Abstract
Background
22q11.2 deletion syndrome (22q11DS) is the most common microdeletion syndrome in humans, characterised by cardiovascular defects such as interrupted aortic arch, outflow tract defects, thymus and parathyroid hypo- or aplasia and cleft palate. Heterozygosity of Tbx1, the mouse homologue of the candidate TBX1 gene, results in mild defects dependent on genetic background, whereas complete inactivation results in severe malformations in multiple tissues.
Results
The loss of function mutations in two Sprouty genes, which encode feedback antagonists of receptor tyrosine kinase (RTK) signaling, phenocopy many defects associated with the syndrome in the mouse. The stepwise reduction of Sprouty gene dosage resulted in different phenotypes emerging at specific steps, suggesting that the threshold up to which a given developmental process can tolerate increased RTK signaling is different. Tbx1 heterozygosity significantly exacerbated the severity of all these defects, which correlated with a substantial increase in RTK signaling.
Conclusions
Our findings suggest that TBX1 functions as an essential component of a mechanism that protects the embryo against perturbations in RTK signaling that may lead to developmental defects characteristic of 22q11.2 deletion syndrome. We propose that genetic factors that enhance RTK signalling ought to be considered as potential genetic modifiers of this syndrome.
Introduction
22q11.2 deletion syndrome (22q11DS), also referred to as DiGeorge syndrome/velocardio-facial syndrome (DGS/VCFS) is characterized by a range of defects, that include congenital heart disease, palate anomalies, hypocalcaemia, T cell mediated immunodeficiency, cognitive difficulties, and dysmorphic faces (Frank et al., 2002). Cardiovascular anomalies include interrupted aortic arch type B (IAA-B) and other aortic arch defects, persistant truncus arteriosus, tetralogy of Fallot and ventricular septal defects (VSD). Most 22q11DS features appear to be due to defects during development of a transient structure in the midgestation embryo, the pharyngeal apparatus (Baldini, 2005; Wurdak et al., 2006). Human genetic studies have shown that 22q11DS is most often associated with a 3Mb microdeletion at chromosome 22q11.2, occurring in approximately 1/4000 births, making it the most common microdeletion syndrome (Scambler, 2000; Botto et al., 2003; Kobrynski and Sullivan, 2007). Some 22q11DS patients have a smaller, nested distal deletion endpoint resulting in a 1.5 Mb deletion (McDermid and Morrow, 2002). Genetic studies in the mouse have shown that haploinsufficiency or complete loss of Tbx1, within the 1.5 Mb region, can recapitulate most of the physical anomalies. 22q11DS patients with mutations in TBX1 in the absence of 22q11 microdeletions have been identified, confirming TBX1 as a prime candidate gene for the syndrome (Gong et al., 2001; Yagi et al., 2003; Zweier et al., 2007). Another gene outside the 1.5 Mb interval, Crkl, which encodes an intracellular RTK signaling adaptor, may modify the phenotype, because inactivation of Crkl results in similar thymus and cardiovascular anomalies in the mouse. These studies have confirmed that combined heterozygosity for Tbx1 and Crkl significantly exacerbated the phenotypes associated with either Tbx1 or Crkl heterozygosity (Guris et al., 2006). These and other studies have indicated that the level of TBX1 is critical for normal development and factors that can alter the intensity of signalling pathways sensitive to TBX1 dosage are of particular interest in understanding the severity of disease.
Indeed, the large clinical variability that typifies this syndrome suggests that factors outside the typically deleted region may modify the phenotype. In order to fully understand this disease, it is vital that these modifiers are identified. Potential modifying mechanisms include differences in breakpoints during deletion, specific modifier genes outside the critical region, allelic variation of genes within the critical region on the non-deleted chromosome, epigenetic phenomena and environmental factors (Scambler et al., 1991; Morrow et al., 1995; Carlson et al., 1997). The search for modifier genes in patients have so far met with limited success (Driscoll et al., 2006; Goldmuntz et al., 2009). Studies in mouse embryos have suggested that Tbx1 functions in a genetic pathway upstream of Fibroblast Growth Factor (FGF) signaling (Abu-Issa et al., 2002; Frank et al., 2002; Vitelli et al., 2002; Brown et al., 2004; Arnold et al., 2006b; Park et al., 2006). Accordingly, loss of function mutations in genes encoding FGF ligands can enhance the mild phenotypes in Tbx1+/- mutants (Vitelli et al., 2002; Aggarwal et al., 2006). Similarly, variants of the Vascular Endothelial Growth Factor (Vegf) gene have been identified as a potential modifier of the pharyngeal arch artery phenotype in mice and humans (Stalmans et al., 2003). A number of other genes have been found to genetically interact with Tbx1 during mouse development, including the BMP antagonist, Chordin, the cardiac transcription factor Pitx2 and the Chd7 gene, which is associated with CHARGE syndrome (Nowotschin et al., 2006; Choi and Klingensmith, 2009; Randall et al., 2009).
The Sprouty genes encode intracellular regulators of RTK signaling and have been shown to function widely to regulate the levels and extent of signaling during embryonic development. Although signaling downstream of many growth factors can be regulated by Sprouty in vitro, in vivo evidence thus far seems to suggest a preference for the FGF and GDNF/RET pathways during mammalian development (Basson et al., 2005; Shim et al., 2005; Klein et al., 2006; Taniguchi et al., 2007). The human and mouse genomes contain four Sprouty genes, Spry1-Spry4. We found that two of these genes, Spry1 and Spry2 were co-expressed at high levels in all key tissues of the developing pharyngeal apparatus of the mouse embryo. We hypothesized that Sprouty genes play important roles in the development of the PA and that these genes may modify the phenotypes in mouse models of 22q11DS. Here we provide experimental evidence in support of this hypothesis, by showing that Sprouty-deficient embryos have many defects in organs derived from the PA and that these genes interact with Tbx1 during development.
Results
Sprouty genes are expressed in the developing pharyngeal apparatus
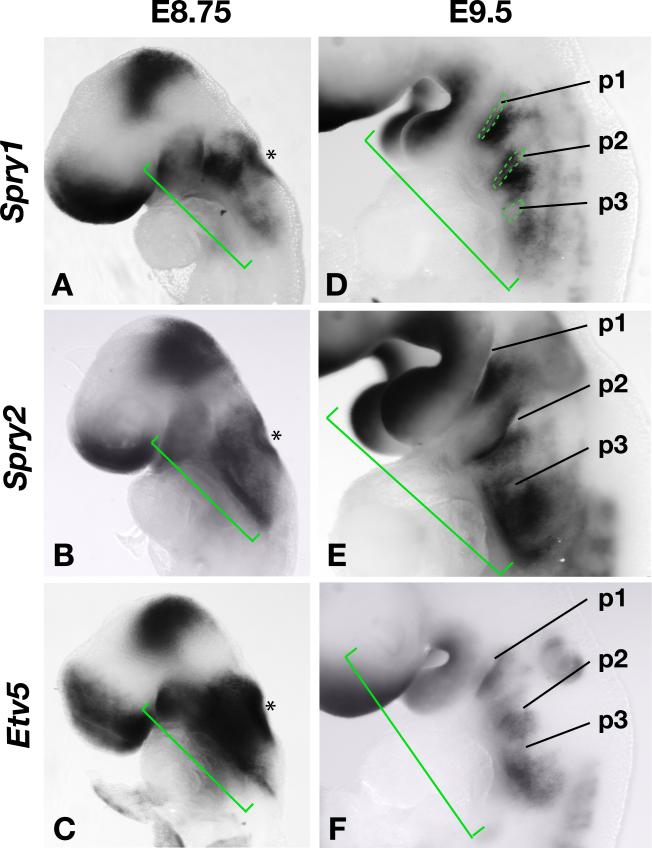
Sprouty genes are feedback antagonists of FGF signalling and previous studies have reported Sprouty gene expression in the developing pharyngeal region of the mouse embryo where several FGF ligands are expressed (Minowada et al., 1999). We confirmed that Spry1 and Spry2 were expressed at high levels in the developing pharyngeal apparatus during the time when the anterior pharyngeal pouches are forming and the otic placode is invaginating to form the otic vesicle (Fig. 1A,B). We recently reported that these genes are expressed in the pharyngeal ectoderm, endoderm and mesoderm (Simrick et al., 2011). Sprouty gene expression is maintained in discrete regions in and around the anterior pharyngeal pouches after their formation and during the formation of the caudal pouches (Fig. 1D,E). As expected, Sprouty gene expression significantly overlapped with the expression of Etv5 (Erm), another gene known to be transcriptionally regulated by FGF signaling (Klein et al., 2008) (Fig. 1C,F).
Figure 1. Sprouty and Etv5 gene expression in the developing pharyngeal apparatus of mouse embryos.
A-C) Spry1, Spry2 and Etv5 gene expression in the developing pharyngeal region (green brackets) and otic vesicle (asterisk) of mouse embryos at E8.75.
D-F) Gene expression in E9.5 embryos with the pharyngeal pouches (p1-p3) indicated.
Spry1-/-;Spry2-/- (Spry1;2dko) embryos present with a range of pharyngeal phenotypes
Given the overlapping expression patterns of Spry1 and Spry2, and the similar activities of SPRY1 and SPRY2 proteins on RTK signaling (Hanafusa et al., 2002; Iatan et al., 2009), we predicted that these genes would function redundantly in the pharyngeal apparatus. To directly test this hypothesis, we produced double Spry1;Spry2 gene knockout embryos (Spry1;2dko) by crossing males homozygous for βactin-Cre and heterozygous for Spry1 and Spry2 null alleles (βactin-Cre;Spry1+/-;Spry2+/-) with females homozygous for conditional (flanked by loxP sites = flox) Spry1 and Spry2 alleles (Spry1flox/flox;Spry2flox/flox) (Basson et al., 2005; Shim et al., 2005). βactincre;Spry1+/- ;Spry2+/- embryos (hereafter referred to as Spry1+/-;Spry2+/-) produced in these crosses were phenotypically normal and were used as littermate controls in all experiments.
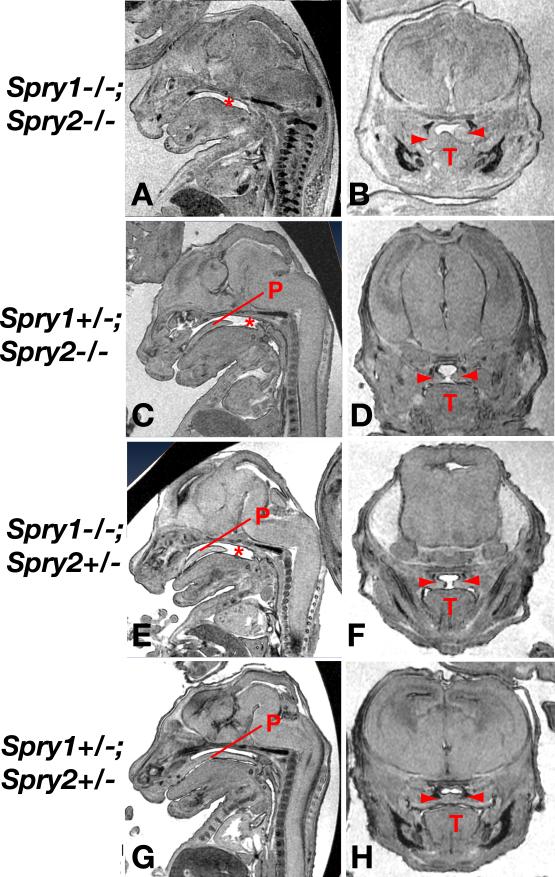
Compound Sprouty gene knockout embryos were screened for the anomalies associated with 22q11DS. Velopharyngeal insufficiency due to cleft palate is a common birth defect associated with 22q11DS. Micro-MRI scanning of E15.5 embryos (Fig. 2) and direct examination of E16.5-E18.5 palates revealed severe clefting of the secondary palate in Spry1;2dko embryos (Fig. 2A, Table 1). Coronal sections through the heads of these embryos indicated that palatal shelves were present bilaterally, but that the shelves on at least one side of the embryo failed to elevate above the tongue (Fig. 2B). Spry1+/-;Spry2-/- (Fig. 2C,D) and Spry1-/-;Spry2+/- (Fig. 2E,F) embryos also presented with cleft palate, although these clefts were generally smaller, only affecting the posterior half of the palate (Fig. 2C,E). Interestingly, coronal sections through these embryos revealed palatal shelves that had elevated above the tongue, but failed to meet at the midline (Fig. 2D,F). Spry1+/-;Spry2+/- embryos (Fig. 2G,H) did not exhibit cleft palate. These data indicated that Sprouty genes controlled palatal development in a gene dosage-dependent manner. Furthermore, we noticed that the incidence of palatal defects did not increase gradually as Sprouty gene dosage was reduced, but that an apparent threshold was crossed between the loss of two and three Sprouty alleles, resulting in an increased incidence from 0% in Spry1+/-;Spry2+/- embryos to 100% in Spry1+/-;Spry2-/- and Spry1-/-;Spry2+/- embryos (Table 1).
Figure 2. Palatal development is sensitive to Sprouty gene dosage.
MRI sections of mutant E15.5 embryos in sagittal (A,C,E,G) or coronal (B,D,F,H) views. The absence of palatal tissue (P) is indicated by an asterisk and the two palatal shelves by red arrowheads. T=tongue.
Table 1.
Sprouty/Tbxl interactions during development of the palate
| Spry1 | Spry2 | Tbx1 | n | Cleft palate |
|---|---|---|---|---|
| +/+ | +/− | 3 | 0 | |
| +/− | +/− | +/+ | 2 | 0 |
| +/− | 21 | 8* | ||
| −/− | +/− | +/+ | 4 | 4 |
| +/− | 16 | 14 | ||
| +/− | −/− | +/+ | 6 | 6 |
| +/− | 15 | 14 | ||
| −/− | −/− | +/+ | 3 | 3 |
| +/− | 7 | 7 |
Shaded cells indicate significant difference between Tbx1 +/+ and Tbx1 +/− embryos:
p=0.03
Next we determined the position and size of the thymus, an organ often affected in DiGeorge syndrome. In Spry1+/-;Spry2+/- control embryos, the thymic lobes had descended completely to their final position at the mediastinum just cranially and ventrally to the heart by E15.5 (see Fig. 5B). The Spry1;2dko embryos had two thymus lobes, similar to controls, but thymi were hypoplastic and located ectopically in the neck of all mutant embryos analysed (Fig. 5C,D). Thymus abnormalities in Spry1+/-;Spry2+/- and all other embryos retaining at least one copy of Spry1 or Spry2 were generally mild and occurred at low frequencies (Table 2).
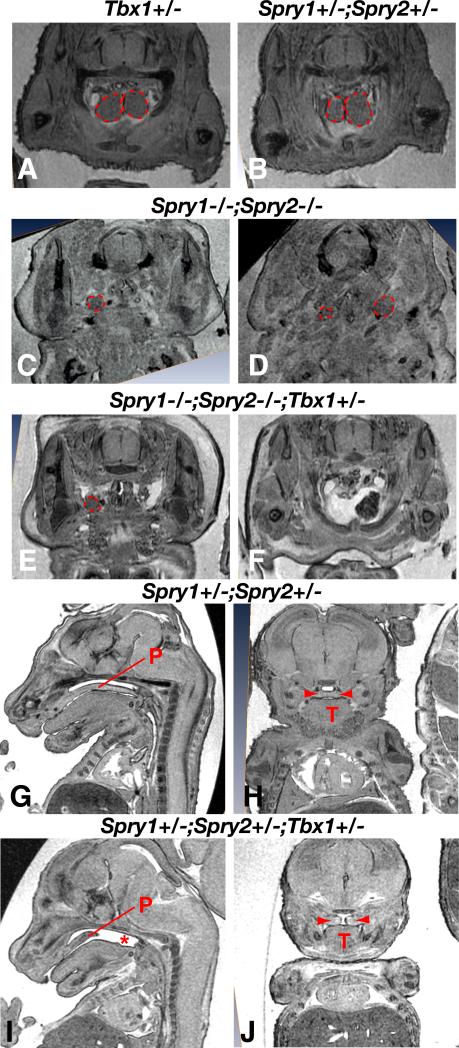
Figure 5. Genetic interaction between Sprouty and Tbx1 genes during thymus and palatal development.
A-F) MRI sections through the thoracic region of E15.5 embryos with the thymus lobes outlined in red. Examples of hypoplasia (C,D,E), hypoplasia with ectopia (C,E) and aplasia (F) can be seen.
G-J) Sagittal (G,I) and frontal (H,J) scans through embryonic heads showing a normal palate fused at the midline in a Spry1+/-;Spry2+/- embryo (G,H) and a cleft palate in a Spry1+/-;Spry2+/-;Tbx1+/- embryo (I,J).
Table 2.
Sprouty/Tbxl interactions during thymus development
| Spry1 | Spry2 | Tbx1 | n | Normal | Hypoplasia | Hypoplasia with ectopia | Aplasia | TOTAL |
|---|---|---|---|---|---|---|---|---|
| +/+ | +/+ | +/− | 35 | 28 | 7 | 0 | 0 | 7 |
| +/− | +/− | +/+ | 23 | 23 | 0 | 0 | 0 | 0 |
| +/− | 35 | 32 | 3 | 0 | 0 | 3 | ||
| −/− | +/− | +/+ | 20 | 18 | 1 | 1 | 0 | 2 |
| +/− | 35 | 30 | 2 | 1 | 2 | 5 | ||
| +/− | −/− | +/+ | 13 | 13 | 0 | 0 | 0 | 0 |
| +/− | 24 | 9 | 9* | 1 | 5 | 15** | ||
| −/− | −/− | +/+ | 7 | 0 | 0 | 7 | 0 | 7 |
| +/− | 16 | 0 | 5 | 0 | 11*^ | 16 |
Shaded cells indicate significant difference between Tbx1+/+ and Tbx1+/− embryos:
p=0.015
p=0.0002
p=0.005
No defects in the outflow tract or major thoracic vessels were detected in Spry1;2dko embryos. However, 2/8 embryos had small ventricular septal defects (VSDs) (Table 3, data not shown).
Table 3.
Sprouty/Tbx1 interactions during cardiovascular development
| E10.5a | E15.5-E17.5b | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Spry1 | Spry2 | Tbx1 | n | 1st PAA | PAA | PAA | Aortic arch | OFT defects | VSD |
| +/+ | +/+ | +/+ | 21 | 0 | 0 | 0 | nd | nd | nd |
| +/− | 17 | 1 | 0 | 0 | 0/38 | 0/38 | 1/17 | ||
| +/− | +/− | +/+ | 8 | 0 | 0 | 0 | 0/30 | 0/30 | 0/3 |
| +/− | 14 | 1 | 0 | 7* | 4/38 | 0/38 | 0/19 | ||
| −/− | +/− | +/+ | 10 | 0 | 0 | 1 | 0/20 | 0/20 | 0/9 |
| +/− | 22 | 1 | 1 | 17** | 9/33** | 0/33 | 0/23 | ||
| +/− | −/− | +/+ | 10 | 1 | 2 | 0 | 0/13 | 0/13 | 0/10 |
| +/− | 14 | 0 | 2 | 7* | 8/29**^ | 0/30 | 5/19 | ||
| −/− | −/− | +/+ | 16 | 0 | 6^ | 5^^ | 0/11 | 0/11 | 2/8 |
| +/− | 12 | 5** | 10* | 10*** | 12/16*** | 2/16 | 4/10 | ||
Aortic arch defects were scored at E10.5 by India ink injections (a) or at later stages by MRI or histology (b).
Outflow tract (OFT) defects and Ventricular Septal defects (VSD) were also scored in E15.5-E17.5 embryos.
Significant incidence compared to wildtype controls (Fisher's exact test, two tailed):
p=0.003
p=0.01
Shaded cells indicate a significant difference between Tbx1+/+ and Tbx1+/− embryos:
p=0.02
p=0.01
p=0.04
p<0.001
In addition to the defects described here, we have also noted abnormalities in the parathyroids (JG, MAB and Nancy Manley, manuscript submitted). Thus, a range of phenotypes associated with 22q11del syndrome is present in embryos with reduced Spry1 and Spry2 gene dosage. These embryos may therefore represent useful mouse models for investigating the development of a subset of organs defective in this syndrome.
Pharyngeal arch artery defects in Spry1;2dko embryos
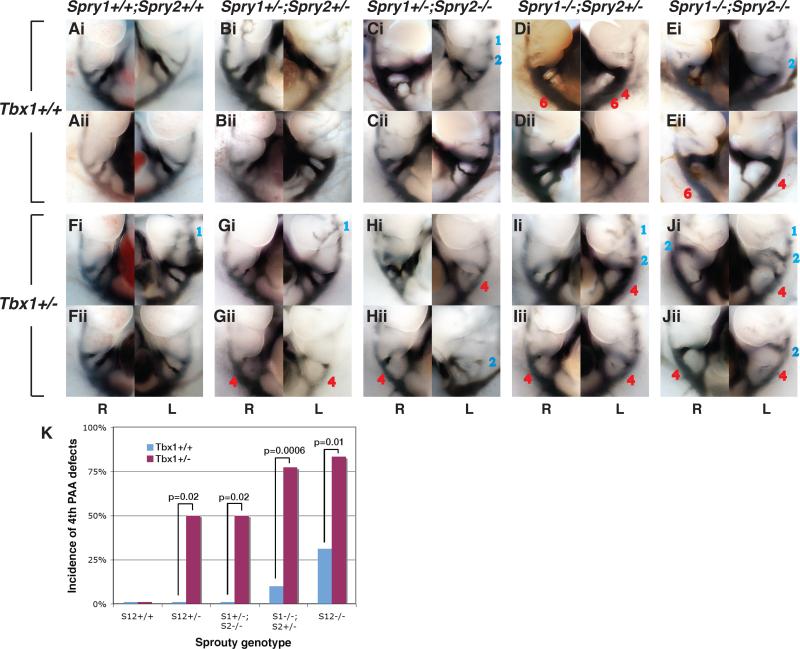
The pharyngeal arch arteries undergo a complex asymmetric remodeling process during development to form the large thoracic vessels, including the aortic arch. Aberrant loss or maintenance of segments of the embryonic PAAs is the primary cause of aortic arch defects (Srivastava and Olson, 2000; Hiruma et al., 2002). For example, the left-sided 4th PAA contributes to the portion of the descending aorta between the left common carotid (LCC) and left subclavian arteries (LSA). The absence of this portion of the aortic arch is referred to as interrupted aortic arch type B (IAA-B) and is a characteristic defect in 22q11del syndrome . The pharyngeal arch arteries can be visualized at E10.5 of development by intracardiac India ink injections, which allowed us to determine the incidence of 4th PAA abnormalities. Wildtype and Spry1+/-;Spry2+/- embryos had normal pharyngeal arch arteries (Fig. 3A,B). A significant proportion (n=7/18) of Spry1;2dko embryos exhibited an abnormal regression of the 4th PAA at E10.5 (Fig. 3Eii, K). 4th PAA defects were also detected in embryos in which three Sprouty genes had been deleted: Spry1-/-;Spry2+/- and Spry1+/-;Spry2-/- (Fig. 3C-D, K, Table 3). Defects in other arch arteries were also observed. By E10.5 of development (35 somite stage), cranial arch arteries (1st and 2nd) had completely regressed in control embryos (Table 3). Similarly, no significant incidence of persistent 1st or 2nd arteries were observed in Spry1-/-;Spry2+/-, Spry1+/-;Spry2-/- embryos. Spry1;2dko embryos did not show a significant 1st PAA phenotype either (Table 3). However, persistent 2nd PAA defects occurred at high frequency in Spry1;2dko embryos (Fig. 3Ei, Table 3). Thus, Spry1;2dko embryos exhibited 2nd and 4th PAA defects at E10.5.
Figure 3. Examples of pharyngeal arch artery defects in Sprouty-deficient embryos on Tbx1 wildtype or heterozygous backgrounds.
Micrographs of right (R) and left (L) sided views of E10.5 embryos with ink-filled aortic arch arteries are depicted. Examples of embryos with wildtype complement (A,F) or loss of 2-4 Spouty alleles (B-E, G-J) on a Tbx1 wildtype (A-E) or Tbx1+/- (F-J) background are shown, two for each genotype (i and ii). Persistant arch arteries are labeled in blue and missing arteries in red.
K) The incidence of 4th PAA defects are indicated for each genotype, note the significant increase in incidence of this phenotype on a Tbx1+/- background for all four Sprouty-deficient genotypes.
Sprouty genes interact with Tbx1 during PA development
Sprouty and Tbx1 genes have been shown to regulate FGF signaling, albeit in apparently opposite ways, with Tbx1 being required for FGF gene expression and Sprouty inhibiting FGF signal transduction. The apparent phenotypic similarities between Sprouty and Tbx1 mutant embryos with respect to craniofacial, pharyngeal arch artery and thymus defects, suggested that Sprouty genes and Tbx1 either affected the FGF (or another) pathway in a similar way, or that hypo- or hyperactivation of the pathway could result in similar phenotypes. We generated embryos with different allelic combinations of Spry1, Spry2 and Tbx1 loss of function alleles and determined their phenotypes in an attempt to distinguish between these possibilities. We reasoned that a phenotypic rescue would indicate that these mutations had opposing effects on signaling, whereas an exacerbation in the phenotype, would indicate that Sprouty and Tbx1 mutation could affect the same pathway in a similar way.
Tbx1+/- embryos exhibit a low incidence of cardiovascular defects on certain genetic backgrounds (Merscher et al., 2001). The sensitivity of these defects to genetic background has also been reported in a mouse model in which the chromosomal region syntenic with the human critical region was deleted on one chromosome (Taddei et al., 2001). With the exception of one embryo (n=1/17), no cardiovascular defects were observed in Tbx1+/- mutants maintained on a mixed genetic background in our colony. This observation is in agreement with the low incidence of cardiovascular (9%) defects in these mutants reported previously (Liao et al., 2004). As all four Spry1 and Spry2 alleles had to be deleted before significant PAA defects were detected (Table 3), we chose to analyse embryos with various combinations of Spry1, Spry2 and Tbx1 null alleles.
To test for genetic interactions between Sprouty and Tbx1 genes, we first visualised the PAAs in E10.5 embryos by ink injection. The incidence of 1st PAA persistence in Tbx1+/- or Spry1;2dko embryos was significantly increased when these mutations were combined in Spry1;2dko;Tbx1+/- embryos (Fig. 3J, Table 3). The incidence of 2nd and 4th PAA defects in Spry1;2dko embryos was also significantly increased on a Tbx1+/- background (Fig. 3, Table 3). First and second PAA defects are not normally observed in Tbx1+/- embryos. Importantly, Tbx1 heterozygosity also increased the incidence of 4th PAA defects significantly in Spry1-/-;Spry2+/-, Spry1+/-;Spry2-/- and even in Spry1+/-;Spry2+/- embryos (Fig. 3F-I, K, Table 3). These data indicate that Sprouty and Tbx1 genes interacted during PAA development and that a reduction in gene dosage significantly increased the incidence of 4th PAA phenotypes. We concluded from these experiments that Sprouty and Tbx1 loss of function mutations had similar effects on a genetic pathway that regulated PAA development.
To determine the consequences of these early PAA defects and to provide a more comprehensive picture of cardiovascular defects, we analysed E15.5-E17.5 embryos by micro-MRI, India ink injections and histology. The only cardiovascular defect observed in Sprouty-deficient embryos was ventricular septal defects that occurred at low frequency (e.g. n=2/8 in Spry1;2dko embryos, Table 3). Despite the clear PAA defects in many Spry1;2dko embryos at E10.5, no large thoracic vessel phenotypes were observed in Sprouty mutants at E15.5 (Fig. 4A,E,G,I and Table 3). A similar discrepancy between early pharyngeal arch artery and later aortic arch defects has been reported previously in Tbx1+/- mutants. Although defects in PAA development is associated with cardiovascular defects such as IAA-B, it has been shown that embryos with PAA defects at E10.5 appear to have the ability to repair/rescue a significant number of these early defects (Lindsay et al., 2001). These data indicated that Sprouty-deficient embryos could also recover from aortic arch defects after E10.5.
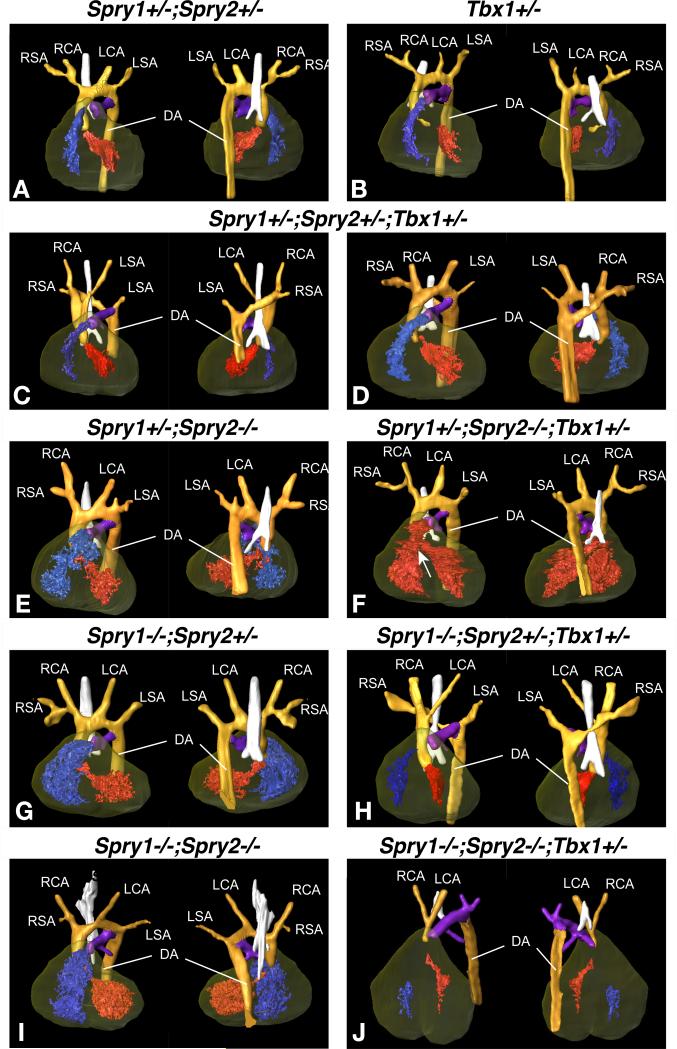
Figure 4. Anatomy of the great arteries as determined by micro MRI scans of E14.5 embryos.
3D reconstruction of high resolution MRI data from E15.5 embryos (Amira® software, Visage Imaging Inc.) showing the great arteries. Ventral (left) and dorsal (right) views are shown for each genotype indicated. Individual structures are coloured as follows: orange for the aortic arch and descending aorta, purple for the pulmonary artery, the trachea is highlighted in white and the heart is shaded in brown with the right ventricle in blue and the left ventricle in red. Where there is a ventricular septal defect (VSD), both ventricles are in red. Arteries are labelled as LSA: Left Subclavian Artery, LCA: Left Common Carotid Artery, RSA: Right Subclavian Artery, RCA: Right Common Carotid Artery and DA: Descending Aorta.
A) Spry1+/-;Spry2+/- control embryo and (B) Tbx1+/- embryo with normal arteries and normal heart.
C, D) Examples of Spry1+/-;Spry2+/-;Tbx1+/- embryos with normal vessels (C) or retroesophageal aortic arch (REAA) and aortic vascular ring (D).
E,G,I) Compound Spry1;Spry2 mutant embryos with normal anatomy.
F) Spry1+/-;Spry2-/-;Tbx1+/- embryo with normal arteries and a ventricular septal defect (VSD, indicated by an arrow). Both ventricles are coloured in red to indicate mixing of oxygenated and de-oxygenated blood due to the VSD.
H) Spry1-/-;Spry2+/-;Tbx1+/- embryo with REAA.
J) Spry1-/-;Spry2-/-;Tbx1+/- embryo with interrupted aortic arch type B (IAA-B).
The increased severity of PAA defects at E10.5 in Sprouty;Tbx1 compound mutants resulted in severe great vessel defects at E15.5 (Fig. 4, Table 3). The loss of three or four Sprouty alleles in combination with one Tbx1 allele was associated with clear aortic arch defects at E15.5. The observed defects included retroesophageal aortic arch (Fig. 4C,H) and interrupted aortic arch type B (Fig. 4J). Both these defects are caused by the absence of a left 4th arch artery, resulting in an interruption between the left common carotid and left subclavian arteries. Embryos with vascular rings were also observed (Fig. 4D), a defect that can arise due to a persistant bilateral 4th aortic arch. The incidence of these defects was significantly increased compared to Tbx1+/+ embryos of the corresponding Sprouty genotype (Table 3). Although a few Spry1+/-;Spry2+/-;Tbx1+/- embryos exhibited aortic arch defects (n=4/38), this low incidence was not statistically different from Spry1+/-;Spry2+/-;Tbx1+/+ embryos. Mild outflow tract rotation defects were only observed in a small subset of Spry1-/-;Spry2-/-;Tbx1+/- embryos and not in any other genotype (Table 3). We concluded from this data that Tbx1 haploinsufficiency significantly exacerbated PAA defects in Sprouty-deficient embryos and that the exacerbated phenotype at E10.5 correlated well with an increased incidence of great vessel anomalies by E15.5.
Sprouty and Tbx1 genes interact during thymus organogenesis
A small fraction (n=7/32) of Tbx1+/- embryos analysed had thymus hypoplasia (defined as <1/3 of the mean of control thymus size) (Fig. 5A, Table 2). A significant incidence of thymus defects was only observed upon the deletion of both Sprouty genes, and all these Spry1;2dko embryos exhibited thymus hypoplasia with thymus lobes located ectopically in the neck (Fig. 5D, Table 2). All of these embryos had two thymus lobes each, although only one could be seen in many sections due to variability in their ectopic locations (Fig. 5C). By contrast, when Tbx1 gene dosage was reduced in Spry1;2dko embryos, a significant number exhibited thymus aplasia (Fig. 5E,F, Table 2). Tbx1 heterozygosity also had significant effects on thymus size in other Sprouty-deficient embryos. Spry1+/-;Spry2-/- embryos that had normal thymus development on a Tbx1+/+ background, showed a significant increase in the incidence of thymus hypoplasia on a Tbx1+/- background (Table 2).
Sprouty genes interact with Tbx1 during palatogenesis
All embryos in which 3 or 4 Sprouty alleles had been deleted have cleft palate (Fig. 2, Table 1). Tbx1 heterozygosity does not significantly alter the incidence of this phenotype (Table 1). No Spry1+/-;Spry2+/- or Tbx1+/- embryos with palatal defects were observed during the course of our study (Fig. 5G,H, Table 1). Intriguingly, many Spry1+/-;Spry2+/-;Tbx1+/- embryos had cleft palate, and the incidence of cleft palate in these mutants was significantly increased compared to Spry1+/-;Spry2+/- embryos (Fig. 5I,J, Table 1). Thus, it appears that Tbx1 heterozygosity also sensitized the embryos to palatal defects.
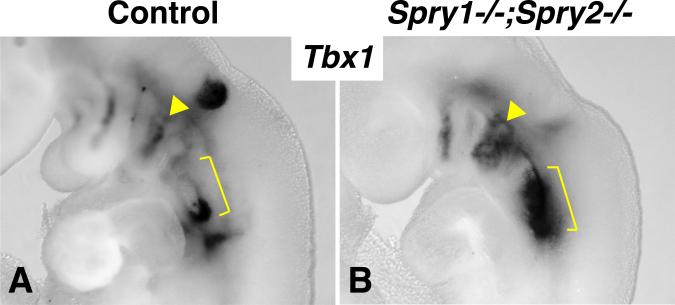
Expanded Tbx1 expression in Spry1;2dko embryos
One possible explanation for the exacerbation of pharyngeal phenotypes by reducing Tbx1 gene dosage in Spry1;2dko embryos, is that Tbx1 gene expression may be downregulated in Spry1;2dko embryos. An examination of Tbx1 expression in embryos between E9.0 and E9.5 revealed an expansion of Tbx1 expression in the caudal (II-VI) arches (Fig. 6). The expansion of Tbx1-expressing core mesoderm in the second arch correlated with the increased size of this arch in these embryos, suggested that the expansion of Tbx1-expressing tissue is responsible for this phenotype (Simrick et al., 2011). This observation argued against a simple model whereby reduced Tbx1 expression could account for the observed genetic interactions.
Figure 6. Expanded Tbx1 expression in Sprouty-deficient embryos.
In situ hybridisation for Tbx1 mRNA in E9.5 control (A) and Spry1-/-;Spry2-/- (B) embryos are compared. Note the expanded Tbx1 expression in the second pharyngeal arch (yellow arrowhead) and caudal arches (yellow bracket) in the mutant embryo.
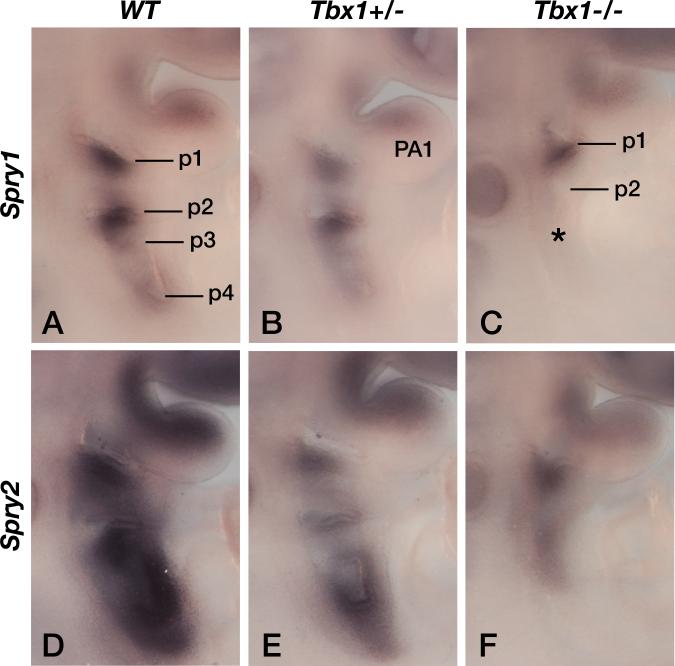
Tbx1 protects the embryos against mild perturbations in RTK signaling
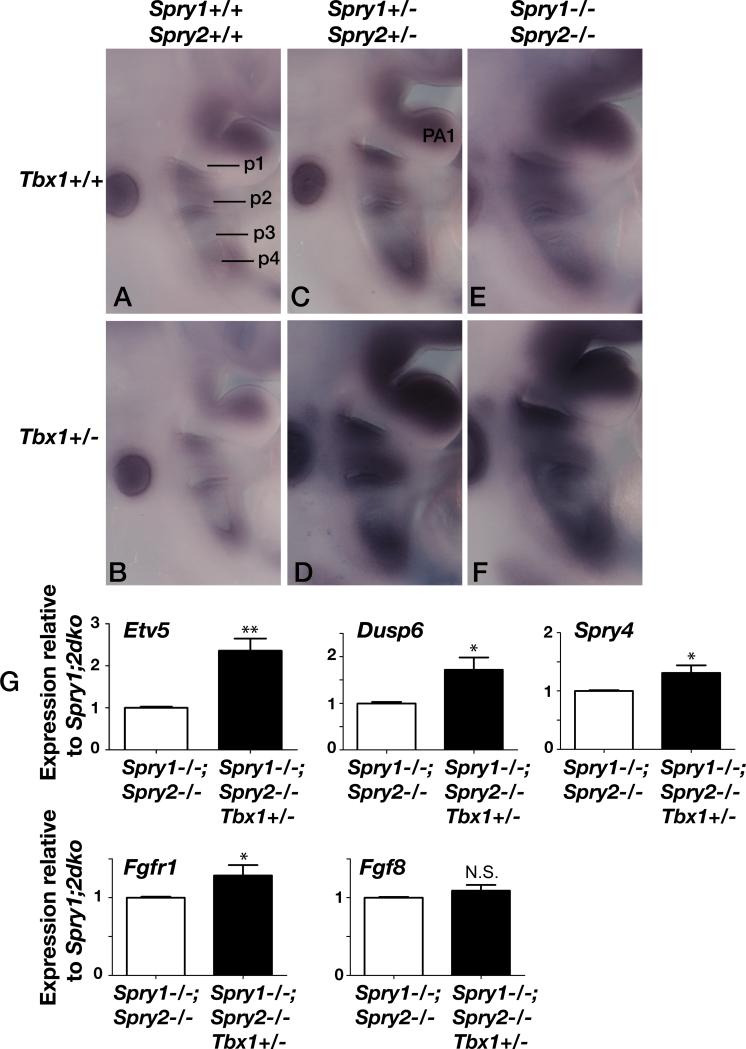
As both TBX1 and Sprouty have been associated with the regulation of RTK signaling, in particular FGF signaling, we investigated to what extent the level of FGF signaling in the developing pharyngeal apparatus was perturbed in the various mutants. As all previous studies that concluded that TBX1 was an upstream, positive regulator of FGF signaling, only reported changes in FGF gene expression in Tbx1 null embryos, we first wanted to determine whether FGF signaling was downregulated in Tbx1+/- embryos. As both Spry1 (Basson et al., 2008) and Spry2 (Chambers et al., 2000) genes are transcriptional read-outs of FGF signaling in the embryo, we monitored their expression in wildtype (WT), Tbx1+/- and Tbx1-/- embryos. Both Spry1 (Fig. 7A-C, n=2/2) and Spry2 (Fig. 7D-F, n=2/2) expression was downregulated in the pharyngeal apparatus of Tbx1 mutant embryos in a manner that was clearly proportionate to the Tbx1 gene dosage. Next, we compared the expression of another FGF read-out, Etv5 (Klein et al., 2006). Etv5 expression appeared slightly downregulated in Tbx1+/- mutants compared to wild type controls (Fig. 8A,B, n=4/4), perhaps clearest in the 1st pharyngeal arch (PA1). As expected, the expression levels of Etv5 was increased in Sprouty mutant embryos, with the extent of upregulation correlating with the number of Sprouty alleles deleted: slight upregulation in Spry1+/-;Spry2+/- (Fig. 8C) and stronger upregulation in Spry1;2dko embryos (Fig. 8E, n=4/4), consistent with these genes functioning as RTK antagonists. To our surprise, Etv5 expression was drastically upregulated in many Spry1+/-;Spry2+/-;Tbx1+/- (Fig. 8D, n=2/3) and Spry1;2dko;Tbx1+/- (Fig 8F, n=2/4) embryos compared to stage-matched Spry1+/-;Spry2+/- and Spry1;2dko embryos. Quantitation of Etv5 mRNA levels in microdissected pharyngeal tissue by quantitative RT-PCR, confirmed a significant upregulation of Etv5 expression in Spry1;2dko;Tbx1+/- embryos compared to stage-matched Spry1;2dko;Tbx1+/+ embryos (Fig. 8G). These observations suggest that the ability of Tbx1 heterozygosity to enhance the phenotypes associated with Sprouty gene deficiency may be due to a significant upregulation of RTK signalling. To find further evidence for this, we also quantified the expression of several other genes that are transcriptionally regulated by RTK signaling. Both Dusp6 and Spry4 were upregulated in Spry1;2dko;Tbx1+/- embryos compared to Spry1;2dko embryos (Fig. 8G). We also measured Fgfr1 transcript levels and found a slight, but significant increase, consistent with the observed increased RTK signaling. By contrast, Fgf8 gene expression was not altered by the loss of one Tbx1 allele, suggesting that increased Fgf8 expression is unlikely to account for our observations (Fig. 8G).
Figure 7. Reduced RTK signaling as measured by Sprouty gene expression in Tbx1-deficient embryos.
A-C) Spry1 gene expression in somite stage-matched E9.5 wildtype (WT), Tbx1+/- and Tbx1-/- embryos. Pharyngeal pouches (p1-p6) and the first pharyngeal arch (PA1) are labeled.
D-F) Spry2 gene expression in somite stage-matched E9.5 wildtype (WT), Tbx1+/- and Tbx1-/- embryos.
Figure 8. Deregulated RTK signaling as measured by Etv5 (Erm) gene expression in Sprouty; Tbx1 -deficient embryos.
Comparative images of the pharyngeal region of somite-matched E9.5 embryos after in situ hybridization for Etv5. Hybridisations were performed simultaneously under identical conditions and at least two independent experiments gave similar results. Pharyngeal pouches (p1-p4) and the first pharyngeal arch (PA1) are labeled. Note the slight reduction in Etv5 expression in B, compared to A, especially in PA1, the gradual increase in Etv5 expression as more Sprouty alleles are deleted (compare A, with C, with E), and the pronounced upregulation in Tbx1+/- embryos (D,F) compared to Tbx1+/+ embryos with the same Sprouty genotype (C,E).
G) Quantitative RT-PCR analysis of RTK target gene (Etv5, Dusp6 and Spry4) expression in the pharyngeal apparatus isolated from Spry1-/-;Spry2-/- and Spry1-/-;Spry2-/-;Tbx1+/- embryos (as in E and F) are shown. Quantitation of Fgfr1 and Fgf8 transcripts are also shown. Expression levels are represented as relative to those in Spry1-/-;Spry2-/- (Spry1;2dko) embryos. *p<0.05; **p<0.01; N.S. = no significant difference.
Discussion
In the present study, we investigated the role of two Sprouty genes in pharyngeal development. Our data indicate that Spry1 and Spry2 function redundantly during PA development and that several functions are only revealed when more than one Sprouty gene is deleted, consistent with observations in other organs (Taniguchi et al., 2007; Klein et al., 2008). The most intriguing observation reported here is that Tbx1 heterozygosity enhanced all these phenotypes, indicating that Sprouty and Tbx1 genes functioned in the same or parallel genetic pathways and that loss of function Sprouty mutations should be considered as potential genetic modifiers of human phenotypes caused by TBX1 haploinsufficiency.
TBX1 is a prime candidate for DiGeorge syndrome and the observation that Tbx1 null mouse embryos exhibited severe defects in the formation of the caudal pharyngeal arches provided strong experimental evidence in support of the idea that Tbx1 is a key player in pharyngeal apparatus development (Jerome and Papaioannou, 2001; Lindsay et al., 2001). Studies demonstrating that the expression of several genes encoding FGF ligands in the pharyngeal apparatus are lost in Tbx1 null embryos placed TBX1 upstream of FGF signaling. Furthermore, Fgf8 heterozygosity enhances the mild phenotypes present in Tbx1+/- embryos, in agreement with TBX1 being a positive regulator of FGF signaling (Vitelli et al., 2002; Aggarwal et al., 2006). The data presented in this manuscript argues against a simple linear model where TBX1 functions strictly as an upstream, positive regulator of FGF gene expression in all contexts. Indeed, a previous study reported that Fgf8 overexpression in Tbx1-expressing cells that were also Tbx1+/-, enhanced the 4th PAA phenotypes (Vitelli et al., 2006). This observation is consistent with our findings. However, other phenotypes (e.g. thymus and palate) were not enhanced in this study and the authors concluded that the PAA phenotype was somehow unique with respect to FGF signalling. Our data now suggests that this phenomenon is applicable to other organs too and that the effect of TBX1 on FGF signalling thresholds may be more universal than previously thought.
A recent study in which the effects of reducing Tbx1 transcript levels by small intervals were assessed, showed that the appearance of developmental defects did not correlate in a linear fashion with Tbx1 levels, but rather that it exhibited an abrupt appearance of defects once Tbx1 levels were reduced below a certain threshold. These results suggested that the maintenance of Tbx1 levels within a certain range was required for the stability of the developmental system and that the sensitivity of an embryo to stochastic or environmental variation may be increased substantially when Tbx1 levels approached this critical threshold. This interpretation is consistent with a model whereby both genetic and stochastic factors could contribute to the severity of the disease, as is indeed suggested by observations of discordant phenotypes in monozygotic twins (Goodship et al., 1995; Yamagishi et al., 1998; Vincent et al., 1999; Hillebrand et al., 2000), and the variation in phenotypic severity in inbred mouse strains (Jerome and Papaioannou, 2001; Taddei et al., 2001). TBX1 therefore appears to function as a central component that is required for the stability of the gene regulatory network(s) that control pharyngeal development (Baldini, 2005).
Our data appear to be in disagreement with two recent reports of cleft palate in Spry2-deficient mice on a C57BL/6J genetic background (Welsh et al., 2007; Matsumura et al., 2011). Mice homozygous for a targeted deletion of the Spry2 open reading frame used in our experiments do not have cleft palate, even on a C57BL/6J background (Gail Martin, personal communication). The reason for this discrepancy is unknown, however, our studies indicate that Sprouty genes are required for normal palatal development, in a dosage-dependent manner.
Our data suggested that increasing RTK signaling by simultaneously deleting Spry1 and Spry2 resulted in phenotypes in many of the same organs typically affected by the loss or gain of Tbx1 function (Funke et al., 2001; Merscher et al., 2001; Liao et al., 2004; Vitelli et al., 2009). The Tbx1+/- embryos used in the present study were generated from the Tbx1tm1.2Bem conditional allele (Arnold et al., 2006a). Most of these embryos appeared phenotypically normal on our mixed genetic background (129Sv;C57BL/6J; FVB/N; CD1), making it ideal to test for exacerbated phenotypes. The analyses of compound Spry1;Spry2;Tbx1 mutants revealed a strong interaction between these mutations during PA development, suggesting that these genes functioned in the same or parallel genetic pathways.
To understand the reason for these genetic effects, we investigated whether RTK signaling was deregulated in unexpected ways in compound mutant embryos at a critical time point for caudal PA development (Xu et al., 2005). To our surprise, we found that the level of RTK signaling was disproportionally enhanced in Sprouty-deficient embryos on a Tbx1+/- background. We conclude that Tbx1 heterozygosity “sensitized” the embryo to perturbations in FGF signaling. Thus, it appears that TBX1 has a crucial role in maintaining the stability of the developmental system and essentially functioned like a buffer against the perturbation of RTK signaling (Fig. 9).
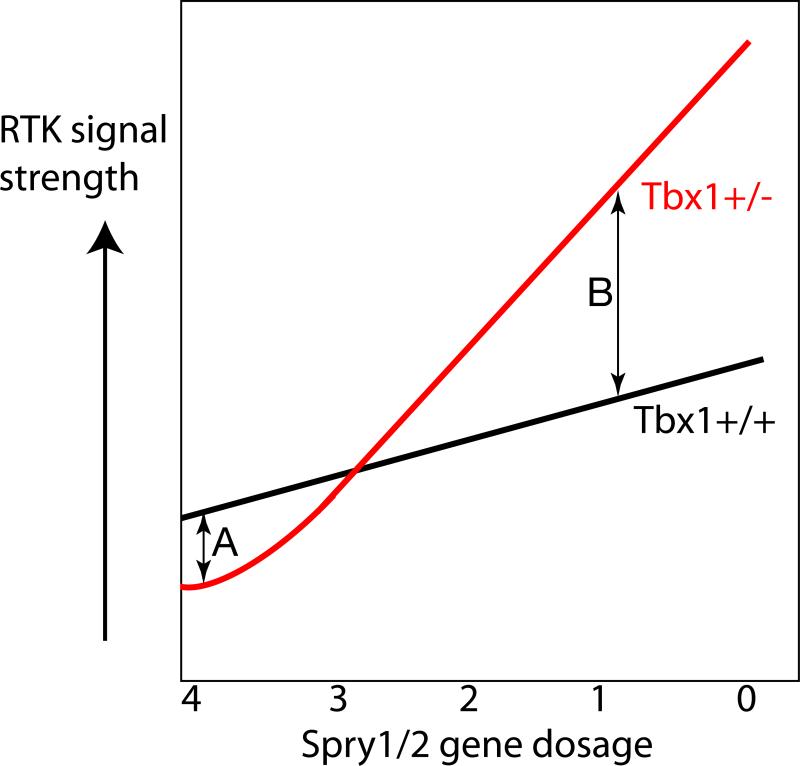
Figure 9. Diagrammatic representation of the effects of Tbx1 haploinsufficiency on RTK signal strength as a function of Sprouty gene dosage.
In embryos with a wild-type complement of Spry1/2 gene dosage (4 wildtype alleles), Tbx1+/- embryos show reduced levels of FGF signalling in the pharyngeal apparatus compared to Tbx1+/+ embryos, presumably due to the downregulation of FGF ligands in different tissues (A). As Sprouty gene dosage is reduced in Tbx1+/+ embryos, the level of RTK signalling increases due to the loss of RTK antagonists (black line). The level of RTK signalling increases disproportionally in Tbx1+/- embryos (red line), suggesting that normal TBX1 levels is required to buffer or protect the pharyngeal apparatus from increased RTK signaling (B).
The exact molecular mechanism whereby TBX1 might maintain the stability of the developmental system is not known. One possibility is that TBX1 does so by interacting with and integrating a large network of signaling pathways. Tbx1 has indeed been shown to interact with a number of major signaling pathways, including those regulated by FGF (Abu-Issa et al., 2002; Vitelli et al., 2002; Aggarwal et al., 2006), VEGF (Stalmans et al., 2003), Retinoic acid (Guris et al., 2006; Roberts et al., 2006), BMP (Fulcoli et al., 2009), SHH (Garg et al., 2001; Yamagishi et al., 2003), and Notch (Mitsiadis et al., 2010). The developing PA is a region in which all these signaling pathways are active and cells presumably integrate signals from this complex milieu. Thus, TBX1 may stabilise this system by functioning as an important integrator of many of these pathways. Although TBX1 may achieve this at the level of transcription, recent studies by the Baldini group have identified roles for TBX1 that appear distinct from its transcriptional activity. For example, TBX1 interacts with Serum Response Factor (SRF) and targets it for degradation (Chen et al., 2009) and interacts with SMAD1 to suppress BMP signaling (Fulcoli et al., 2009). Future studies are required to identify and characterize the individual interactions further, as well as integrate these interactions to understand the role of TBX1 at a “systems” level. Intriguingly, TBX1 has also been shown to be required for a mixed population of embryonic fibroblasts to respond to FGF8 in vitro, suggesting that TBX1 may have a more direct role in regulating FGF-responsiveness in cells (Herbst et al., 2010). Whether Sprouty and Tbx1 genes function in the same cell type e.g. the pharyngeal ectoderm to control neural crest migration will require the systematic analysis of mutants in which all these genes had been ablated in a cell type-specific manner (Eichenauer et al., 2009).
We found that different organs exhibited different sensitivities to alterations in RTK signaling and Tbx1 levels. The most sensitive developmental system to Sprouty gene dosage appears to be the palate, with the arch arteries and thymus being relatively insensitive. Interestingly, we observed an extremely low incidence of heart (outflow tract and ventricular septum) defects in the Sprouty mutants and no significant genetic interaction with Tbx1. Zhang et al. also found that these structures were the least sensitive to the reduction in Tbx1 transcript level (Zhang and Baldini, 2008). Vitelli et al. also found no exacerbation of outflow tract defects in Tbx1+/- embryos when overexpressing Fgf8 (Vitelli et al., 2006). Two groups recently reported that the inhibition of FGF signaling in the secondary heart field by overexpressing Spry1 or Spry2 resulted in OFT defects (Park et al., 2008; Yang et al., 2010). Our data suggest that increased FGF signaling in the absence of Spry1 and Spry2 is still compatible with normal OFT development.
The observations reported here have important implications for our understanding of the aetiology underlying 22q11DS. One implication of our findings is that genetic, epigenetic or environmental factors that perturb RTK signaling during the critical time of development of the PA may have disproportionally large effects on Tbx1+/- embryos. Our data suggest that environmental insults enhance RTK signalling, in addition to those that can potentially reduce Fgf8 expression, are likely to exacerbate phenotypes in Tbx1-deficient embryos (Vitelli et al., 2002; Sparrow et al., 2012). These interactions may account for the large clinical variability observed in 22q11DS patients.
Experimental Procedures
Mouse strains, breeding and genotyping
All mutant lines used in this study have been described previously and were maintained on a mixed genetic background; βactin-cre (Lewandoski et al., 1997), Spry1 flox/null allele (Basson et al., 2005), Spry2 flox/null allele (Shim et al., 2005) and Tbx1 flox/null allele (Arnold et al., 2006a). Mice homozygous for β-actin cre and heterozygous for Spry1 and Spry2 null alleles (βcre2;Spry1+/-;Spry2+/-) were mated with mice homozygous for Spry1 and Spry2 flox alleles (Spry1flox/flox;Spry2 flox/flox). Sprouty;Tbx1 compound mutants were generated either by mating βcre2;Spry1+/-;Spry2+/-;Tbx1+/- mice with Spry1flox/flox;Spry2 flox/flox mice, or by mating βcre2;Spry1+/-;Spry2+/- mice with Spry1flox/flox;Spry2flox/flox;Tbx1flox/flox mice. Embryos were harvested from timed matings, where noon on the day of the vaginal plug was taken as E0.5. Harvested embryos were fixed overnight in 4% paraformaldehyde at 4°C, then either ink injected or dehydrated and stored in 100% methanol at -20°C for whole mount or section in situ hybridisation. E15.5 embryos collected for MRI scanning, were bled out into warm Hank's Balanced Salt solution before fixing in 4% formaldehyde at 4°C. In some cases, the chest cavities of E16.5-E18.5 embryos were opened and examined for the presence, position and size of thymus lobes and hearts removed for histological analysis. All experiments involving mice were approved by the BSU ethical review board, King's College London and were performed as specified by a UK Home Office Project Licence 70/9904.
Genotypes were determined by PCR amplification using embryonic yolk sac or tail DNA as appropriate. PCR primers for βactin-cre were CCT GGA AAA TGC TTC TGT CCG and CAG GGT GTT ATA AGC AAT CCC, which gave a 390bp product in cre positive embryos and no product in wild type embryos. Spry1 genotyping primers were GGG AAA ACC GTG TTC TAA GGA GTA GC, GTT CTT TGT GGC AGA CAC TCT TCA TTC and CTC AAT AGG AGT GGA CTG TGA AAC TGC; which produced a 342bp product with a flox allele, a 150bp product with a null allele and a 311bp product with a wild type allele. Genotyping primers for Spry2 were GGA TGG CTC TGA TCT GAT CC, TTG AGA ACA TGC CTC GAC C and GCA TGG GCT ATT CAC AAA C, resulting in PCR products of 500bp with a flox allele, a 225bp product with a null allele and a 350bp product with a wild type allele. Tbx1 genotyping primers were TGA CTG TGC TGA AGT GCA TC, TCT TCT TGG GGC TGT AGA CT and AGC GCA ATG GCT TTT AAG GG, which generated a 580bp product with a flox allele, a 415bp product with a null allele and a 532 product with a wild type allele.
India ink injections
The pharyngeal arch arteries and the great arteries were visualized by Indian ink (Pelikan) injections into the outflow tract with a glass microinjection needle.
MRI
E15.5 embryos were dissected, exsanguinated and fixed in 4% PFA with Gd-DTPA for at least three days. Fixed embryos were embedded in agarose, also containing Gd-DTPA and MRI was performed as previously described in Schneider et al. (Schneider and Bhattacharya, 2004). The analysis and 3D reconstructions of MRI data were done using the Amira® software (Visage Imaging Inc.).
Histology
Embryos were embedded in paraffin and sectioned at 7μm thickness in preparation for section immunohistochemistry or H&E staining. H&E (hematoxylin and eosin) staining was performed by Dr. Alasdair Edgar according to standard protocols.
In situ hybridisation
Whole mount RNA in situ hybridisation was performed according to standard protocols. Spry1 and Spry2 RNA probes were produced from constructs as described by Minowada et al. (Minowada et al., 1999). The Etv5 (Erm) RNA probe has previously been detailed by Klein et al. (Klein et al., 2006).
Quantitative RT-PCR
The pharyngeal apparatus from stage-matched embryos (21-23ss) was microdissected and all adjacent neural and heart tube tissues removed. Total RNA was extracted and genomic DNA removed using the Absolutely RNA Microprep Kit (Agilent Technologies) . A total of 200ng of RNA was used for first-strand DNA synthesis with nanoScript Precision RT kit (PrimerDesign Ltd.) according to the manufacturer's specifications. cDNA synthesis reactions without reverse transcriptase enzyme (no RT) were used as controls for q-RT-PCR. qRT-PCR was performed on a RotorGene Q cycler (Qiagen) using Precision qPCR MasterMix kit (PrimerDesign Ltd.). Appropriate normalising genes were detected using qbasePLUS software (Biogazelle) with geNorm reference kits (PrimerDesign Ltd.) All reactions were performed twice in duplicate each time and normalised to GAPDH. Cq threshold values were determined manually and all were at least 5 Cq values below no RT controls. ΔΔCq values were calculated relative to S12dko samples using Microsoft Excel software and graphs were produced using GraphPad Prism software. Primers were as follows: Etv5 (5’-TGCCCACTTCATCGCCTGGAC-3’ and 5’-TAGCGGAGAGAGCGGCTCAG-3’), Dusp6 (5’- TCTGTTTGAGAATGCGGGCGAG-3’ and 5’- GCCAAGCAATGCACCAGGACAC-3’), Spry4 (5’- TGCGACTTCAACGGCGACTG-3’ and 5’- ACTGCACCAAGGGACAGGCTTC-3’), Fgfr1 (5’- GGCAGCGATACCACCTACTTCTCC-3’ and 5’- GGCCTACGGTTTGGTTTGGTGTTG-3’), Fgf8 (5’- AGGTCTCTACATCTGCATGAAC-3’ and 5’- TGTTCTCCAGCACGATCTCT-3’), Tbx1 (5’- CGACAAGCTGAACTGACCA-3’ and 5’- CAATCTTAAGCTGCGTGATCC-3’) and Gapdh as a normaliser (F, 5’-AGGTCGGTGTGAACGGATTTG– 3’ and R, 5’- TGTAGACCATGTAGTTGAGGTCA-3’).
Supplementary Material
Key findings.
Sprouty gene deletion results in multiple developmental phenotypes characteristic of 22q11.2 deletion syndrome
Contrary to expectations, reducing the Tbx1 gene dosage significantly exacerbates these phenotypes
Tbx1 haploinsufficiency in the context of Sprouty gene deletion is associated with increased Receptor Tyrosine Kinase (RTK) signalling, suggesting that TBX1 can prevent excessive RTK signalling levels during development
Acknowledgements
We thank Gail Martin in whose laboratory this project was initiated, for her support and for providing βactin-cre and Spry2flox mouse lines, Gail Martin (Spry1, Spry2), Silvia Arber (Etv5) and Virginia Papaioannou (Tbx1) for generously provided in situ probes, Hagen Schmidt and Samantha Martin for technical assistance, Raj Thakker, Pete Scambler, Denny Monks and our laboratory colleagues for suggestions, discussions and comments on the manuscript. All animal experiments were approved by and performed according to a Project Licence from the UK Home Office. This work was supported by a grant from the Medical Research Council (G0601104) to MAB, a British Heart Foundation Chair Award (CH/09/003), EU FP7-HEALTH B Program Grant (223463) and Cardiogenet BHF Program Grant (RG/10/17/28553) to SB, and NIH grants (R01 HL088698 and P01 HD070454) to BM.
References
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- Aggarwal VS, Liao J, Bondarev A, Schimmang T, Lewandoski M, Locker J, Shanske A, Campione M, Morrow BE. Dissection of Tbx1 and Fgf interactions in mouse models of 22q11DS suggests functional redundancy. Hum Mol Genet. 2006;15:3219–3228. doi: 10.1093/hmg/ddl399. [DOI] [PubMed] [Google Scholar]
- Arnold JS, Braunstein EM, Ohyama T, Groves AK, Adams JC, Brown MC, Morrow BE. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Hum Mol Genet. 2006a;15:1629–1639. doi: 10.1093/hmg/ddl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JS, Werling U, Braunstein EM, Liao J, Nowotschin S, Edelmann W, Hebert JM, Morrow BE. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development. 2006b;133:977–987. doi: 10.1242/dev.02264. [DOI] [PubMed] [Google Scholar]
- Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev. 2005;15:279–284. doi: 10.1016/j.gde.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Basson MA, Echevarria D, Ahn CP, Sudarov A, Joyner AL, Mason IJ, Martinez S, Martin GR. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development. 2008;135:889–898. doi: 10.1242/dev.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O'Leary LA, Wong LY, Elixson EM, Mahle WT, Campbell RM. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112:101–107. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- Brown CB, Wenning JM, Lu MM, Epstein DJ, Meyers EN, Epstein JA. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev Biol. 2004;267:190–202. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Carlson C, Papolos D, Pandita RK, Faedda GL, Veit S, Goldberg R, Shprintzen R, Kucherlapati R, Morrow B. Molecular analysis of velo-cardio-facial syndrome patients with psychiatric disorders. Am J Hum Genet. 1997;60:851–859. [PMC free article] [PubMed] [Google Scholar]
- Chambers D, Medhurst AD, Walsh FS, Price J, Mason I. Differential display of genes expressed at the midbrain - hindbrain junction identifies sprouty2: an FGF8-inducible member of a family of intracellular FGF antagonists. Mol Cell Neurosci. 2000;15:22–35. doi: 10.1006/mcne.1999.0801. [DOI] [PubMed] [Google Scholar]
- Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res. 2009;105:842–851. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Klingensmith J. Chordin is a modifier of tbx1 for the craniofacial malformations of 22q11 deletion syndrome phenotypes in mouse. PLoS Genet. 2009;5:e1000395. doi: 10.1371/journal.pgen.1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DA, Boland T, Emanuel BS, Kirschner RE, LaRossa D, Manson J, McDonald-McGinn D, Randall P, Solot C, Zackai E, Mitchell LE. Evaluation of potential modifiers of the palatal phenotype in the 22q11.2 deletion syndrome. Cleft Palate Craniofac J. 2006;43:435–441. doi: 10.1597/05-070R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenauer DA, Bredenfeld H, Haverkamp H, Muller H, Franklin J, Fuchs M, Borchmann P, Muller-Hermelink HK, Eich HT, Muller RP, Diehl V, Engert A. Hodgkin's lymphoma in adolescents treated with adult protocols: a report from the German Hodgkin study group. J Clin Oncol. 2009;27:6079–6085. doi: 10.1200/JCO.2008.20.2655. [DOI] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcoli FG, Huynh T, Scambler PJ, Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS One. 2009;4:e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Funke B, Epstein JA, Kochilas LK, Lu MM, Pandita RK, Liao J, Bauerndistel R, Schuler T, Schorle H, Brown MC, Adams J, Morrow BE. Mice overexpressing genes from the 22q11 region deleted in velo-cardio-facial syndrome/DiGeorge syndrome have middle and inner ear defects. Hum Mol Genet. 2001;10:2549–2556. doi: 10.1093/hmg/10.22.2549. [DOI] [PubMed] [Google Scholar]
- Garg V, Yamagishi C, Hu T, Kathiriya IS, Yamagishi H, Srivastava D. Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol. 2001;235:62–73. doi: 10.1006/dbio.2001.0283. [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Driscoll DA, Emanuel BS, McDonald-McGinn D, Mei M, Zackai E, Mitchell LE. Evaluation of potential modifiers of the cardiac phenotype in the 22q11.2 deletion syndrome. Birth Defects Res A Clin Mol Teratol. 2009;85:125–129. doi: 10.1002/bdra.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Gottlieb S, Collins J, Blescia A, Dietz H, Goldmuntz E, McDonald-McGinn DM, Zackai EH, Emanuel BS, Driscoll DA, Budarf ML. Mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects. J Med Genet. 2001;38:E45. doi: 10.1136/jmg.38.12.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodship J, Cross I, Scambler P, Burn J. Monozygotic twins with chromosome 22q11 deletion and discordant phenotype. J Med Genet. 1995;32:746–748. doi: 10.1136/jmg.32.9.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- Herbst C, Rehan FA, Brillant C, Bohlius J, Skoetz N, Schulz H, Monsef I, Specht L, Engert A. Combined modality treatment improves tumor control and overall survival in patients with early stage Hodgkin's lymphoma: a systematic review. Haematologica. 2010;95:494–500. doi: 10.3324/haematol.2009.015644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillebrand G, Siebert R, Simeoni E, Santer R. DiGeorge syndrome with discordant phenotype in monozygotic twins. J Med Genet. 2000;37:E23. doi: 10.1136/jmg.37.9.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma T, Nakajima Y, Nakamura H. Development of pharyngeal arch arteries in early mouse embryo. J Anat. 2002;201:15–29. doi: 10.1046/j.1469-7580.2002.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatan I, Dastani Z, Do R, Weissglas-Volkov D, Ruel I, Lee JC, Huertas-Vazquez A, Taskinen MR, Prat A, Seidah NG, Pajukanta P, Engert JC, Genest J. Genetic variation at the proprotein convertase subtilisin/kexin type 5 gene modulates high-density lipoprotein cholesterol levels. Circ Cardiovasc Genet. 2009;2:467–475. doi: 10.1161/CIRCGENETICS.109.877811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet. 2007;370:1443–1452. doi: 10.1016/S0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Taketomi T, Yoshizaki K, Arai S, Sanui T, Yoshiga D, Yoshimura A, Nakamura S. Sprouty2 controls proliferation of palate mesenchymal cells via fibroblast growth factor signaling. Biochem Biophys Res Commun. 2011;404:1076–1082. doi: 10.1016/j.bbrc.2010.12.116. [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE. Genomic disorders on 22q11. Am J Hum Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010 doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow B, Goldberg R, Carlson C, Das Gupta R, Sirotkin H, Collins J, Dunham I, O'Donnell H, Scambler P, Shprintzen R, et al. Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am J Hum Genet. 1995;56:1391–1403. [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall V, McCue K, Roberts C, Kyriakopoulou V, Beddow S, Barrett AN, Vitelli F, Prescott K, Shaw-Smith C, Devriendt K, Bosman E, Steffes G, Steel KP, Simrick S, Basson MA, Illingworth E, Scambler PJ. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J Clin Invest. 2009;119:3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Hum Mol Genet. 2006;15:3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet. 2000;9:2421–2426. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- Scambler PJ, Carey AH, Wyse RK, Roach S, Dumanski JP, Nordenskjold M, Williamson R. Microdeletions within 22q11 associated with sporadic and familial DiGeorge syndrome. Genomics. 1991;10:201–206. doi: 10.1016/0888-7543(91)90501-5. [DOI] [PubMed] [Google Scholar]
- Schneider JE, Bhattacharya S. Making the mouse embryo transparent: identifying developmental malformations using magnetic resonance imaging. Birth Defects Res C Embryo Today. 2004;72:241–249. doi: 10.1002/bdrc.20017. [DOI] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Simrick S, Lickert H, Basson MA. Sprouty genes are essential for the normal development of epibranchial ganglia in the mouse embryo. Dev Biol. 2011;358:147–155. doi: 10.1016/j.ydbio.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, O'Reilly VC, Saga Y, Zackai EH, Dormans JP, Alman BA, McGregor L, Kageyama R, Kusumi K, Dunwoodie SL. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306. doi: 10.1016/j.cell.2012.02.054. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Stalmans I, Lambrechts D, De Smet F, Jansen S, Wang J, Maity S, Kneer P, von der Ohe M, Swillen A, Maes C, Gewillig M, Molin DG, Hellings P, Boetel T, Haardt M, Compernolle V, Dewerchin M, Plaisance S, Vlietinck R, Emanuel B, Gittenberger-de Groot AC, Scambler P, Morrow B, Driscol DA, Moons L, Esguerra CV, Carmeliet G, Behn-Krappa A, Devriendt K, Collen D, Conway SJ, Carmeliet P. VEGF: a modifier of the del22q11 (DiGeorge) syndrome? Nat Med. 2003;9:173–182. doi: 10.1038/nm819. [DOI] [PubMed] [Google Scholar]
- Taddei I, Morishima M, Huynh T, Lindsay EA. Genetic factors are major determinants of phenotypic variability in a mouse model of the DiGeorge/del22q11 syndromes. Proc Natl Acad Sci U S A. 2001;98:11428–11431. doi: 10.1073/pnas.201127298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Ayada T, Ichiyama K, Kohno R, Yonemitsu Y, Minami Y, Kikuchi A, Maehara Y, Yoshimura A. Sprouty2 and Sprouty4 are essential for embryonic morphogenesis and regulation of FGF signaling. Biochem Biophys Res Commun. 2007;352:896–902. doi: 10.1016/j.bbrc.2006.11.107. [DOI] [PubMed] [Google Scholar]
- Vincent MC, Heitz F, Tricoire J, Bourrouillou G, Kuhlein E, Rolland M, Calvas P. 22q11 deletion in DGS/VCFS monozygotic twins with discordant phenotypes. Genet Couns. 1999;10:43–49. [PubMed] [Google Scholar]
- Vitelli F, Huynh T, Baldini A. Gain of function of Tbx1 affects pharyngeal and heart development in the mouse. Genesis. 2009;47:188–195. doi: 10.1002/dvg.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002;129:4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Zhang Z, Huynh T, Sobotka A, Mupo A, Baldini A. Fgf8 expression in the Tbx1 domain causes skeletal abnormalities and modifies the aortic arch but not the outflow tract phenotype of Tbx1 mutants. Dev Biol. 2006;295:559–570. doi: 10.1016/j.ydbio.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh IC, Hagge-Greenberg A, O'Brien TP. A dosage-dependent role for Spry2 in growth and patterning during palate development. Mech Dev. 2007;124:746–761. doi: 10.1016/j.mod.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdak H, Ittner LM, Sommer L. DiGeorge syndrome and pharyngeal apparatus development. Bioessays. 2006;28:1078–1086. doi: 10.1002/bies.20484. [DOI] [PubMed] [Google Scholar]
- Xu H, Cerrato F, Baldini A. Timed mutation and cell-fate mapping reveal reiterated roles of Tbx1 during embryogenesis, and a crucial function during segmentation of the pharyngeal system via regulation of endoderm expansion. Development. 2005;132:4387–4395. doi: 10.1242/dev.02018. [DOI] [PubMed] [Google Scholar]
- Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Ishii C, Maeda J, Kojima Y, Matsuoka R, Kimura M, Takao A, Momma K, Matsuo N. Phenotypic discordance in monozygotic twins with 22q11.2 deletion. Am J Med Genet. 1998;78:319–321. [PubMed] [Google Scholar]
- Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, Meyers EN, Yamagishi C, Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kilgallen S, Andreeva V, Spicer DB, Pinz I, Friesel R. Conditional expression of Spry1 in neural crest causes craniofacial and cardiac defects. BMC Dev Biol. 2010;10:48. doi: 10.1186/1471-213X-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2008;17:150–157. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- Zweier C, Sticht H, Aydin-Yaylagul I, Campbell CE, Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am J Hum Genet. 2007;80:510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.