Abstract
Amyotrophic lateral sclerosis (ALS) is caused by the progressive degeneration of motor neurons. Mutations in the Cu/Zn superoxide dismutase (SOD1) are found in about 20% of patients with familial ALS. Mutant SOD1 causes motor neuron death through an acquired toxic property. Although the molecular mechanism underlying this toxic gain-of-function remains unknown, evidence support the role of mutant SOD1 expression in non-neuronal cells in shaping motor neuron degeneration. We have previously found that in contrast to non-transgenic cells, SOD1G93A-expressing astrocytes induced apoptosis of co-cultured motor neurons. This prompted us to investigate whether the effect on motor neuron survival was related to a change in the gene expression profile. Through high-density oligonucleotide microarrays, we found changes in the expression of genes involved in transcription, signaling, cell proliferation, extracellular matrix synthesis, response to stress and steroid and lipid metabolism. The most up-regulated gene was decorin (Dcn), a small multifunctional extracellular proteoglycan. Down-regulated genes included the insulin-like growth factor-1 receptor (Igf-1r) and the RNA binding protein ROD1. Rod1 was also found downregulated in purified motor neurons expressing SOD1G93A. Changes in the expression of Dcn, Igf-1r and Rod1 were found in the spinal cord of asymptomatic animals, suggesting these changes occur before overt neuronal degeneration and potentially influence astrocyte-motor neuron interaction in the course of the disease. The astrocyte-specific gene expression profile might contribute to the identification of possible candidates for cell type-specific therapies in ALS.
Keywords: motor neurons, decorin, IGF-1R, ROD1, microarray
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease, caused by the progressive degeneration of motor neurons in the spinal cord, brainstem and motor cortex. Motor neuron death leads to muscle weakness and paralysis causing death in one to five years from symptom onset (Rowland & Schneider, 2001). About 10% of ALS shows a familial inheritance and 10–20% of these familial ALS are caused by mutations of the Cu/Zn superoxide dismutase (SOD1) (Rosen et al., 1993). A toxic gain-of-function of mutant SOD1 has been considered the cause of neurodegeneration, based on the observation that mice lacking endogenous SOD1 fail to show motor neuron degeneration, while rodents over-expressing human mutant SOD1 generally develop an ALS-like phenotype (Gurney et al., 1994; Reaume et al., 1996; Howland et al., 2002). Several hypotheses, including oxidative stress, glutamate excitotoxicity, formation of high molecular weight aggregates, and mitochondrial dysfunction have been proposed to explain the toxic effect of mutated SOD1 (Beckman et al., 2001; Cleveland & Rothstein, 2001; Bruijn et al., 2004; Manfredi & Xu, 2005).
Although the molecular mechanism underlying this toxic gain-of-function remains unknown, several lines of evidence suggest that ALS is not a cell-autonomous disease. Transgenic mice expressing mutated forms of SOD1 exclusively in neurons (Pramatarova et al., 2001; Lino et al., 2002) or astrocytes (Gong et al., 2000) fail to develop the disease. Moreover, toxicity to motor neurons requires damage caused by mutant SOD1 expression in non-neuronal cells (Clement et al., 2003). A large proportion of non-neuronal cells in the ventral horn of the spinal cord are astrocytes, which closely interact with neurons to provide structural, metabolic and trophic support and actively participate in modulating neuronal excitability and neurotransmission (Volterra & Meldolesi, 2005). Following injury, astrocytes respond by proliferating and adopting a reactive phenotype characterized morphologically by hypertrophic nuclei and cell bodies and by elaboration of distinct long and thick processes with increased content of glial fibrillary acidic protein (GFAP). In addition, morphological alterations are accompanied by changes in the expression of cytoskeleton proteins, cell surface and matrix molecules, proteases, protease inhibitors, several growth factors and cytokines (Ridet et al., 1997). Therefore, functional alterations in activated astrocytes can shape the interaction with surrounding cells such as damaged neurons, microglia and immune cells, and consequently could modulate the survival of motor neurons.
Reactive glial changes occur in most neurodegenerative diseases. In ALS patients, a strong glial reaction typically surrounds both upper and lower motor neurons (Hirano 1996; Kushner et al., 1991; Nagy et al., 1994). Astrocytes in ALS show increased immunoreactivity for GFAP and the calcium binding protein S100β and express inflammatory makers such as COX-2, inducible nitric oxide synthase (NOS) and neuronal NOS (Migheli et al., 1999; Sasaki et al., 2000; Anneser et al., 2001; Maihofner et al., 2003). In addition, astrocyte pathology is accompanied by markers of oxidative and nitrative stress (Abe et al., 1997; Sasaki et al., 2000). A similar pattern of astrocytic changes has been described in mice and rats models of ALS (Gurney et al., 1994; Bruijn et al., 1997; Hall et al., 1998; Almer et al., 1999; Nagai et al., 2001; Howland et al., 2002). This evidence has prompted us to investigate the role of astrocytes in the disease. Reactive astrocytes in ALS may promote the elimination of damaged motor neurons re-expressing the p75 neurotrophin receptor, while increasing the support for remaining healthy neurons (Pehar et al., 2004, Cassina et al., 2005; Barbeito et al., 2004, Vargas et al., 2005). The involvement of these cells in pathogenesis offers a potential target for developing novel strategies to prevent neurodegeneration occurring in ALS.
Primary spinal cord astrocytes monolayers support the survival of highly purified embryonic motor neurons in the absence of any added trophic factor. However, when astrocytes are pre-treated with LPS, peroxynitrite or FGF-1, activated astrocytes cause a 40% neuronal loss over the next 48–72 h (Cassina et al., 2002; Pehar et al., 2004, Cassina et al., 2005). We have reported that in contrast to the trophic support provided by non-transgenic astrocytes, 40% of motor neurons were lost when co-cultured on untreated astrocytes isolated from SOD1G93A rats (Vargas et al., 2006). Recently, Nagai et al. (2007) have confirmed our previous results and extended this observation to astrocytes expressing other ALS-linked mutant SOD1. Because the restricted expression of mutant hSOD1 to astrocytes causes the death of non-transgenic motor neurons in our co-culture model, we investigated whether this observation could be related to changes in gene expression induced by SOD1G93A in astrocytes. The differences in gene expression were compared between resting non-transgenic and SOD1G93A spinal cord astrocytes using the Affymetrix microarray system (Rat 230 2.0) to monitor the expression profile of about 28,000 rat genes in astrocytes. The results obtained may help to understand the role of astrocytes and their interaction with motor neurons in ALS.
Materials and Methods
Materials
Culture media and serum were obtained from Gibco-Invitrogen (CA, USA). Primers were obtained from Integrated DNA Technologies, Inc (IA, USA). Anti-Decorin antibody (LF-114) was kindly provided by Dr. Larry W. Fisher (Fisher et al., 1995) and anti-IGF-1Rβ was from Santa Cruz Biotech. (CA, USA). Antibodies to GFAP, tubulin and actin were from Sigma (MO, USA). All other reagents were from Sigma unless otherwise specified.
Cell cultures
Primary astrocyte cultures were prepared from spinal cords of 1–2 day old rat pups according to the procedures of Saneto and De Vellis (1987) with minor modifications (Cassina et al., 2002). Purified astrocyte monolayers were obtained after 14 days in culture. At this point, astrocytes were plated at a density of 2×104 cells/cm2 and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, HEPES (3.6 g/L), penicillin (100 IU/mL) and streptomycin (100 μg/mL) for additional 7 days. Thus, all experiments were performed with astrocytes cultured for a total of 21 days. The same paradigm was used in our previous co-culture studies where altered astrocyte-motor neuron interaction was observed (Vargas et al., 2006). Astrocyte monolayers were >98% pure as determined by GFAP immunoreactivity and devoid of OX42-positive microglial cells. In order to have astrocyte cultures of both genotypes in each preparation, cultures were prepared from transgenic SOD1G93A and non-transgenic littermates genotyped by PCR. Sprague-Dawley SOD1G93A L26H rats were kindly provided by Dr. David S. Howland (Wyeth Research, Princeton, NJ) (Howland et al., 2002). Motor neurons were isolated from embryonic day 15 (E15) rat spinal cord by a combination of metrizamide-gradient centrifugation and immunopanning with the monoclonal antibody IgG192 against p75NTR (Henderson et al., 1995).
RNA isolation
Confluent astrocytes monolayers were changed to supplemented L15 medium (Vargas et al., 2006) for 24h before RNA isolation. Total RNA was isolated with RNeasy kit/RNase-Free DNase Set (Qiagen, CA, USA). RNA quality was assessed with the A260/280 ratio and the 2100 Bioanalyzer (Agilent Technologies, CA, USA) to ensure the integrity of the samples used for microarray analysis.
Animals
Sprague-Dawley SOD1G93A L26H rats were observed daily for onset of disease symptoms as well as progression to death. Onset of motor neuron disease was scored as the first observation of an abnormal gait or evidence of hind limb weakness (typically after 135d). Five asymptomatic (60d) and five at the age of symptoms onset SOD1G93A animals and the same number of age-matched nontransgenic (NonTG) rats were used. Animal procedures followed the guidelines for Care and Use of Laboratory Animals established by the National Institute of Health. Lumbar spinal cords were immediately frozen in liquid nitrogen followed by mortar fragmentation; total RNA was isolated with RNeasy kit (Qiagen) after discarding the upper lipid layer from the lysate in the RLT buffer.
Microarray analysis
Double-stranded cDNA was synthesized from 5 μg of total RNA and used for microarray analysis with the Rat Genome 230 2.0 array (Affymetrix, CA, USA). A total of six arrays divided in three control and three transgenic samples were used. Labeling was performed with one-cycle target labeling assay according to Affymetrix, including the eukaryotic poly-A RNA control and the eukaryotic hybridization control kit. Hybridization, washing and scanning were carried out as described in the Affymetrix GeneChip expression analysis technical manual at the Oregon State University’s Center for Genome Research and Biocomputing. Image processing was done using Affymetrix GCOS 1.4 software. The quality of hybridization and overall chip performance was determined by visual inspection of the raw scanned data and the GCOS-generated report file. Microarray data (.CEL files) was normalized using GC-RMA probe-level analysis in ArrayAssist 4.0 (Stratagene, CA, USA). Following variance stabilization and Log transformation, the fold-change versus p-value was calculated using nontransgenic samples as base values. Selected probe sets were annotated using NetAffx (www.affymetrix.com/analysis/index.affx) and Gene Ontology Consortium (www.geneontology.org) database in early 2006. Affymetrix probe set identifiers were replaced with official gene nomenclature using NCBI and NetAffx databases. In compliance with MIAME standards, Affymetrix data files (CHP and CEL) have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (GSE7441).
Relative quantitative RT-PCR
2 μg of total RNA were randomly reverse-transcribed using SuperScript II reverse transcriptase (Invitrogen, CA, USA) according to the manufacturer’s protocol. Specific primers pairs and QuantumRNA Classic 18S Internal Standards primers (Ambion, TX, USA) were used together in the reaction to amplify the target cDNA and a 489 bp from 18S rRNA cDNA. PCR primers were as follows, Dcn: 5’-ATCTCCGAAGTTGGGCAGCATG-3’, and 5’-CTGAAGGTGTGTGGGTGAATTTGC-3’ (125 bp); Igf-1r: 5’-CGCAGGATGGCTATCTGTTCCG-3’, 5’-GGTCTGGGCACAAAGATGGAGTTG-3’ (250 bp); Rod1: 5’-ACATCTTCCCTCCGTCAGCCAC-3’, 5’-TTGCTTCCTCCACAGATCCCAAC-3’ (171 bp). PCRs were carried out in a 50 μl reaction volume containing 1 μl of cDNA, 20 pmoles of each specific primer, 18S primers (1:9 ratio), 200 μM dNTPs, 1.5 mM MgCl2, 1.5 U of PlatiniumTaq and 1X PCR buffer (Invitrogen, CA, USA). The cycling parameters were as follows: 95°C, 30 sec; 55°C, 30 sec; 72°C, 30 sec, for 19 to 22 cycles depending on the specific linear range of each amplicon. Minus RT controls were included in each assay. The amplification products were run in non-denaturing 6% polyacrylamide gels and stained with SYBR Gold Nucleic Acid Gel Stain (Molecular Probes, OR, USA). Densitometric analyses were performed using the NIH Image program and gene expression levels were normalized against the 18S levels.
Western blot analysis
Confluent astrocyte culture were washed with cold PBS and whole cell extracts were prepared in 50 mM HEPES, pH 7.5, 50 mM NaCl, 1% Triton X-100, and Complete protease inhibitor cocktail (Roche) and sonicated 3 times for 3 s. Spinal cords were removed and processed in SDS 1% plus Complete protease inhibitor cocktail (Roche) for protein extraction. Protein concentration was measured by the bicinchoninic acid method (Pierce). Protein samples were resolved on 12% SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Amersham Biosciences). Membranes were blocked for 1 h in Tris-buffered saline, 0.1% Tween-20, and 5% nonfat dry milk, followed by an overnight incubation with primary antibody diluted in the same buffer. After washing with 0.1% Tween in Tris-buffered saline the membrane was incubated with peroxidase-conjugated secondary antibody (Bio-Rad) for 1 h, and then washed and developed using the ECL chemiluminescent detection system (Amersham Biosciences). Primary antibodies were used at dilutions recommended by the manufacturers.
Immunofluorescence
Astrocyte cultures on Lab-Tek (Nunc, IL, USA) slides were fixed in ice-cold 4% paraformaldehyde in PBS. Briefly, cultures were permeabilized with 0.1% Triton X-100 in PBS for 15 min and blocked for 1h with 10% goat serum, 2% bovine serum albumin, and 0.1% Triton X-100 in PBS. Primary antibodies diluted in blocking solution were incubated overnight at 4 °C. After washing with PBS, fluorophore-conjugated anti-rabbit or anti-mouse secondary antibodies diluted in blocking solution were incubated for 1h at room temperature. The slides were mounted using ProLong anti-fade kit (Molecular Probes, OR, USA). Primary antibodies used were anti-GFAP polyclonal antibody (1:400) and anti-tubulin monoclonal antibody (1:2000). Secondary antibodies were Alexa Fluor488-conjugated goat anti-rabbit (5 μg/mL; Molecular Probes) and AlexaFluor594-conjugated goat anti-mouse (5 μg/mL; Molecular Probes). Nuclei were stained with DAPI (1 μg/mL; Molecular Probes, OR, USA).
Statistics
Each experiment was repeated at least three times and data are reported as mean ± SD. Comparison of the means was performed by one-way analysis of variance. Pairwise contrasts between means utilized the Student-Newman-Keuls test and differences were declared statistically significant if p<0.05. All statistical computations were performed using the SigmaStat Software (Jandel Scientific).
Results
Morphological analysis
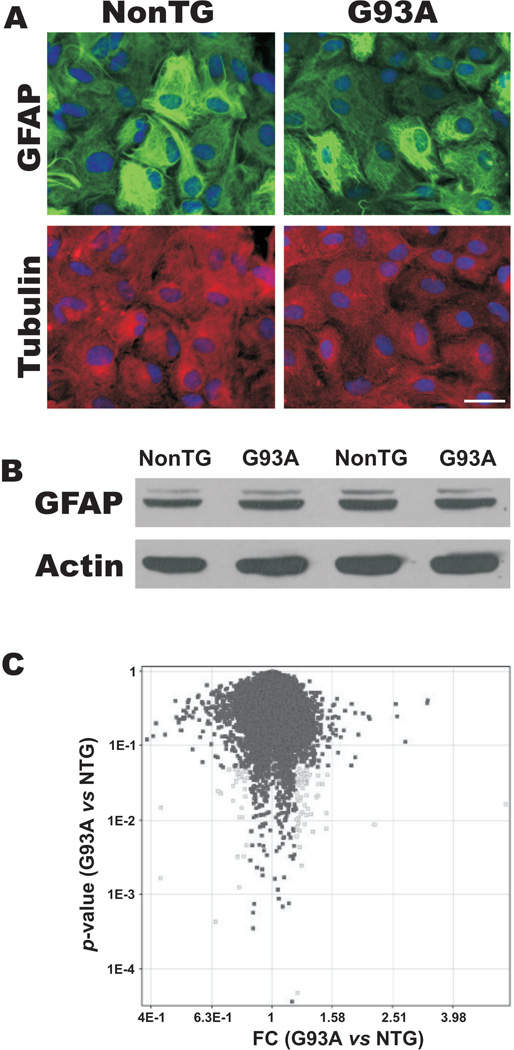
Primary spinal cord astrocytes expressing hSOD1G93A are morphologically equivalent to non-transgenic astrocytes in culture, as determined by immunofluorescence for GFAP and α-tubulin (Fig. 1A). SOD1G93A astrocytes did not develop the typical thick and long processes with increased immunoreactivity for GFAP that characterize reactive astrocytes. Accordingly, no increase on GFAP expression was observed by western blot (Fig. 1B). Thus, in vitro the expression of mutant SOD1 alone did not cause the typical astrocyte activation found in ALS. However, under the same conditions, SOD1G93A astrocytes induced apoptosis of co-cultured motor neurons (Vargas et al., 2006). This prompted us to investigate whether the effect on motor neuron survival was related to changes in the gene expression pattern of astrocytes affecting the interaction with motor neurons.
Figure 1.
A) Microphotography showing morphological aspect of non-transgenic (NonTG) and SOD1G93A (G93A) spinal cord astrocyte monolayers. Cultures were stained for GFAP (green) and α-tubulin (red). Nuclei were stained with DAPI. Scale bar: 30 μm. B) GFAP protein expression analyzed by western blot in non-transgenic (NonTG) and SOD1G93A spinal cord astrocytes. β-Actin is shown as a loading control. C) Scatter plot of the fold-change (FC G93A vs. NonTG) versus the result p-value of significance analysis. Probe intensity data were normalized using GC-RMA. Following variance stabilization and Log transformation, the fold-change versus the p-value was calculated using non-transgenic samples as base values. Probe sets with significant change (p<0.05) of at least 1.2 fold are marked in gray.
Gene expression profiling
Microarrays analyses were performed with RNA samples from 3 independent preparations of primary astrocytes obtained from SOD1G93A animals and non-transgenic littermates. We used the Affymetrix Rat Genome 230 2.0 array, which contains over 31,000 probe sets comprising about 28,000 rat genes, followed by GC-RMA normalization. We selected genes with at least 1.2-fold change in non-transgenic and SOD1G93A astrocytes with a statistical significance of p<0.05 (Gibson, 2003). The analyses identified a total of 81 transcripts that were statistically different in SOD1G93A astrocytes: 55 transcripts were up-regulated and 26 were down-regulated (Table 1 and 2). The genes were categorized taking into account their molecular function, biological process and cellular component. Approximately half of the genes in each category have no functional information available.
Table I.
Genes increased in SOD1G93A astrocytes.
| Accession Number | Gene | Fold | p-value |
|---|---|---|---|
| Extracellular matrix/Cell migration | |||
| NM_024129 | Decorin | 5.95 | 0.01676 |
| NM_080394 | Reelin | 1.52 | 0.04025 |
| NM_017089 | Ephrin B1 | 1.30 | 0.03252 |
| AI406747 | Neuron-glia-CAM-related cell adhesion molecule | 1.27 | 0.03799 |
| Signaling/Receptor activity | |||
| NM_080586 | GABA A receptor, gamma 1 | 1.42 | 0.03069 |
| AA859669 | Neuropilin 2 | 1.28 | 0.01612 |
| BC087578 | Arrestin, beta 2 | 1.28 | 0.00902 |
| NM_053856 | Secretogranin III | 1.28 | 0.04208 |
| NM_001012061 | Rattus norvegicus Cnksr family member 3 (Cnksr3) | 1.25 | 0.00248 |
| NM_001013192 | Similar to olfactomedin-like 1 | 1.25 | 0.04174 |
| Transcription | |||
| XM_232488 | Atf7 interacting protein (predicted) | 1.27 | 0.03522 |
| NM_057211 | Basic transcription element binding protein 1 | 1.24 | 0.01243 |
| XM_237754 | Zinc finger protein 213 (predicted) | 1.23 | 0.01902 |
| NM_012866 | Nuclear transcription factor-Y gamma | 1.20 | 0.00762 |
| Cell Proliferation | |||
| BF408114 | Breast cancer metastasis-suppressor 1 (predicted) | 1.41 | 0.02159 |
| Response to stress | |||
| NM_148889 | Aprataxin | 1.22 | 0.04865 |
| NM_021863 | Heat shock protein 2 | 1.22 | 0.01087 |
| Oxidoreductase activity/Metabolism/Transport | |||
| NM_053896 | Aldehyde dehydrogenase family 1, subfamily A2 | 2.18 | 0.00889 |
| AA997683 | Aldehyde dehydrogenase 1 family, member B1 (predicted) | 1.47 | 0.03085 |
| NM_017217 | Solute carrier family 7, member 3 (Slc7a3) | 1.36 | 0.00714 |
| XM_224713 | Abhydrolase domain containing 8 (predicted) | 1.34 | 0.04100 |
| XM_220931 | RAD52 homolog B (S. cerevisiae) (predicted) | 1.31 | 0.03727 |
| L21698 | UDP-glucuronosyltransferase 8 | 1.28 | 0.04557 |
| AI408770 | Villin (predicted) | 1.25 | 0.04092 |
| NM_017268 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | 1.21 | 0.00005 |
| Catalytic activity | |||
| NM_001025017 | Similar to 4921507I02Rik protein (predicted) | 1.23 | 0.00441 |
| BI285949 | Guanosine monophosphate reductase 2 | 1.22 | 0.01854 |
| Unknown function | |||
| BI283971 | Transcribed locus | 1.51 | 0.00783 |
| BE103036 | cDNA clone | 1.51 | 0.01256 |
| BF400747 | Transcribed locus | 1.42 | 0.03988 |
| AW522471 | Transcribed locus | 1.40 | 0.02077 |
| BF290416 | Transcribed locus | 1.34 | 0.01223 |
| XM_341321 | Similar to HCDI protein (predicted) | 1.31 | 0.01455 |
| AI043758 | Transcribed locus | 1.30 | 0.02167 |
| AI511280 | Similar to DNA segment, Chr 3, ERATO Doi 250, (predicted) | 1.30 | 0.01190 |
| AW528715 | Transcribed locus | 1.30 | 0.04675 |
| AA943385 | Similar to hypothetical protein FLJ13273 (predicted) | 1.29 | 0.03120 |
| XM_575385 | Similar to PHD finger protein 14 isoform 1 | 1.28 | 0.03260 |
| BF402699 | Transcribed locus | 1.26 | 0.01305 |
| BI276075 | Transcribed locus | 1.26 | 0.02746 |
| AI409701 | Transcribed locus | 1.24 | 0.04551 |
| AI409069 | Similar to MEGF11 protein | 1.24 | 0.02068 |
| BE108716 | Similar to MGC68837 protein | 1.23 | 0.03232 |
| XM_343161 | MOB1, Mps One Binder kinase activator-like 2B (predicted) | 1.23 | 0.03461 |
| XM_229106 | Oculocerebrorenal syndrome of Lowe | 1.23 | 0.00738 |
| BF390720 | cDNA clone | 1.22 | 0.00933 |
| AI104921 | Transcribed locus | 1.22 | 0.03004 |
| BI303858 | Transcription factor Pur-beta | 1.21 | 0.02020 |
| AI101553 | Transcribed locus | 1.21 | 0.03558 |
| BI291954 | Transcribed locus | 1.21 | 0.04310 |
| AW528635 | Transcribed locus | 1.21 | 0.00823 |
| AI070952 | Transcribed locus | 1.21 | 0.03260 |
| XM_233227 | Similar to KIAA1573 protein (predicted) | 1.20 | 0.00620 |
| XM_227208 | Similar to Hypothetical protein MGC57096 | 1.20 | 0.01042 |
| NM_001037656 | Similar to lactation elevated 1 | 1.20 | 0.02391 |
Table II.
Genes decreased in SOD1G93A astrocytes.
| Accession Number |
Gene | Fold | p-value |
|---|---|---|---|
| Cell proliferation/Signaling | |||
| NM_022282 | Discs, large homolog 2 (Drosophila) | −1.52 | 0.02524 |
| BI289840 | Fibroblast growth factor 1 | −1.29 | 0.03758 |
| NM_052807 | Insulin-like growth factor 1 receptor | −1.25 | 0.04427 |
| BI289676 | LIM domains containing 1 (predicted) | −1.21 | 0.03650 |
| Nucleic acid binding | |||
| BE118358 | ROD1 regulator of differentiation 1 (S. pombe) | −2.34 | 0.00167 |
| BE104395 | Rearranged L-myc fusion sequence (predicted) | −1.31 | 0.04095 |
| BC088338 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 (predicted) | −1.25 | 0.00335 |
| Oxidoreductase activity/Metabolism | |||
| BF408445 | Glutathione peroxidase 7 (predicted) | −1.35 | 0.01854 |
| NM_012940 | Cytochrome P450, family 1, subfamily b, polypeptide 1 | −1.35 | 0.02966 |
| NM_001031662 | Coenzyme Q4 homolog (yeast) (predicted) | −1.26 | 0.01484 |
| Transport | |||
| AI411753 | Synaptobrevin-like 1 | −1.32 | 0.03394 |
| X57523 | Transporter 1, ATP-binding cassette, (MDR/TAP) | −1.24 | 0.02683 |
| Actin binding | |||
| XM_341540 | Supervillin (predicted) | −1.32 | 0.04734 |
| Unknown function | |||
| AA957343 | Transcribed locus | −1.55 | 0.00044 |
| XM_217233 | Similar to 5-3 exonuclease (predicted) | −1.39 | 0.04751 |
| BF281893 | LOC502928 | −1.48 | 0.02350 |
| AI177465 | cDNA clone | −1.31 | 0.00325 |
| AW532489 | Transcribed locus | −1.30 | 0.00128 |
| XM_574821 | Similar to MADS box transcription enhancer factor 2, C | −1.30 | 0.04758 |
| AI227948 | Similar to RIKEN cDNA 1200009B18; EST AA408438 | −1.29 | 0.04216 |
| NM_134419 | Hspb associated protein 1 (Hsp27 binding protein) | −1.29 | 0.01102 |
| BM385061 | Transcribed locus | −1.27 | 0.03868 |
| NM_138975 | WW domain binding protein 2 | −1.26 | 0.01114 |
| AI556531 | Transcribed locus, strongly similar to NP_060595.2 | −1.24 | 0.03425 |
The scatter plot of the fold-change versus the result p-value of significance analysis (Fig. 1C), shows that no major changes in the expression pattern were observed in these samples. We applied several probe-level analyses (PLIER, MBEI and MAS5) and obtained similar results, indicating that although these astrocytes displayed a strikingly functional difference, there is not a prominent difference at the transcriptional level. Interestingly, only the small proteoglycan decorin (Dcn) is greatly induced in SOD1G93A astrocytes, whereas only one gene displayed a down-regulation greater than 2-fold in expression (regulator of differentiation, Rod1).
Expression of genes previously implicated in ALS
Increased oxidative stress, as a consequence of enhanced reactive oxygen and nitrogen species generation, is known to be involved in ALS. We found increased expression of the stress response DNA-repair protein, aprataxin, which has been associated with increased resistance to oxidative stress (Hirano et al., 2006). However, we did not find changes in genes directly associated with increased production of pro-oxidant species or decreased antioxidant defenses. Our study detected no change in the expression of any of the three nitric oxide synthases, although we have previously observed a modest increased accumulation of nitrite and nitrate in the media of transgenic astrocyte-motor neuron co-cultures (Vargas et al., 2006).
In addition, no modification in the expression level of the astroglial glutamate transporter EAAT2 were detected, although diminished EAAT2 protein levels reported in ALS could be due to an aberrant splicing of EAAT2 messenger (Lin et al., 1998). In contrast to previous reports (Yoshihara et al., 2002; Dangond et al., 2004), we found an increased expression of GABA-A receptor in SOD1G93A astrocytes. However, the decreased GABA-A receptor expression previously reported might be linked to a late stage of the disease, since this studies were performed in 17-week old ALS-mice and post-mortem human tissue. Within the genes with unknown function, we found increased expression of the gene linked to the oculocerebrorenal syndrome of Lowe. Mutations in this gene have been linked to altered phosphoinositide metabolism and have also been reported to be increased in the spinal cord of sporadic ALS patients (Jiang et al., 2005).
Four genes related to cell proliferation were decreased in transgenic astrocytes (Table 2). Among them, the fibroblast growth factor-1, previously reported to be down-regulated in the anterior horn of spinal cord from ALS patients (Kage et al., 2001), was found decreased in primary SOD1G93A astrocytes. Our study also detected a decreased expression of IGF-1 receptor gene (Igf-1r).
RT-PCR verification
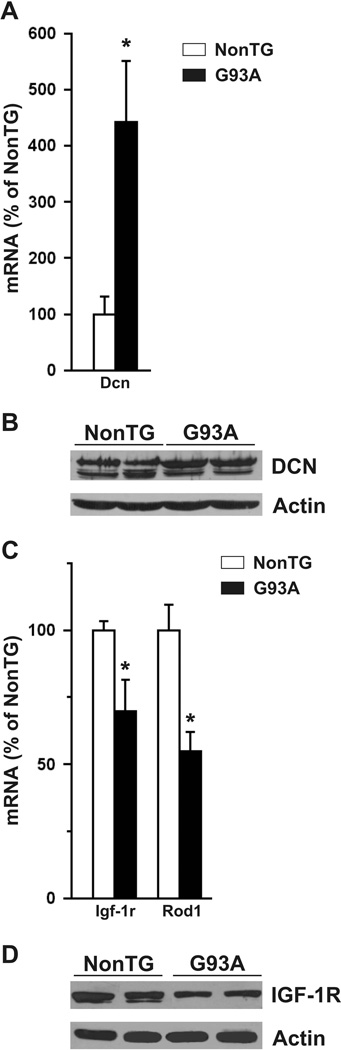
We confirmed the level of expression of Dcn, Rod1 and Igf-1r based on the largest fold-change and its putative relevance for in ALS. We used four independent astrocyte samples to assess differential expression by RT-PCR. We confirmed the up-regulation of decorin in SOD1G93A astrocytes at messenger and protein level (Fig. 2A, 2B). We also confirmed the reduced expression of the most down-regulated gene, Rod1 and the down-regulation of Igf-1r obtained from microarray data in SOD1G93A astrocytes (Fig. 2C). IGF-1R protein was also down-regulated in SOD1G93A astrocytes (Fig. 2D).
Figure 2.
Verification of selected differentially expressed genes in SOD1G93A astrocytes. Up-regulation of decorin (Dcn) mRNA (A) and protein (B) was confirmed by relative quantitative reverse transcriptase-PCR and Western blot as described under Materials and Methods. C) insulin-like growth factor-1 receptor (Igf-1r) and regulator of differentiation (Rod1) mRNA down-regulation in SOD1G93A astrocytes. D) Decreased IGF-1R expression in SOD1G93A astrocytes as determined by Western blot. In (A) and (C) mRNA levels are expressed as % of non-transgenic (NonTG) controls and corrected by 18S rRNA cDNA amplification. * Significantly different from control (p<0.05).
Expression in spinal cord of SOD1G93A rats and in purified motor neurons
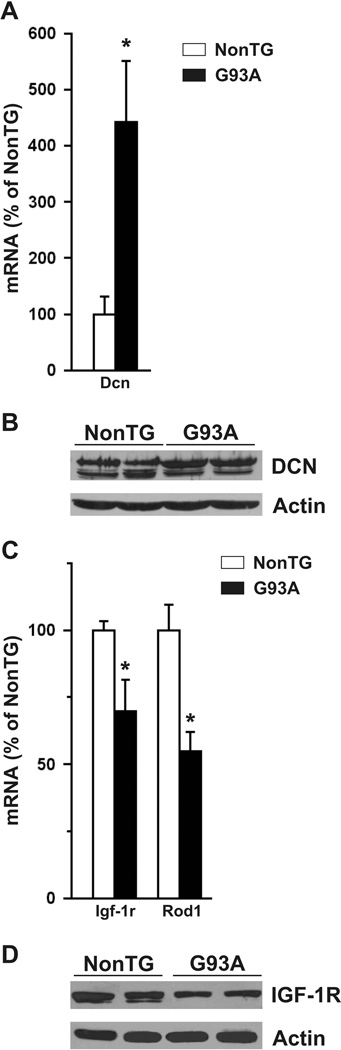
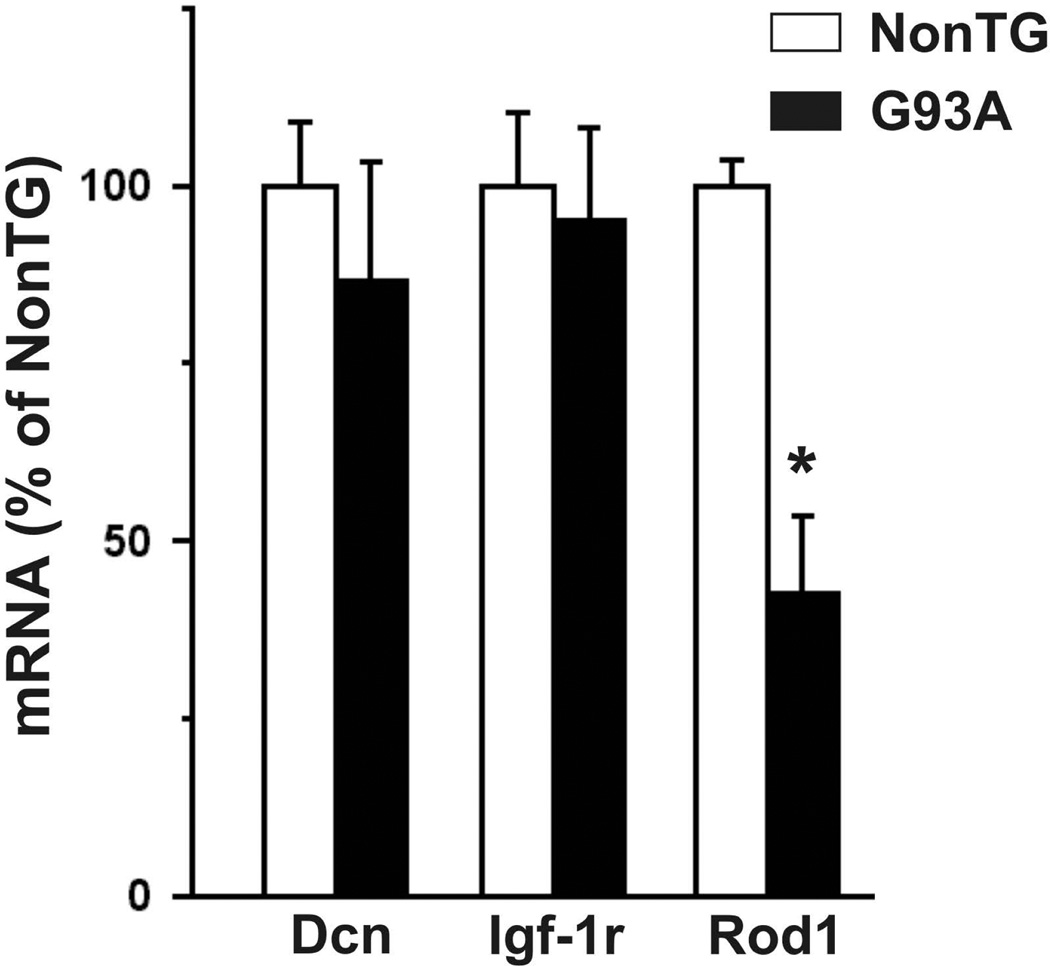
We examined the expression of Dcn, Igf-1r, and Rod1 in the spinal cord of SOD1G93A rats (Fig. 3). Dcn was found to be up-regulated, while Igf-1r and Rod1 were down-regulated in both asymptomatic (60 days old) and early symptomatic (~135 days old) transgenic animals. The increase in decorin messenger correlated with an increase in protein content in the spinal cord (Fig. 3C), whereas no apparent change in IGF-1R protein was observed (Fig. 3D). When assessed in purified embryonic motor neurons, we observed down-regulation in the expression of Rod1, while no changes in the expression of Dcn or Igf-1r were found (Fig. 4).
Figure 3.
Expression of decorin (Dcn), insulin-like growth factor-1 receptor (Igf-1r) and regulator of differentiation (Rod1) in the spinal cord of SOD1G93A rats. Comparative mRNA levels from spinal cord of age-matched non-transgenic (NonTG) rats and A, asymptomatic or B, early symptomatic SOD1G93A rats (G93A). mRNA levels were corrected by 18S rRNA cDNA amplification. Data for each stage were expressed as % of age-matched non-transgenic littermates. * Significantly different from control (p<0.05). Decorin (C) and IGF-1R (D) expression levels analysed by Western blot in the spinal cord of asymptomatic (AS) and early symptomatic (ES) SOD1G93A rats compared to age-matched non-transgenic (NonTG) animals.
Figure 4.
Expression of decorin (Dcn), insulin-like growth factor-1 receptor (Igf-1r) and regulator of differentiation (Rod1) in isolated spinal cord motor neurons from non-transgenic (NonTG) and SOD1G93A-expressing (G93A) E15 embryos. The mRNA levels were corrected by 18S rRNA cDNA amplification and expressed as % of non-transgenic (NonTG) controls. * Significantly different from control (p<0.05).
Discussion
Although the toxic effect of the gain of function of mutated SOD1 is still under intensive research, the interplay between different cell types has emerged as a major determinant in the pathophysiological mechanisms of ALS (Clement et al., 2003, Barbeito et al., 2004, Yamanaka et al., 2008). In this context, purified primary cultures represent useful tools to identify the primary molecular events of pathological importance. Astrocytes play a pivotal role in nervous system homeostasis and represent a promising target to provide neuroprotection. We have previously reported that the expression of SOD1G93A in astrocytes causes apoptosis in about 40% of co-cultured motor neurons (Vargas et al., 2006). However, the present microarray analysis of SOD1G93A astrocytes showed changes in the expression of less than 0.5% of the genes examined even though a low fold change (1.2) cutoff was used. The overall analysis showed a marked degree of transcriptional activation, as has been observed in many genome-wide analyses of tissues from ALS patients or animal models (Malaspina & de Belleroche, 2004). Despite the fact that functional categorization periodically changes and differentially expressed genes are expected to be involved in several pathways, altered expression of genes related to transcription, signaling, cell proliferation, stress responses and steroid and lipid metabolism were observed in SOD1G93A astrocytes. Although we could not directly link genes changes to the toxicity that astrocytes exert towards co-cultured motor neurons, several genes previously implicated in ALS were identified, as well as new candidates that might play a role in the disease.
Rod1 displayed a consistent decreased expression in transgenic astrocytes, motor neurons, asymptomatic and early symptomatic spinal cords. Rod1 is the functional mammalian homologue of fission yeast’s nrd1+, encoding a RNA binding protein (Yamamoto et al., 1999). Although scarce functional data have been reported for ROD1, its overexpression effectively blocks differentiation in human cells without affecting their ability to proliferate. Arrays data also revealed the reduced expression of LIM-domains-containing-1, a gene reported to be down-regulated in human lung cancer and which expression inhibits tumor growth in vitro and in vivo (Sharp et al., 2004). Together with the concomitant reduction in the expression of FGF-1 gene, the results give rise to the possibility that astrocytes in ALS have an altered pattern of proliferation and/or differentiation that may affect its interaction with surrounding cells.
Another interesting implication of Rod1 down-regulation is its potential participation in RNA processing. Rod1 is a paralog gene of the polypyrimidine tract binding protein (PTB1) (Yamamoto et al., 1999). PTB1 has diverse roles in pre-mRNA and mRNA function, including the regulation of alternative splicing. Several examples of aberrant RNA splicing exist in neurodegenerative processes (Gallo et al., 2005). In particular, the loss of the astroglial glutamate transporter EAAT2 in ALS and the occurrence of an isoform of peripherin that cause motor neuron toxicity has been linked with altered RNA processing (Lin et al., 1998; Robertson et al., 2003). In this context, the functional consequences of down-regulation of Rod1 in both astrocytes and motor neurons expressing SOD1G93A mutation deserve further investigation.
Changes in the expression of extracellular matrix-related genes in CNS affect several biological processes including proliferation, neuronal migration and axon guidance. Decorin, a multifunctional proteoglycan controlling matrix construction, was increased more than 4-fold in SOD1G93A astrocytes. Ephrin B1 and Reelin, two genes associated with neuronal migration and axon guidance (Flanagan & Vanderhaeghen, 1998; Jossin et al., 2004) were also up-regulated in transgenic astrocytes. Additionally, Dcn expression was increased in the spinal cord of SOD1G93A animals. Decorin is up-regulated in astrocytes after CNS injury and attenuates glial scar formation and inflammation (Stichel et al., 1995; Logan et al., 1999, Davies et al., 2004, 2006). These effects are mediated through the capacity of decorin to neutralize all isoforms of transforming growth factor-β (Yamaguchi et al., 1990) and modulate epidermal growth factor receptor tyrosine kinase (Santra et al., 2002). Thus, up-regulation of decorin may influence matrix remodeling, glial reactivity and inflammation associated to neurodegeneration occurring in ALS. Moreover, decorin has been shown to inhibit neuronal differentiation of adult multipotent neuronal stem/progenitor cells (Barkho et al., 2006). Therefore, the observed up-regulation of decorin may hinder cell replacement therapies in ALS.
Up-regulation of decorin has a potential functional link to the IGF-1R down regulation found in primary astrocytes and spinal cords from SOD1G93A rats. Decorin binds to the IGF-1R (Kd = 18 nM) with an affinity comparable to that of the IGF-1 itself (Kd = 1.2 nM). Decorin binding to IGF-1R causes its activation followed by receptor down-regulation (Schonherr et al., 2005). Conversely, increased IGF-1R immunoreactivity and IGF-1 binding sites has been previously observed in the spinal cord of ALS patients and ALS-mice (Doré et al., 1996; Chung et al., 2003; Wilczak et al., 2003). However, other genome-scale analysis have not detected increased expression of Igf-1r mRNA in spinal cord of SOD1G93A mice (Olsen et al., 2001) or ALS patients (Dangond et al., 2004). In addition, we could not observe changes in Igf-1r mRNA expression in isolated embryonic motor neurons expressing SOD1G93A. The remarkable effect of IGF-1 therapy in prolonging lifespan and in delaying disease onset in SOD1G93A mice (Kaspar et al., 2003) contrasts with the limited success of IGF-1 trial in ALS patients (Lai et al., 1997; Borasio et al., 1998). These results could be consequence of reduced IGF-1 bioavailability due to increased expression of IGF-1 binding proteins in ALS spinal cord (Wilczak et al., 2003). Decorin can also bind IGF-1, though with much lower affinity than IGF-1 binding proteins (Schonherr et al., 2005). However, in situations in which decorin is strongly over-expressed in a well-delimited region, it may compete for IGF-1 and thus might contribute to the increased IGF-1 binding capacity observed in spinal cord of ALS patients (Doré et al., 1996; Wilczak et al., 2003). In this context, increased decorin and decreased IGF-1R expression in astrocytes could have several implications for ALS, because i) decorin is able to reduce IGF-1R expression and eventually reduced IGF-1 bioavailability and ii) at least some of the beneficial effects of IGF-1 on motor neurons are mediated through astrocytes (Ang et al., 1992, 1993).
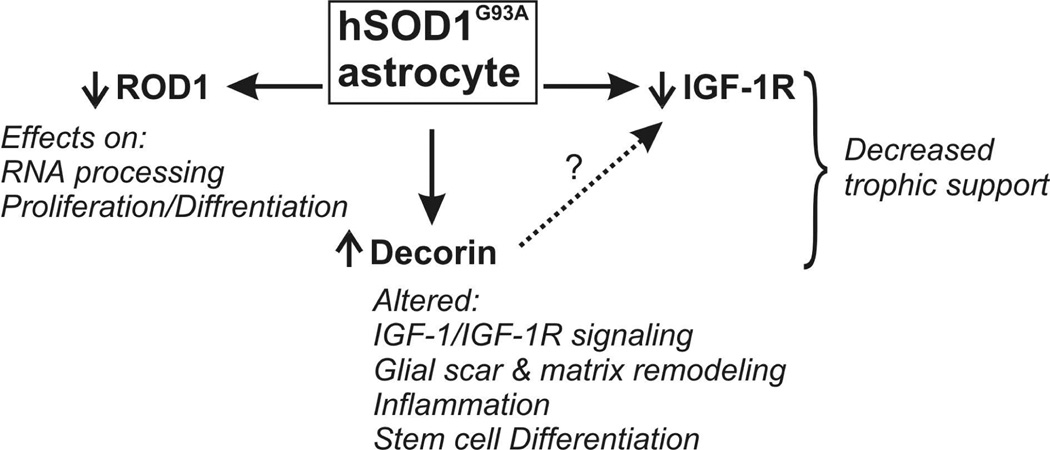
Primary cultures and transgenic animal models cannot fully recapitulate human pathology. However, candidates genes derived from transcriptional profiles at genome-wide scale can improve our knowledge concerning the pathogenic clues of ALS and allow the identification of targets for cell type-specific therapies. Here we showed that SOD1G93A expression might have functional consequences in RNA processing, in both astrocytes and motor neurons, by down-regulating Rod1. In addition, we showed that SOD1G93A expression in astrocytes causes changes in the expression of Decorin and IGF-1R. These changes were also observed in the spinal cord of SOD1G93A animals but not in cultured motor neurons expressing SOD1G93A, suggesting a cell type-specific effect. In particular, Decorin up-regulation might interfere with IGF-1/IGF-1R system by limiting IGF-1 bioavailability and hindering IGF-1R signaling in astrocytes affecting its interaction with surrounding motor neurons and other cell types (Fig. 5).
Figure 5.
Schematic representation of some of the changes in gene expression found in SOD1G93A-astrocytes and its putative consequences.
Acknowledgments
We thank Anne-Marie Girard (Center for Genome Research and Biocomputing at the Oregon State University) for her technical support with microarrays analysis and Dr. Larry W Fisher (NIH/NIDCR) for providing the decorin antibody.
Grants
This work was supported by grant FIRCA RO3TW006482 and AT002034 to J.S.B and L.B; PEDECIBA, the Linus Pauling Institute and the Environmental Health Sciences Center (ES00210) at Oregon State University provided additional support.
The abbreviations used are
- ALS
amyotrophic lateral sclerosis
- SOD1
Cu/Zn superoxide dismutase
- GFAP
glial fibrillary acidic protein
- Dcn
decorin
- Igf-1r
insulin-like growth factor-1 receptor and Rod1, regulator of differentiation
References
- Abe K, Pan LH, Watanabe M, Konno H, Kato T, Itoyama Y. Upregulation of protein-tyrosine nitration in the anterior horn cells of amyotrophic lateral sclerosis. Neurol Res. 1997;19:124–128. doi: 10.1080/01616412.1997.11740784. [DOI] [PubMed] [Google Scholar]
- Almer G, Vukosavic S, Romero N, Przedborski S. Inducible nitric oxide synthase up-regulation in a transgenic mouse model of familial amyotrophic lateral sclerosis. J Neurochem. 1999;72:2415–2425. doi: 10.1046/j.1471-4159.1999.0722415.x. [DOI] [PubMed] [Google Scholar]
- Ang LC, Bhaumick B, Juurlink BH. Neurite promoting activity of insulin, insulin-like growth factor I and nerve growth factor on spinal motoneurons is astrocyte dependent. Brain Res Dev Brain Res. 1993;74:83–88. doi: 10.1016/0165-3806(93)90086-p. [DOI] [PubMed] [Google Scholar]
- Ang LC, Bhaumick B, Munoz DG, Sass J, Juurlink BH. Effects of astrocytes, insulin and insulin-like growth factor I on the survival of motoneurons in vitro. J Neurol Sci. 1992;109:168–172. doi: 10.1016/0022-510x(92)90164-g. [DOI] [PubMed] [Google Scholar]
- Anneser JM, Cookson MR, Ince PG, Shaw PJ, Borasio GD. Glial cells of the spinal cord and subcortical white matter up-regulate neuronal nitric oxide synthase in sporadic amyotrophic lateral sclerosis. Exp Neurol. 2001;171:418–421. doi: 10.1006/exnr.2001.7756. [DOI] [PubMed] [Google Scholar]
- Barbeito LH, Pehar M, Cassina P, Vargas MR, Peluffo H, Viera L, Estévez AG, Beckman JS. A role for astrocytes in motor neuron loss in amyotrophic lateral sclerosis. Brain Res Brain Res Rev. 2004;47:263–274. doi: 10.1016/j.brainresrev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Barkho BZ, Song H, Aimone JB, Smrt RD, Kuwabara T, Nakashima K, Gage FH, Zhao X. Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 2006;15:407–421. doi: 10.1089/scd.2006.15.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman JS, Estevez AG, Crow JP, Barbeito L. Superoxide dismutase and the death of motoneurons in ALS. Trends Neurosci. 2001;24:S15–S20. doi: 10.1016/s0166-2236(00)01981-0. [DOI] [PubMed] [Google Scholar]
- Borasio GD, Robberecht W, Leigh PN, Emile J, Guiloff RJ, Jerusalem F, Silani V, Vos PE, Wokke JH, Dobbins T. A placebo-controlled trial of insulin-like growth factor-I in amyotrophic lateral sclerosis. European ALS/IGF-I Study Group. Neurology. 1998;51:583–586. doi: 10.1212/wnl.51.2.583. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, Borchelt DR, Price DL, Cleveland DW. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- Cassina P, Pehar M, Vargas MR, Castellanos R, Barbeito AG, Estévez AG, Thompson JA, Beckman JS, Barbeito L. Astrocyte activation by fibroblast growth factor-1 and motor neuron apoptosis: implications for amyotrophic lateral sclerosis. J Neurochem. 2005;93:38–46. doi: 10.1111/j.1471-4159.2004.02984.x. [DOI] [PubMed] [Google Scholar]
- Cassina P, Peluffo H, Pehar M, Martinez-Palma L, Ressia A, Beckman JS, Estevez AG, Barbeito L. Peroxynitrite triggers a phenotypic transformation in spinal cord astrocytes that induces motor neuron apoptosis. J Neurosci Res. 2002;67:21–29. doi: 10.1002/jnr.10107. [DOI] [PubMed] [Google Scholar]
- Chung YH, Joo KM, Shin CM, Lee YJ, Shin DH, Lee KH, Cha CI. Immunohistochemical study on the distribution of insulin-like growth factor I (IGF-I) receptor in the central nervous system of SOD1(G93A) mutant transgenic mice. Brain Res. 2003;994:253–259. doi: 10.1016/j.brainres.2003.09.047. [DOI] [PubMed] [Google Scholar]
- Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillee S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, Brown RH, Jr, Julien JP, Goldstein LS, Cleveland DW. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- Dangond F, Hwang D, Camelo S, Pasinelli P, Frosch MP, Stephanopoulos G, Stephanopoulos G, Brown RH, Jr, Gullans SR. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol Genomics. 2004;16:229–239. doi: 10.1152/physiolgenomics.00087.2001. [DOI] [PubMed] [Google Scholar]
- Davies JE, Tang X, Bournat JC, Davies SJ. Decorin promotes plasminogen/plasmin expression within acute spinal cord injuries and by adult microglia in vitro. J Neurotrauma. 2006;23:397–408. doi: 10.1089/neu.2006.23.397. [DOI] [PubMed] [Google Scholar]
- Davies JE, Tang X, Denning JW, Archibald SJ, Davies SJ. Decorin suppresses neurocan, brevican, phosphacan and NG2 expression and promotes axon growth across adult rat spinal cord injuries. Eur J Neurosci. 2004;19:1226–1242. doi: 10.1111/j.1460-9568.2004.03184.x. [DOI] [PubMed] [Google Scholar]
- Dore S, Krieger C, Kar S, Quirion R. Distribution and levels of insulin-like growth factor (IGF-I and IGF-II) and insulin receptor binding sites in the spinal cords of amyotrophic lateral sclerosis (ALS) patients. Brain Res Mol Brain Res. 1992;41:128–133. doi: 10.1016/0169-328x(96)00081-2. [DOI] [PubMed] [Google Scholar]
- Fisher LW, Stubbs JT, 3rd, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl. 1995;266:61–65. [PubMed] [Google Scholar]
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- Gallo JM, Jin P, Thornton CA, Lin H, Robertson J, D'Souza I, Schlaepfer WW. The role of RNA and RNA processing in neurodegeneration. J Neurosci. 2005;25:10372–10375. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Microarray analysis: genome-scale hypothesis scanning. PLoS Biol. 2003;1:E15. doi: 10.1371/journal.pbio.0000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong YH, Parsadanian AS, Andreeva A, Snider WD, Elliott JL. Restricted expression of G86R Cu/Zn superoxide dismutase in astrocytes results in astrocytosis but does not cause motoneuron degeneration. J Neurosci. 2000;20:660–665. doi: 10.1523/JNEUROSCI.20-02-00660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hall ED, Oostveen JA, Gurney ME. Relationship of microglial and astrocytic activation to disease onset and progression in a transgenic model of familial ALS. Glia. 1998;23:249–256. doi: 10.1002/(sici)1098-1136(199807)23:3<249::aid-glia7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Henderson CE, Bloch-Gallego E, Camu W. Purification and culture of embryonic motorneurons. In: Cohen J, Wilkin G, editors. Neural Cell Culture: a Practical Approach. Oxford, England: IRL Press; 1995. pp. 61–81. [Google Scholar]
- Hirano A. Neuropathology of ALS: an overview. Neurology. 1996;47:S63–S66. doi: 10.1212/wnl.47.4_suppl_2.63s. [DOI] [PubMed] [Google Scholar]
- Hirano M, Furiya Y, Asai H, Yasui A, Ueno S. ALADINI482S causes selective failure of nuclear protein import and hypersensitivity to oxidative stress in triple A syndrome. Proc Natl Acad Sci U S A. 2006;103:2298–2303. doi: 10.1073/pnas.0505598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J, DeVito L, Psaltis G, DeGennaro LJ, Cleveland DW, Rothstein JD. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS) Proc Natl Acad Sci U S A. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, Niwa J, Tanaka F, Doyu M, Yoshida M, Hashizume Y, Sobue G. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- Jossin Y. Neuronal migration and the role of reelin during early development of the cerebral cortex. Mol Neurobiol. 2004;30:225–251. doi: 10.1385/MN:30:3:225. [DOI] [PubMed] [Google Scholar]
- Kage M, Yang Q, Sato H, Matsumoto S, Kaji R, Akiguchi I, Kimura H, Tooyama I. Acidic fibroblast growth factor (FGF-1) in the anterior horn cells of ALS and control cases. Neuroreport. 2001;12:3799–3803. doi: 10.1097/00001756-200112040-00040. [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Llado J, Sherkat N, Rothstein JD, Gage FH. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- Kushner PD, Stephenson DT, Wright S. Reactive astrogliosis is widespread in the subcortical white matter of amyotrophic lateral sclerosis brain. J Neuropathol Exp Neurol. 1991;50:263–277. doi: 10.1097/00005072-199105000-00008. [DOI] [PubMed] [Google Scholar]
- Lai EC, Felice KJ, Festoff BW, Gawel MJ, Gelinas DF, Kratz R, Murphy MF, Natter HM, Norris FH, Rudnicki SA. Effect of recombinant human insulin-like growth factor-I on progression of ALS. A placebo-controlled study. The North America ALS/IGF-I Study Group. Neurology. 1997;49:1621–1630. doi: 10.1212/wnl.49.6.1621. [DOI] [PubMed] [Google Scholar]
- Lin CL, Bristol LA, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein JD. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- Lino MM, Schneider C, Caroni P. Accumulation of SOD1 mutants in postnatal motoneurons does not cause motoneuron pathology or motoneuron disease. J Neurosci. 2002;22:4825–4832. doi: 10.1523/JNEUROSCI.22-12-04825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan A, Baird A, Berry M. Decorin attenuates gliotic scar formation in the rat cerebral hemisphere. Exp Neurol. 1999;159:504–510. doi: 10.1006/exnr.1999.7180. [DOI] [PubMed] [Google Scholar]
- Maihofner C, Probst-Cousin S, Bergmann M, Neuhuber W, Neundorfer B, Heuss D. Expression and localization of cyclooxygenase-1 and -2 in human sporadic amyotrophic lateral sclerosis. Eur J Neurosci. 2003;18:1527–1534. doi: 10.1046/j.1460-9568.2003.02879.x. [DOI] [PubMed] [Google Scholar]
- Malaspina A, de Belleroche J. Spinal cord molecular profiling provides a better understanding of amyotrophic lateral sclerosis pathogenesis. Brain Res Brain Res Rev. 2004;45:213–229. doi: 10.1016/j.brainresrev.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Manfredi G, Xu Z. Mitochondrial dysfunction and its role in motor neuron degeneration in ALS. Mitochondrion. 2005;5:77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Migheli A, Cordera S, Bendotti C, Atzori C, Piva R, Schiffer D. S-100beta protein is upregulated in astrocytes and motor neurons in the spinal cord of patients with amyotrophic lateral sclerosis. Neurosci Lett. 1999;261:25–28. doi: 10.1016/s0304-3940(98)01001-5. [DOI] [PubMed] [Google Scholar]
- Nagai M, Aoki M, Miyoshi I, Kato M, Pasinelli P, Kasai N, Brown RH, Jr, Itoyama Y. Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J Neurosci. 2001;21:9246–9254. doi: 10.1523/JNEUROSCI.21-23-09246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy D, Kato T, Kushner PD. Reactive astrocytes are widespread in the cortical gray matter of amyotrophic lateral sclerosis. J Neurosci Res. 1994;38:336–347. doi: 10.1002/jnr.490380312. [DOI] [PubMed] [Google Scholar]
- Olsen MK, Roberds SL, Ellerbrock BR, Fleck TJ, McKinley DK, Gurney ME. Disease mechanisms revealed by transcription profiling in SOD1-G93A transgenic mouse spinal cord. Ann Neurol. 2001;50:730–740. doi: 10.1002/ana.1252. [DOI] [PubMed] [Google Scholar]
- Pehar M, Cassina P, Vargas MR, Castellanos R, Viera L, Beckman JS, Estevez AG, Barbeito L. Astrocytic production of nerve growth factor in motor neuron apoptosis: implications for amyotrophic lateral sclerosis. J Neurochem. 2004;89:464–473. doi: 10.1111/j.1471-4159.2004.02357.x. [DOI] [PubMed] [Google Scholar]
- Pramatarova A, Laganiere J, Roussel J, Brisebois K, Rouleau GA. Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J Neurosci. 2001;21:3369–3374. doi: 10.1523/JNEUROSCI.21-10-03369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH, Jr, Scott RW, Snider WD. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- Ridet JL, Malhotra SK, Privat A, Gage FH. Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci. 1997;20:570–577. doi: 10.1016/s0166-2236(97)01139-9. [DOI] [PubMed] [Google Scholar]
- Robertson J, Doroudchi MM, Nguyen MD, Durham HD, Strong MJ, Shaw G, Julien JP, Mushynski WE. A neurotoxic peripherin splice variant in a mouse model of ALS. J Cell Biol. 2003;160:939–949. doi: 10.1083/jcb.200205027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung WY, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- Saneto RP, De Vellis J. Neuronal and glial cells: cell culture of the central nervous system. In: Turner AJ, Brachelard HS, editors. Neurochemistry: a Practical Approach. Washington, DC: IRL Press; 1987. pp. 27–63. [Google Scholar]
- Santra M, Reed CC, Iozzo RV. Decorin binds to a narrow region of the epidermal growth factor (EGF) receptor, partially overlapping but distinct from the EGF-binding epitope. J Biol Chem. 2002;277:35671–35681. doi: 10.1074/jbc.M205317200. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shibata N, Komori T, Iwata M. iNOS and nitrotyrosine immunoreactivity in amyotrophic lateral sclerosis. Neurosci Lett. 2000;291:44–48. doi: 10.1016/s0304-3940(00)01370-7. [DOI] [PubMed] [Google Scholar]
- Schonherr E, Sunderkotter C, Iozzo RV, Schaefer L. Decorin, a novel player in the insulin-like growth factor system. J Biol Chem. 2005;280:15767–15772. doi: 10.1074/jbc.M500451200. [DOI] [PubMed] [Google Scholar]
- Sharp TV, Munoz F, Bourboulia D, Presneau N, Darai E, Wang HW, Cannon M, Butcher DN, Nicholson AG, Klein G, Imreh S, Boshoff C. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc Natl Acad Sci U S A. 2004;101:16531–16536. doi: 10.1073/pnas.0407123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichel CC, Kappler J, Junghans U, Koops A, Kresse H, Muller HW. Differential expression of the small chondroitin/dermatan sulfate proteoglycans decorin and biglycan after injury of the adult rat brain. Brain Res. 1995;704:263–274. doi: 10.1016/0006-8993(95)01131-5. [DOI] [PubMed] [Google Scholar]
- Vargas MR, Pehar M, Cassina P, Beckman JS, Barbeito L. Increased glutathione biosynthesis by Nrf2 activation in astrocytes prevents p75NTR-dependent motor neuron apoptosis. J Neurochem. 2006;97:687–696. doi: 10.1111/j.1471-4159.2006.03742.x. [DOI] [PubMed] [Google Scholar]
- Vargas MR, Pehar M, Cassina P, Martinez-Palma L, Thompson JA, Beckman JS, Barbeito L. Fibroblast growth factor-1 induces heme oxygenase-1 via nuclear factor erythroid 2-related factor 2 (Nrf2) in spinal cord astrocytes: consequences for motor neuron survival. J Biol Chem. 2005;280:25571–25579. doi: 10.1074/jbc.M501920200. [DOI] [PubMed] [Google Scholar]
- Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- Wilczak N, de Vos RA, De Keyser J. Free insulin-like growth factor (IGF)-I and IGF binding proteins 2, 5, and 6 in spinal motor neurons in amyotrophic lateral sclerosis. Lancet. 2003;361:1007–1011. doi: 10.1016/S0140-6736(03)12828-0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Mann DM, Ruoslahti E. Negative regulation of transforming growth factor-b by the proteoglycan decorin. Nature. 1990;346:281–284. doi: 10.1038/346281a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol Cell Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T, Ishigaki S, Yamamoto M, Liang Y, Niwa J, Takeuchi H, Doyu M, Sobue G. Differential expression of inflammation- and apoptosis-related genes in spinal cords of a mutant SOD1 transgenic mouse model of familial amyotrophic lateral sclerosis. J Neurochem. 2002;80:158–167. doi: 10.1046/j.0022-3042.2001.00683.x. [DOI] [PubMed] [Google Scholar]