Abstract
Papillary urothelial neoplasia of low malignant potential (PUNLMP) recurs in approximately 35% of patients. Conventional histopathological assessment does not distinguish non-recurrent from recurrent PUNLMP. The aim of the study was to explore the differences in global histone acetylation and global DNA methylation between non-recurrent and recurrent PUNLMP. Acetylated histone H3 lysine 9 (AcH3K9) and 5-methylcytosine (5MeC) were investigated by immunohistochemistry (IHC) in 20 PUNLMP cases (10 non-recurrent and 10 recurrent), in 5 cases of normal urothelium (NU) and in 5 cases of muscle invasive pT2 urothelial carcinoma (UC). The total optical density of the nuclear staining was measured photometrically in at least 40 nuclei separately for the basal, intermediate and luminal positions in each case. Concerning the total optical density values for both acetylation and methylation, a decrease in staining is observed from non-recurrent PUNLMP to recurrent PUNLMP, at all nuclear locations. For acetylation the mean value in non-recurrent. PUNLMP, intermediate between NU and UC, is closer to the former than to latter. The mean value in recurrent PUNLMP is closer to UC than to NU. In NU, non-recurrent and recurrent PUNLMP the acetylation to methylation ratio decreased from the nuclei in basal position to those in the surface, the average for the above groups being 1.491, 1.611 and 1.746, respectively. Setting the observed values for NU at each sampling location to unity, acetylation shows a steady decrease, the percentages of changes in this nuclear location compared to NU being − 5% in non-recurrent PUNLMP, − 15% in recurrent PUNLMP and − 24% in UC. Concerning methylation, there is slight increase in non-recurrent PUNLMP (+ 5%), a decrease in recurrent PUNLMP (− 19%) followed by a sharp rise for the UC (+ 61%). In conclusion there are differences in global histone acetylation and DNA methylation patterns between non-recurrent and recurrent PUNLMP. Further studies are needed to elucidate the complex interplay between chromatin structure, its modifications and recurrence of PUNLMP.
Keywords: Urothelium, papillary urothelial neoplasm of low malignant potential, recurrence, global histone acetylation, global DNA methylation
INTRODUCTION
The 2004 WHO classification of the non-invasive papillary urothelial tumors subdivides the morphologic spectrum of the non-invasive urothelial papillary neoplasia into urothelial papilloma, papillary urothelial neoplasm of low malignant potential (PUNLMP), low-grade papillary carcinoma, and high-grade papillary carcinoma (1). It replaces the 1973 WHO system which included urothelial papilloma, and papillary carcinoma of grade 1, grade 2 and grade 3.
PUNLMP is a papillary urothelial lesion with an orderly arrangement of cells with minimal architectural abnormalities and minimal nuclear atypia (1). PUNLMP recurs in approximately 35% of patients (2). Conventional histopathological assessment does not identify those PUNLMP cases that will recur. Karyometry detects subvisual differences in chromatin organization status between non-recurrent and recurrent PUNLMP, and provides a valuable biomarker for the prediction of recurrence (3).
The status of chromatin organization depends on epigenetic events, such as histone modifications and DNA methylation (4–6). Two copies of histones H2A, H2B, H3 and H4, assembled in an octameric core with 146–147 bp of DNA tightly wrapped around it, form a nucleosome. This is the first level of chromatin organization (5). Core histones are characterized by the presence of N-terminal tails of variable length that are reversibly modified by acetylation of lysines and methylation of lysines and argynines among others (6–11). In particular, acetylation of specific residues such as lysine 9 (K9) of histone 3 (H3K9) has been associated with an open chromatin configuration and a permissive transcription state (6).
Methylation of DNA, which occurs at the cytosine residues of cytosine-phospho-guanine (CpG) dinucleotides by an enzymatic reaction that produces 5-methylcytosine (5MeC), is a well characterized mechanism for epigenetic gene regulation (4,6). Normal and neoplastic cells may simultaneously harbor widespread (global) genomic hypomethylation, regional areas of hypermethylation, and increased DNA-methyltransferase activity (4).
In two previous immunohistochemical studies, in which our group was involved, antibodies raised against acetylated H3K9 (AcH3K9) and 5MeC allowed evaluation of the overall epigenetic status, i.e., global histone acetylation and global DNA methylation, in pre-neoplastic and neoplastic lesions of the prostate (6) and in urothelial papillary neoplasms (6). Global histone acetylation and overall DNA methylation were investigated immunohistochemically by others in oral squamous cell carcinoma and its precursors, thyroid neoplasia, lung cancer, and colon cancer (12–25).
The present immunohistochemistry- and photometry-based feasibility study was designed to quantify global histone acetylation and DNA methylation in non-recurrent and recurrent PUNLMP. Similarly to our previous investigations, antibodies against AcH3K9 and 5MeC were used (26).
MATERIAL AND METHODS
Twenty cases of PUNLMP were retrieved from the Pathology Services associated with Ancona Polytechnic University-United Hospitals. All the cases were diagnosed by one of our group (RM). Ten were from patients who had a solitary lesion, less than 1 cm in diameter, and were disease-free during a follow-up period of at least 10 years. This group was defined as “non-recurrent”. The other ten were from patients with a unifocal lesion, less than 1 cm in diameter, who experienced one or more recurrences in the follow-up (in most of the cases the first recurrence was seen six months to one year after the removal of the primary tumor). This group was defined as “recurrent”. The recurrent lesions showed a histological appearance identical to that seen in the first presentation, i.e., none of the these cases progressed to a higher grade and/or became invasive. From this group only the primary or initial tumors were included in the investigation.
The initial tumors and the recurrences were treated by trans-urethral resection of the bladder. None of the patients received adjuvant therapy, e.g., BCG or intravesical chemotherapy. As far as sex and age of the patients (their mean age was 61.5 years) were concerned, there were no differences when the non-recurrent and recurrent groups were compared.
The study also included 5 cases of NU obtained from patients with benign prostatic hyperplasia and no history of bladder and prostate cancer and 5 cases of bladder muscle invasive (pT2) UC. The procedure for this research project conforms to the provisions of the Declaration of Helsinki.
All the cases had been fixed in 4% buffered formaldehyde for approximately 24 hours before processing. For the purpose of this study, serial five-micron thick serial sections were cut from the paraffin blocks.
Immunohistochemistry
The sections were dewaxed in xylene and rehydrated through a graded series of ethanol. Antigen retrieval was done by microwave treatment for 20 min at 98°C using 0.01 M Citric Acid buffer pH 6.0. Endogenous peroxidase activity was quenched by incubating the sections in 3% hydrogen peroxide for 10 min at room temperature. Non-specific binding sites were blocked through pre-incubation with 5% normal goat serum in PBS for 10 min at room temperature. Reacted tissue sections were then incubated with a rabbit polyclonal antibody raised against AcH3K9 (Cell Signalling Technology Laboratories, Danvers, MA, USA, dilution 1:100) for 18 hours at 4°C and a mouse monoclonal antibody raised against 5MeC (Aviva Systems Biology, San Diego, CA, USA, dilution 1:200) for 1 hour at room temperature Antigen-antibody complex was subsequently visualized using the Envision™ Detection System kit peroxidase/DAB (DAKO, Glustrop, Denmark) and counterstained with hematoxylin. Negative controls were used for the tested antibodies; the primary antibody was replaced by either mouse or rabbit non-immune serum, as appropriate.
Evaluation of Immunohistochemistry
The total optical density (reported in arbitrary, relative units) of the nuclear staining was measured photometrically at the Arizona Cancer Center, Tucson, AZ. For the NU and PUNLMP cases the nuclei were separately evaluated in the following three compartments: cell layer adjacent to the stroma, i.e., basal cells; superficial or luminal cell layer; and intermediate cells, i.e., those between the basal cells and superficial cells. Since superficial cells are not present in UC, the nuclei were evaluated in the cell layer adjacent to the stroma, the nuclei in all the other cell layers being considered equivalent to the intermediate nuclei of NU and PUNLMP. At least 40 nuclei per location were measured in each case. Details of the statistical analysis applied in this study are reported in two our previous papers (27,28).
RESULTS
Total optical density values for acetylation and methylation
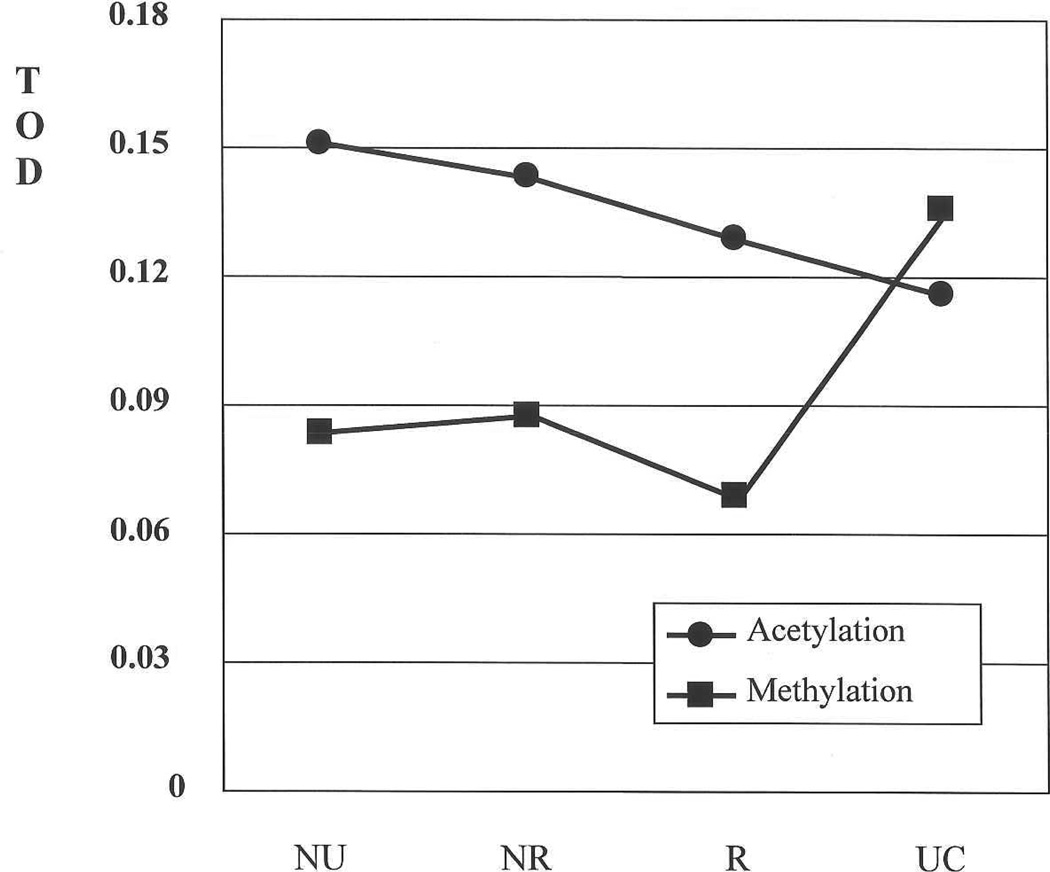
Table I presents the mean total optical density (TOD) values for acetylation and methylation (Figures 1 A and 1B). Figure 2, based on the values of the first column of Table I, shows graphically the changes for the nuclei in basal position for both markers. Acetylation decreases steadily from NU to UC. There is a steep rise in methylation in UC, into the range of the acetylation level in NU.
Table I.
Mean total optical density values for acetylation and methylation
| Nuclear location | |||
|---|---|---|---|
| basal | intermediate | superficial | |
| Acetylation | |||
| NU | 0.1510 | 0.1413 | 0.1351 |
| Non-recurrent PUNLMP | 0.1431 | 0.1645 | 0.1684 |
| Recurrent PUNLMP | 0.1283 | 0.1379 | 0.1435 |
| UC | 0.1154 | 0.1339 | NA* |
| Methylation | |||
| NU | 0.0835 | 0.0926 | 0.1185 |
| Non-recurrent PUNLMP | 0.0874 | 0.0970 | 0.1181 |
| Recurrent PUNLMP | 0.0679 | 0.0782 | 0.0904 |
| UC | 0.1349 | 0.1339 | NA* |
NA, not applicable. Superficial cells are not present in UC.
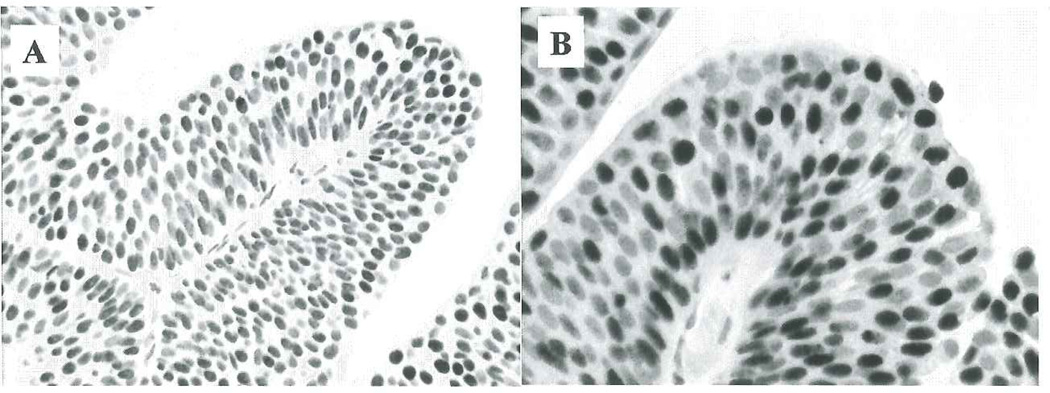
Figure 1.
Nuclear staining for acetylation (A) and methylation (B) in papillary urothelial neoplasia of low malignant potential (Note that the nuclei vary in staining intensity).
Figure 2.
Total optical density (TOD) values. Changes for the nuclei in basal position for acetylation and methylation.
For both acetylation and methylation, a decrease in staining is observed from non-recurrent PUNLMP to recurrent PUNLMP, at all nuclear locations. For acetylation the mean value in non-recurrent PUNLMP, intermediate between NU and UC, is closer to the former than to latter. The mean value in recurrent PUNLMP is closer to UC than to NU. For the measurements taken at the basal, intermediate and superficial locations the decreases are 10%, 18% and 19%, respectively. For methylation the decreases are 24%, 20% and 23%, respectively.
Taking as example the intermediate location of acetylation, the mean values are 0.1645 for non-recurrent PUNLMP vs. 0.1379 for recurrent PUNLMP. The standard deviation is 0.0787 for non-recurrent PUNLMP and 0.0632 for recurrent PUNLMP. For sample sizes of 497 vs. 403 nuclei, the 95 % confidence limits become 0.16381 < 0.1645 < 0.16519 for non-recurrent PUNLMP and 0.1318 < 0.138 < 0.1442 for recurrent PUNLMP, the differences being statistically significant. For the methylation we have, at this nuclear location, 0.0970 for non-recurrent PUNLMP and 0.0782 for recurrent PUNLMP. The confidence limits overlap, i.e. there are no statistically significant differences. The tolerance regions for the value distributions for recurrent PUNLMP and recurrent PUNLMP cases overlap widely. This means that the differences are too small to allow a prediction for an individual case.
Acetylation to methylation ratio
Table II shows the acetylation to methylation ratio (A/M ratio) for the four diagnostic groups in the three nuclear locations. It is based on the assumption of similar or equal stoechiometry for the acetylation and the methylation markers. In NU, non-recurrent and recurrent PUNLMP, the A/M ratio decreases from the nuclei in the basal position to those in the surface. The average for the above groups are 1.491, 1.611 and 1.746, respectively. For UC the A/M is 0.855 for the nuclei in the basal position and 1.0 for the nuclei in the intermediate position, with an average of 0.927. The low value of 0.927 is due to a notable increase in methylation.
Table II.
Acetylation to methylation ratio
| Nuclear locations | ||||
|---|---|---|---|---|
| Diagnostic groups | basal | intermediate | superficial | average |
| NU | 1.808 | 1.525 | 1.140 | 1.491 |
| Non-recurrent PUNLMP | 1.713 | 1.696 | 1.425 | 1.611 |
| Recurrent PUNLMP | 1.889 | 1.763 | 1.587 | 1.746 |
| UC | 0.855 | 1.0 | NA* | 0.927 |
See footnote of Table 1
Comparison of the relative changes of acetylation and methylation
To compare the relative changes of acetylation and methylation directly a normalization is useful.
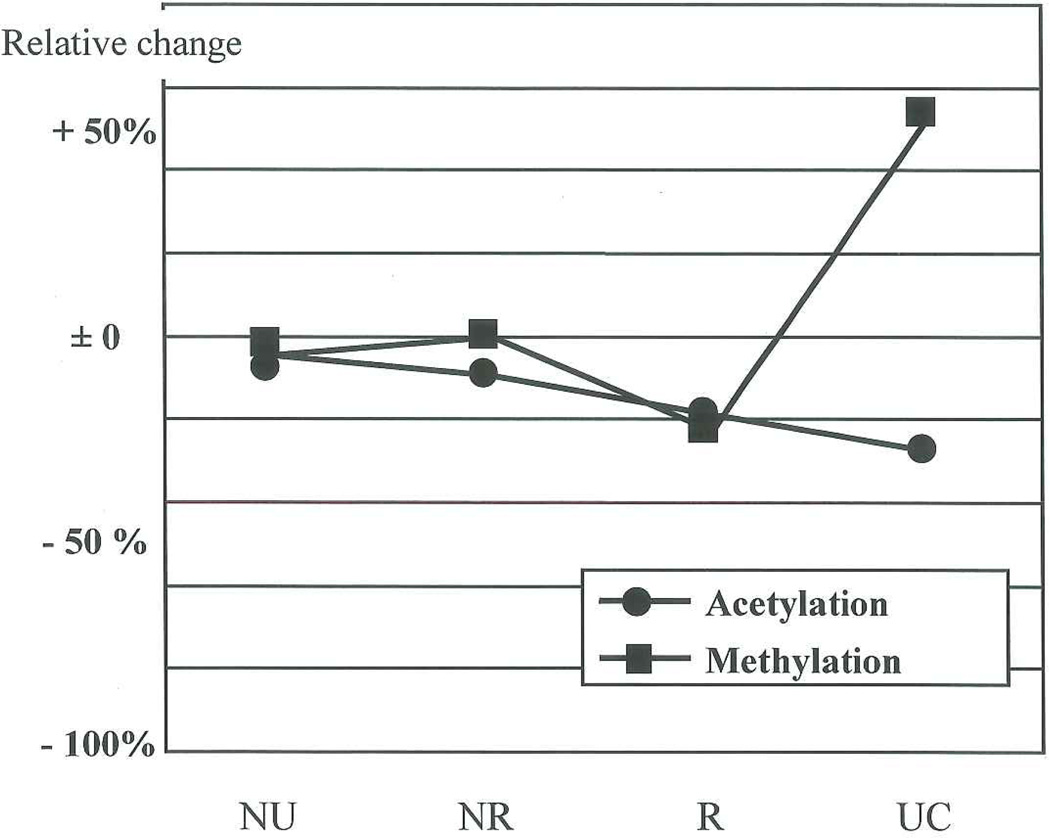
Setting the observed values (see Table I) for NU at each sampling location to unity, the normalized data are shown in Table III. This table also reports the percentages of changes in non-recurrent and recurrent PUNLMP and UC in relation to NU. Figure 3 shows graphically the relative change for the nuclei in basal position. Acetylation shows a steady decrease, the percentages of changes in this nuclear location compared to NU, being − 5% in non-recurrent PUNLMP, − 15% in recurrent PUNLMP and − 24% in UC. Concerning methylation, there is slight increase in non-recurrent PUNLMP (+ 5%), a decrease in recurrent PUNLMP (− 19%) followed by a sharp rise for the UC (+ 61%).
Table III.
Relative changes separately for acetylation and methylation (The values in parentheses indicate the percentages of changes in non-recurrent and recurrent PUNLMP and UC in relation to NU)
| Diagnostic groups | ||||
|---|---|---|---|---|
| NU | NR PUNLMP | R PUNLMP | UC | |
| Nuclei in basal position | ||||
| Acetylation | 100% | 95% (− 5%) | 85% (− 15%) | 76% (− 24%) |
| Methylation | 100% | 105% (+ 5%) | 81% (− 19%) | 161% (+ 61%) |
| Nuclei in intermediate position | ||||
| Acetylation | 100% | 116% (+ 16%) | 98% (− 2%) | 99% (− 1%) |
| Methylation | 100% | 105% (+ 5%) | 85% (− 15%) | 141% (+ 41%) |
| Nuclei in superficial position | ||||
| Acetylation | 100% | 125% (+ 25%) | 106% (+ 6%) | NA* |
| Methylation | 100% | 99% (− 1%) | 76% (− 24%) | NA* |
See footnote of Table 1
Figure 3.
Relative changes of acetylation and methylation for the nuclei in basal position. Percentages of changes in non-recurrent and recurrent PUNLMP and UC in relation to NU.
Setting the observed values (see Table I) for the basal location in the four diagnostic groups to unity, Table IV shows the percentages of changes for acetylation and methylation in the intermediate and superficial nuclear location. For acetylation a decrease is seen in both nuclear locations in NU (intermediate − 7%; superficial − 10%), whereas a steady increase is seen in non-recurrent PUNLMP (intermediate + 15%; superficial + 18%) and in recurrent PUNLMP (intermediate + 7%; superficial + 12%) as well as in UC (intermediate + 12%). For methylation the increase in the superficial location is greater than in the intermediate location in NU (intermediate + 10%; superficial + 42%), non-recurrent PUNLMP (intermediate + 11%; superficial + 13%) and recurrent PUNLMP (intermediate + 15%; superficial + 33%), whereas a small reduction is seen in the intermediate position in UC (− 3.2%), compared to the basal position.
Table IV.
Relative changes in immunohistochemical staining for acetylation and methylation in the intermediate and superficial epithelium, as a function of diagnostic category (The values in parentheses indicate the percentages of changes)
| Nuclear location | |||
|---|---|---|---|
| basal | intermediate | superficial | |
| Acetylation | |||
| NU | 100% | 93.6% (− 7%) | 89.5% (− 10%) |
| Non-recurrent PUNLMP | 100% | 114.9% (+ 15%) | 117.7% (+ 18%) |
| Recurrent PUNLMP | 100% | 107.4% (+ 7%) | 111.8% (+ 12%) |
| UC | 100% | 111.6% (+ 12%) | NA* |
| Methylation | |||
| NU | 100% | 110.8% (+ 10%) | 141.8% (+ 42%) |
| Non-recurrent PUNLMP | 100% | 111.0% (+ 11%) | 113.5% (+ 13 %) |
| Recurrent PUNLMP | 100% | 114.9% (+ 15%) | 132.9% (+ 33%) |
| UC | 100% | 96.8% (− 3.2%) | NA* |
See footnote of Table 1
DISCUSSION
Histones are subject to a variety of post-translational modifications, including acetylation of lysines. Such modifications play fundamental roles in gene regulation and other chromatin-based processes. Histone-modifying enzymes affect histones either locally, through targeted recruitment by sequence specific transcription factors (29), or globally throughout the genome in an untargeted manner, affecting virtually all nucleosomes (30). Such widespread functions that occur independently of apparent sequence-specific DNA binding proteins are referred to as “global histone modifications”. Like their targeted effects, the global activity of the histone modifying enzymes can modulate gene activity (30). Therefore, histones are modified locally and globally through multiple histone-modifying enzymes with different substrate specificities, generating hierarchical patterns of modifications from single promoters to large regions of chromosomes and even single cells.
Since histone modifications occur throughout the genome, any potential change in the activity of the histone modifying enzymes results in changes in specific histone patterns detectable at the level of individual nuclei by IHC. While the immunohistochemical approach provides information on global histone acetylation, it does not give information on the genomic, gene–gene differences in distribution of histone modifications. However, it reveals novel cell–cell heterogeneity in histone modification levels that would be hidden in molecular approaches such as chromatin immunoprecipitation (31).
Cancer epigenetics includes DNA methylation (32). Compared to normal cells, DNA of cancer cells is generally hypomethylated, while promoters of certain genes are hypermethylated, in the context of CpG islands. Such promoter-specific increase in methylation leads to silencing of the affected gene that might have functioned as, for instance, a tumor suppressor. Transcriptional repression by DNA methylation is mediated by a class of methyl DNA binding proteins which, by virtue of recognizing specifically methylated DNA sequences, recruit repressive protein complexes including histone deacetylases to gene promoters (33). The combination of CpG island methylation, proteins that binds to them, and repressive histone modifications generates localized regions of specialized chromatin, which can inhibit transcription. Despite a growing list of genes including tumor suppressors and DNA repair genes that are aberrantly hypermethylated in different cancers, only a limited number of the identified hypermethylated genes have demonstrated any potential utility in clinical decision making. As opposed to single-gene analysis, the integrated information on methylation patterns of multiple genes may reflect the functional status of several cellular pathways (4,33).
Global methylation of DNA is usually quantified by chromatography (34), whereas the methylation status of specific genes is studied by molecular hybridization or genomic sequencing (35). These techniques require extraction of DNA and do not allow the cell integrity to be preserved. An immunohistochemical approach was developed using monoclonal antibodies that recognize the presence of a methyl group on the carbon 5 of cytosine, to investigate DNA methylation is situ, which allowed the analysis of global methylation to be performed on interphase nuclei in several cell types, on a cell by cell basis by microscopy (4).
This study shows that it is feasible to quantify the change in global acetylation and methylation in the urinary bladder tissues as a function of diagnostic category, i.e., NU, non-recurrent PUNLMP, recurrent PUNLMP and UC, and as a function of sampling site, i.e., basal, intermediate and superficial cell layers. In particular, the current investigation shows that there are differences in global histone acetylation and DNA methylation patterns between non-recurrent and recurrent PUNLMP and at the same time helps to understand the chromatin organization status in non-recurrent and recurrent PUNLMP.
The limitation of our study is represented by the fact that there is no information available defining the stoechiometry of the immunohistochemical reactions for acetylation or for methylation. Neither is there information concerning linearity of the reactions. The assumption is made that the decadic molar coefficients of absorbance of the dye labels for the two probes is comparable, and not a function of concentration. The results suggest that the overall reproducibility of the photometric measurements was quite good. Under the assumption of equal stoechiometry for the two markers, the absolute measured values reflect the relationship of acetylation to methylation, its changes as a function of diagnostic category and as a function of nuclear location in the tissue.
In conclusion, the present study shows that there are differences in global histone acetylation and DNA methylation patterns between non-recurrent and recurrent PUNLMP in comparison with NU and UC. The above results represent averages over all cases in the study. For their interpretation of one should keep in mind that the values recorded for the individual nuclei are highly dispersed. Not only have the distributions of TOD values a very large coefficient of variation, there is also some heterogeneity in the nuclear populations. The limitation of the study is represented by the small number of cases investigated. We are planning to expand the study including a lager number of cases as well as an increased number of nuclear makers.
Acknowledgment
The study was supported by a Research grant by Fondazione G Berlucchi per la Ricerca sul Cancro, Brescia, Italy, and, in part, by a grant from the National Institutes of Health, NCI, Bethesda, MD, P01 CA - 27502, to David S. Alberts.
References
- 1.Sauter G, Algaba F, Amin MB, Busch C, Cheville J, Gasser T, Grignon DG, Hofstädter F, Lopez-Beltran A, Epstein JI. Tumours of the urinary system. In: Epstein T1, Eble JN, Sesterhenn I, Sauter G, editors. World Health Organization Classification of Tumours Pathology and Genetics: Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press; 2004. pp. 89–157. [Google Scholar]
- 2.Montironi R, Mazzucchelli R, Scarpelli M, Lopez-Beltran A, Cheng L. Morphological diagnosis of the urothelial neoplasms. J Clin Pathol. 2008;61:3–10. doi: 10.1136/jcp.2007.049312. [DOI] [PubMed] [Google Scholar]
- 3.Montironi R, Scarpelli M, Lopez-Beltran A, Mazzucchelli R, Alberts D, Ranger-Moore J, Bartels HG, Hamilton PW, Einspahr J, Bartels PH. Chromatin phenotype karyometry can predict recurrence in papillary urothelial neoplasms of low malignant potential. Cell Oncol. 2007;29:47–58. doi: 10.1155/2007/356464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herranz M, Esteller M. DNA methylation and histone modifications in patients with cancer: potential prognostic and therapeutic targets. Methods Mol Biol. 2007;361:25–62. doi: 10.1385/1-59745-208-4:25. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed MA, Greif PA, Diamond J, Sharaf O, Maxwell P, Montironi R, Young RA, Hamilton PW. Epigenetic events, remodelling enzymes and their relationship to chromatin organization in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. BJU Int. 2007;99:908–915. doi: 10.1111/j.1464-410X.2006.06704.x. [DOI] [PubMed] [Google Scholar]
- 7.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 8.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 9.Marino-Ramirez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev Proteomics. 2005;2:719–729. doi: 10.1586/14789450.2.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orr JA, Hamilton PW. Histone acetylation and chromatin pattern in cancer. A review. Anal Quant Cytol Histol. 2007;29:17–31. [PubMed] [Google Scholar]
- 11.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mel Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 13.Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, Epstein JI, Isaacs WB, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7:531–536. [PubMed] [Google Scholar]
- 14.Galusca B, Dumollard JM, Lassandre S, Niveleau A, Prades JM, Estour B, Peoc'h M. Global DNA methylation evaluation: potential complementary marker in differential diagnosis of thyroid neoplasia. Virchows Arch. 2005;447:18–23. doi: 10.1007/s00428-005-1268-5. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Blazquez FJ, Habib M, Dumollard JM, Barthelemy C, Benchaib M, de Capoa A, Niveleau A. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–693. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marte B. Cancer: a changing global view. Nature. 2005;435:1172. doi: 10.1038/4351172b. [DOI] [PubMed] [Google Scholar]
- 17.Piyathilake CJ, Bell WC, Jones J, Henao OL, Heimburger DC, Niveleau A, Grizzle WE. Patterns of global DNA and histone methylation appear to be similar in normal, dysplastic and neoplastic oral epithelium of humans. Dis Markers. 2005;21:147–151. doi: 10.1155/2005/285134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piyathilake CJ, Frost AR, Bell WC, Oelschlager D, Weiss H, Johanning GL, Niveleau A, Heimburger DC, Grizzle WE. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol. 2001;32:856–862. doi: 10.1053/hupa.2001.26471. [DOI] [PubMed] [Google Scholar]
- 19.Piyathilake CJ, Henao O, Frost AR, Macaluso M, Bell WC, Johanning GL, Heimburger DC, Niveleau A, Grizzle WE. Race- and age-dependent alterations in global methylation of DNA in squamous cell carcinoma of the lung (United States) Cancer Causes Control. 2003;14:37–42. doi: 10.1023/a:1022573630082. [DOI] [PubMed] [Google Scholar]
- 20.Piyathilake CJ, Johanning GL. Cellular vitamins, DNA methylation and cancer risk. J Nutr. 2002;132:2340S–3818S. doi: 10.1093/jn/132.8.2340S. [DOI] [PubMed] [Google Scholar]
- 21.Piyathilake CJ, Johanning GL, Frost AR, Whiteside MA, Manne U, Grizzle WE, Heimburger DC, Niveleau A. Immunohistochemical evaluation of global DNA methylation: comparison with in vitro radiolabeled methyl incorporation assay. Biotech Histochem. 2000;75:251–258. doi: 10.3109/10520290009085128. [DOI] [PubMed] [Google Scholar]
- 22.Qian DZ, Wei YF, Wang X, Kato Y, Cheng L, Pili R. Antitumor activity of the histone deacetylase inhibitor MS-275 in prostate cancer models. Prostate. 2007;67:1182–1193. doi: 10.1002/pros.20611. [DOI] [PubMed] [Google Scholar]
- 23.Rougier N, Bourchis D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 25.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 26.Sasco AJ, Rey F, Reynaud C, Bobin JY, Clavel M, Niveleau A. Breast cancer prognostic significance of some modified urinary nucleosides. Cancer Lett. 1996;108:157–162. doi: 10.1016/s0304-3835(96)04393-5. [DOI] [PubMed] [Google Scholar]
- 27.Barbisan F, Mazzucchelli R, Santinelli A, Stramazzotti D, Scarpelli M, Lopez-Beltran A, Cheng L, Montironi R. Immunohistochemical evaluation of global DNA methylation and histone acetylation in papillary urothelial neoplasm of low malignant potential. Int J Immunopathol Pharmacol. 2008;21:615–623. doi: 10.1177/039463200802100315. [DOI] [PubMed] [Google Scholar]
- 28.Montironi R, Scarpelli M, Lopez-Beltran A, Mazzucchelli R, Alberts D, Ranger-Moore J, Bartels HG, Hamilton PW, Einspahr J, Bartels PH. Chromatin phenotype karyometry can predict recurrence in papillary urothelial neoplasms of low malignant potential. Cell Oncol. 2007;29:47–58. doi: 10.1155/2007/356464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 30.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 31.Kurdistani SK. Histone modifications as markers of cancer prognosis: a cellular view. Br J Cancer. 2007;97:1–5. doi: 10.1038/sj.bjc.6603844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 33.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Gama-Sosa MA, Wang RY, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11:3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakkarainen M, Wahlfors J, Myöhänen S, Hiltunen MO, Eskelinen M, Johansson R, Jänne J. Hypermethylation of calcitonin gene regulatory sequences in human breast cancer as revealed by genomic sequencing. Int J Cancer. 1996;69:471–474. doi: 10.1002/(SICI)1097-0215(19961220)69:6<471::AID-IJC9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]