross AH. Ann Neurol. 1991;30:694–700. doi: 10.1002/ana.410300510. [DOI] [PubMed] [Google Scholar]
- 74.Itagaki S, McGeer PL, Akiyama H. Neurosci Lett. 1988;91:259–264. doi: 10.1016/0304-3940(88)90690-8. [DOI] [PubMed] [Google Scholar]
- 75.Melzer N, Meuth SG, Wiendl H. FASEB J. 2009;23:3659–73. doi: 10.1096/fj.09-136200. [DOI] [PubMed] [Google Scholar]
- 76.Zlokovic B. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 77.Jesse S, Brettschneider J, Süssmuth S, Landwehrmeyer B, Arnim CF, Ludolph A, Tumani H, Otto M. J Neurol. 2011;258:1034–1041. doi: 10.1007/s00415-010-5876-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen DS, Soen Y, Stuge TB, Lee PP, Weber JS, Brown PO, Davis MM. PLoS Med. 2005;2:e265. doi: 10.1371/journal.pmed.0020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W. Anal Chem. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W. Anal Chem. 2008;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng K, Wang Xin, Lee Shuit-Tong. Appl Phys Lett. 2008;92:163103. [Google Scholar]
- 82.Kim DJ, Seol JK, Lee MR, Hyung JH, Kim GS, Ohgai T, Lee SK. Appl Phys Lett. 2012;100:163703. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.