Abstract
Background
Tissue handling can alter global gene expression potentially affecting the analytical performance of genomic signatures, but such effects have not been systematically evaluated.
Methods
Tissue samples from 11 previously untreated breast tumors were minced and aliquots were either snap frozen or placed in RNAlater immediately or after 20, 40, 60, 120 or 180 min at room temperature. RNA was profiled on Affymetrix HG‐U133A arrays. We used probe‐set‐wise hierarchical models to evaluate the effect of preservation method on transcript expression and linear mixed effects models to assess the effect of cold ischemic delay on the expression of individual probe sets. Gene set enrichment analysis identified pathways overrepresented in the affected transcripts. We combined the levels of 41 most sensitive transcripts to develop an index of ischemic stress.
Results
Concordance in global gene expression between the baseline and 40 min delay was higher for samples preserved in RNAlater (average concordance correlation coefficient CCC = 0.92 compared to 0.88 for snap frozen). Overall, 481 transcripts (3%) were significantly affected by the preservation method, most of them involved in processes important in cancer. Prolonged cold ischemic delay of up to 3 h induced marginal global gene expression changes (average CCC = 0.90 between baseline and 3 h delay). However 41 transcripts were significantly affected by cold ischemic delay. Among the induced transcripts were stress response genes, apoptotic response genes; among the downregulated were genes involved in metabolism, protein processing and cell cycle regulation. An index combining the expression levels of these genes was proportional to the cold ischemic delay.
Conclusions
Prolonged cold ischemia induces significant transcriptional changes in a small subset of transcripts in the tissue. Furthermore, the expression level of about 3% of the transcripts is affected by the preservation method. These sensitive transcripts should not be included in genomic signatures for more reliable analytical performance.
Keywords: Cold ischemia, Microarrays, Genomic index, Hypoxia, RNAlater, Snap freezing, Ischemic stress index
Highlights
We evaluated the effects of prolonged cold ischemia at on global gene expression.
Simulated cold ischemia by delaying preservation for up to 3 h at room temperature.
481 transcripts were differentially preserved in RNAlater vs snap freezing.
18 transcripts were significantly induced and 23 reduced by prolonged ischemia.
Ischemic Stress Index based on these 41 genes estimates the length of cold ischemia.
1. Introduction
Biomarker assessment from cancer tissue is integral in the clinical management of breast cancer and is becoming even more important with the advent of molecular targeted therapies. Precise and reproducible measurement of the amount of a specific protein or RNA species in a tissue specimen is therefore necessary for reliable administration of therapeutic course for individual cancer patients. Preanalytic factors, related to the tumor itself but associated with the procedures used for specimen collection, handling and preservation, may introduce variation and potentially bias analytical results obtained via immunohistochemistry or gene expression assays (Abdullah‐Sayani et al., 2006; Ioannidis, 2007; Sparano and Solin, 2010). In an effort to address this issue, guidelines have been established for standardizing the handling of clinical specimens with the goal to minimize the potential impact of preanalytical factors on the quality of the biospecimen (Hammond et al., 2010; Wolff et al., 2007).
Several recent studies have reported on the effects of cold ischemic time, defined as the time from tumor specimen removal to sample preservation, on the immunohistochemical (IHC) measurement of protein expression for key breast cancer biomarkers. With the exception of the progesterone receptor that showed varied sensitivity in one study (Yildiz‐Aktas et al., 2012), the antigenicity of the key biomarkers was remarkably stable for up to 4 h of cold ischemic time (Li et al., 2013; Neumeister et al., 2012). We have recently reported that delays of up to 3 h at room temperature until sample stabilization significantly reduces the yield and quality of RNA extracted from breast cancer specimens, but had only marginal effects on ESR1, ERBB2, MKI67 expression and on two multi‐gene indices, the endocrine sensitivity (SET) index (Symmans et al., 2010) and the genomic grade index (GGI) (Sotiriou et al., 2006), measured on DNA microarrays (Hatzis et al., 2011). To what extent these effects are specific to the genes and signatures evaluated in that study or whether they can be generalized to other genes and signatures is unknown. A previous study reported that 121 genes were differentially expressed after 2 h of delayed freezing of breast cancer specimens, and 657 genes were altered after a 24‐hour delay at room temperature (De Cecco et al., 2009). Another study reported that 1788 mRNAs and 56 miRNAs were differentially expressed in breast cancer samples following an additional 6 h of cold ischemic delay after surgery (Borgan et al., 2011), but in colorectal cancer samples the effects of a 6‐hour cold ischemic delay at room temperature on gene expression were minimal (Musella et al., 2013).
In the present study, we used the same design employed in our previous study (Hatzis et al., 2011) to assess global gene expression changes that may be induced by prolonged cold ischemia and specimen preservation and to characterize the biological processes overrepresented in the affected transcripts. We also developed an index based on the expression level of the most sensitive mRNA transcripts that could be used to assess cold ischemic stress in breast cancer biospecimens, which might be useful in meta‐analysis studies. We presume that excluding genes identified in this study as being overly sensitive to ischemic stress should improve the overall analytical performance of microarray‐based biomarkers.
2. Materials and Methods
2.1. Study design and sample handling
Breast cancer tissue samples of 11 previously untreated tumors were resected and intraoperative pathological assessments were performed at the University of Texas M.D. Anderson Cancer Center (MDACC). The patient clinical‐pathologic characteristics and sample collection parameters were as previously reported (Hatzis et al., 2011) and are summarized in Table 1. Details of the study design have been provided in the previous publication. Briefly, resected specimens were immediately transported to pathology at room temperature. After the standard pathological evaluation, a piece of tumor tissue of at least 2.0 cm in diameter was minced into fragments of 1–2 mm in a Petri dish to minimize intratumoral variation and divided into eight grossly equal portions. Two pieces of the tissue fragments were snap frozen in a vial with dry ice immediately (baseline‐T0) and after 40 min at room temperature. The other tissue samples were placed into RNAlater (Life Technologies, Carlsbad, CA) at room temperature either immediately after mincing or after being held at room temperature for 20, 40, 60, 120, or 180 min. Lids of the Petri dishes were closed during the additional time to prevent excessive drying or contamination of tissue. All stabilized tissue samples were stored at −80 °C until RNA extraction.
Table 1.
Patient clinical and pathological characteristics and details of sample collection.a
| Patient ID | Age at diagnosis, y | Histology at diagnosis | ER status | PR status | HER2 status | Pathological lymph node status | Tumor size, cm | Histological grade | AJCC stageb |
|---|---|---|---|---|---|---|---|---|---|
| MD40 | 78 | IDC | N | N | N | P | 2.8 | 3 | IIB |
| MD49 | 52 | IDC, ILC | P | P | N | P | 2.8 | 2 | IIB |
| MD50 | 47 | IDC | P | P | N | P | 2.3 | 2 | IIB |
| MD52 | 49 | IDC | P | P | N | N | 2.3 | 2 | IIA |
| MD53 | 57 | IDC | N | N | N | N | 3 | 3 | IIA |
| MD57 | 64 | ILC | P | P | N | N | 4.6 | 2 | IIA |
| MD64 | 82 | IDC | N | N | P | N | 2.8 | 3 | IIA |
| MD66 | 57 | IDC | P | P | N | N | 2.6 | 2 | IIA |
| MD67 | 44 | IDC, ILC | P | P | N | P | 3 | 2 | IIB |
| MD69 | 60 | IDC, ILC | P | P | N | N | 2.9 | 2 | IIA |
| MD71 | 48 | IDC | P | P | N | P | 2.9 | 3 | IIB |
IDC = invasive ductal carcinoma; ILC = invasive lobular carcinoma; N = negative; P = positive.
AJCC Stage = American Joint Committee on Cancer staging classification, 6th edition (Fleige and Pfaffl, 2006).
2.2. RNA Extraction and quality assessment
Total RNA was extracted from tissue samples using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The concentration of total RNA (ng/μL) and total RNA yield (μg) were measured with a NanoDrop ND‐1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and RNA purity was assessed by the absorbance ratio at 260 nm and 280 nm. An electrophoresis trace was obtained using the Agilent 2100 Bioanalyzer RNA 6000 Nano LabChip (Agilent Technologies, Palo Alto, CA) and the RNA integrity number (RIN) was calculated using the 2100 Expert Software (Agilent Technologies). RIN values greater than 6 are generally considered to represent RNA integrity acceptable for gene expression measurement (Schroeder et al., 2006).
2.3. Gene expression profiling and microarray processing
RNA extracted from 11 tumor samples (MD40–MD71) was processed for microarray hybridization as previously described and hybridized to Affymetrix human genome U133A gene chips (Affymetrix Inc, Santa Clara, CA)(Hatzis et al., 2011). Raw gene expression data files are available at the Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/) under accession ID GSE25011 and in the Array Express repository (http://www.ebi.ac.uk/arrayexpress/) under accession ID E‐GEOD‐25011. Raw intensity (CEL) files were processed using MAS5.0 (R/Bioconductor, www.bioconductor.org) (Gentleman et al., 2004) to generate probe‐level intensities, normalized to a median array intensity of 600, transformed to log2 values, and then scaled to reference values of 1322 breast cancer reference genes. Microarray quality control was assessed based on the similarity in the distribution of intensities for the 1322 reference genes compared to a reference distribution for these genes (Hatzis et al., 2011). Prior to gene‐based analysis, non‐specific filtering was employed to remove probe sets that had low binding specificity (extensions _xrif_ in the probe set name), were housekeeping probe sets (starting with AFFX), or were not adequately expressed (log2‐transformed intensity of at least 5 in at least 75% of the arrays). A total of 16,588 probe sets were retained for further analysis.
2.4. Statistical analysis
To assess the effect of cold ischemic delay on the expression level of individual probe sets we used linear mixed effects models (LME) with fixed slope and random within‐group intercept to account for biological variation among tumors. Probe sets were ranked according to marginal t‐statistic for the slope. The beta‐uniform mixture model (BUM) was used to control a false discovery rate (FDR) of 0.01 for selecting significantly affected probe sets (Pounds and Cheng, 2004). We used a probe‐set‐wise hierarchical model with empirical Bayes to estimate the effects of preservation method, ischemic delay and their interaction on probe set expressions. We used the software package limma (Smyth, 2004) and accounted for inter‐tumor correlation in the expression of each probe set through a consensus correlation coefficient. This approach is equivalent to using LME models, except that it utilizes empirical Bayes to pool information across probe sets (Smyth et al., 2005). Probe sets with significant effects at an FDR <0.05 were selected. We used R 3.0.0 software for all computations (R Core Team, 2013).
2.5. Gene‐based ischemic stress index
We developed a gene‐based ischemic stress index (ISI) as an indicator of cold ischemic stress of a tissue specimen. The index combined the expression levels of 41 probe sets that were significantly affected by cold ischemic delay weighted by the absolute value of the fixed‐effect slope estimated from the LME model for each probe set:
where g i and β i are gene expression level and slope for probe set i, superscripts + and – denote the sets of 18 and 23 probe sets having positive or negative response to cold ischemic stress and |.| denotes the absolute value. The multiplier constant was used to expand the range of the index. The performance of the index was evaluated under a complete random cross‐validation (CRCV) scheme in which 15% (10) of the data points were randomly withheld across tumor samples and time points and LME models for each of the 16,588 probe sets were fit using the remaining 85% (56) of the data. The ISI was defined as above from probe sets selected at FDR <0.02. Separate linear regression models for each tumor sample were then fit to predict cold ischemic time from ISI. These linear models were then used to estimate the cold ischemic time associated with each of the held out specimens from the ISI of the specimen. A prediction was scored as accurate if the actual cold ischemic time was within the 95% prediction interval of the linear model. The entire process was repeated 200 times.
3. Results
3.1. mRNA transcripts affected by specimen preservation method
We compared gene expression from 10 different breast cancer specimens – MD40 was excluded because snap frozen specimens were not collected. The specimens were either preserved in RNAlater or by snap freezing, either immediately after mincing (defined as 0 min or baseline) or after a 40 min delay at room temperature. As shown in Figure 1 for specimens from one tumor, overall concordance in gene expression was generally high. However, there is a visible group of probe sets with higher expression in the RNAlater‐preserved sample at baseline (T0) compared to the other treatments, suggesting that snap freezing or a 40 min delay affects the stability of some mRNA transcripts. Overall, the average concordance correlation coefficient (CCC) in expression levels between specimens from the same tumor sample that were stabilized in RNAlater or by snap freezing at baseline was 0.871 (range 0.690–0.927), and it was similar after a 40 min delay (average 0.860, range 0.729–0.921). The 40 min delay had a smaller effect in samples preserved in RNAlater (average CCC 0.918, range 0.879–0.942) compared to the snap frozen samples (average CCC 0.877, range 0.831–0.926), and this difference was significant by a paired t‐test (mean difference in CCC 0.041, t‐statistic = 4.61, P = 0.001).
Figure 1.
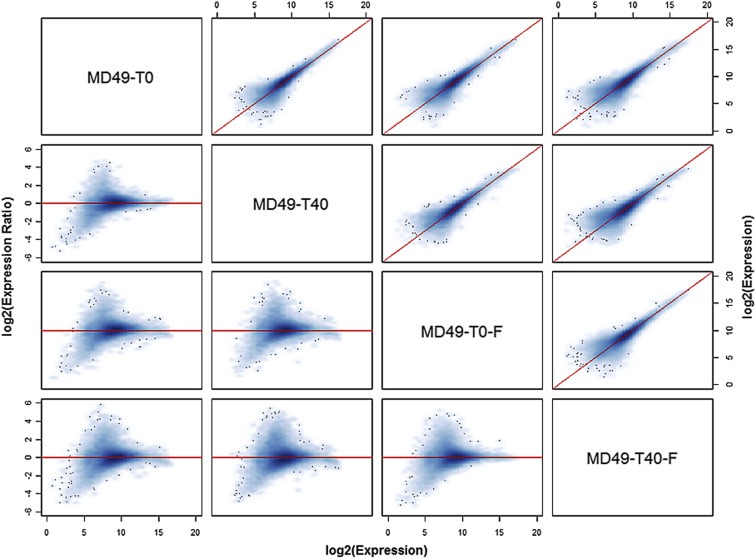
Concordance matrix of global gene expression measurements obtained from specimens derived from the same tumor. Specimens from tumor MD49 were preserved in RNAlater immediately after mincing (T0) or after a 40 min delay at room temperature (T40), or were snap frozen immediately (T0‐F) or after a 40 min delay at room temperature (T40‐F). The concordance plots above the diagonal compare the expression levels of all transcripts in each pair of specimens, with the diagonal line indicating perfect concordance. The corresponding log ratio vs. average (MA) plot for each pair of specimens is shown below the diagonal, where for each transcript the difference in log2 expression values is plotted against their mean. Here, the horizontal line through zero indicates perfect concordance; points above the horizontal line represent transcripts expressed higher in the left‐most sample, whereas points below the line are expressed higher in the second sample (e.g. the M‐A plot in the second row, first column shows expression in T0 vs. T40; points above the zero line are transcripts expressed higher in T0 and those below are transcripts expressed higher in T40).
Combined analysis of the 2 × 2 preservation by ischemic delay study using a main effects plus interaction model with inter‐tumor correlation and empirical Bayes estimation identified 481 probe sets that had a significant preservation method effect at FDR <0.05. Of these, 238 (49.5%) were expressed higher in specimens preserved in RNAlater and 243 (50.5%) were higher in specimens preserved by snap freezing (Supplementary Tables S1 and S2). Interestingly, 19 endogenous control probe sets (prefix AFFX) were among the probe sets expressed higher in snap frozen specimens. The transcripts that are significantly affected by the RNA preservation method have a broad distribution of expression levels (Figure 2). In particular, several transcripts overexpressed in snap frozen specimens have very high expression levels (>15) (Figure 2B), suggesting that biologically important signals may be attenuated or lost by preservation in RNAlater. However, no probe sets showed a significant ischemic delay effect (0 vs. 40 min delay) or a significant preservation by delay interaction (FDR < 0.05). Gene ontology annotation of the affected transcripts indicates that these genes are mainly involved in metabolic processes and are categorized under protein binding function (Supplementary Figure S1). Gene set enrichment analysis (Luo et al., 2009) of the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways revealed that several key pathways that are highly relevant in cancer are better in RNAlater, but that some pathways involved in drug metabolism appear to be better preserved in snap frozen specimens (Table 2).
Figure 2.
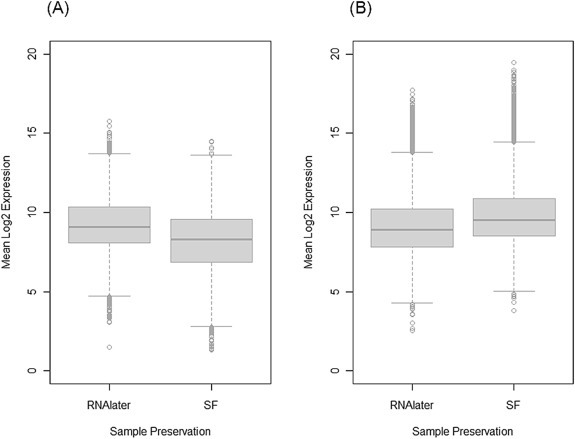
Boxplots showing the distribution of expression levels of transcripts with higher expression in aliquots preserved in RNAlater (238 probe sets) (A), and those with higher expression in snap frozen (SF) (243 probe sets) aliquots (B). Snap frozen samples appear to preserve highly expressed transcripts more effectively, as indicated by the large number of outlier transcripts with log2 expression levels above 15.
Table 2.
Gene set enrichment analysis for KEGG pathways overrepresented in transcripts affected by sample preservation.
| KEGG pathway | P‐Value | Adjusted P‐Valuea | Number of genes |
|---|---|---|---|
| Transcripts Higher in RNAlater | |||
| Focal adhesion | 2.53E‐08 | 4.28E‐06 | 184 |
| ECM‐receptor interaction | 1.14E‐05 | 9.62E‐04 | 78 |
| B cell receptor signaling pathway | 6.41E‐05 | 3.61E‐03 | 59 |
| TGF‐beta signaling pathway | 1.90E‐04 | 8.03E‐03 | 76 |
| Citrate cycle (TCA cycle) | 1.16E‐03 | 3.80E‐02 | 27 |
| Chondroitin sulfate biosynthesis | 1.59E‐03 | 3.80E‐02 | 15 |
| Colorectal cancer | 1.99E‐03 | 3.80E‐02 | 79 |
| Pyruvate metabolism | 2.01E‐03 | 3.80E‐02 | 33 |
| Proteasome | 2.03E‐03 | 3.80E‐02 | 41 |
| Cell cycle | 2.52E‐03 | 4.26E‐02 | 101 |
| p53 signaling pathway | 5.72E‐03 | 8.55E‐02 | 58 |
| Transcripts Higher in Snap Frozen | |||
| Neuroactive ligand–receptor interaction | 1.70E‐20 | 2.88E‐18 | 180 |
| Olfactory transduction | 9.79E‐15 | 8.27E‐13 | 54 |
| Retinol metabolism | 2.53E‐07 | 1.42E‐05 | 34 |
| Ribosome | 4.65E‐07 | 1.83E‐05 | 65 |
| Drug metabolism – cytochrome P450 | 5.40E‐07 | 1.83E‐05 | 45 |
| Taste transduction | 7.43E‐07 | 2.09E‐05 | 29 |
| Linoleic acid metabolism | 9.11E‐05 | 2.20E‐03 | 22 |
| Metabolism of xenobiotics by cytochrome P450 | 1.19E‐04 | 2.52E‐03 | 40 |
| Maturity onset diabetes of the young | 6.87E‐04 | 1.29E‐02 | 11 |
| Cytokine–cytokine receptor interaction | 1.71E‐03 | 2.89E‐02 | 190 |
| Nitrogen metabolism | 3.59E‐03 | 5.51E‐02 | 22 |
| Autoimmune thyroid disease | 4.60E‐03 | 6.46E‐02 | 38 |
| Calcium signaling pathway | 4.97E‐03 | 6.46E‐02 | 141 |
Adjusted P‐value by the Benjamini Hochberg approach to account for multiple testing (Benjamini and Hochberg, 1995).
3.2. Global transcriptional changes induced by prolonged cold ischemic stress
For each tumor sample, we evaluated the overall concordance in global expression profiles between the baseline specimen (preserved in RNAlater immediately after mincing) and those preserved after a 20, 40, 60, 120 and 180 min delay at room temperature. The CCC between baseline and delayed samples slightly decreased with increasing cold ischemic delay, but the trend was not consistent for all samples. The average CCC across tumor samples ranged from 0.916 (baseline vs. 20 min) to 0.890 (baseline vs. 180 min), suggesting that the effect of prolonged cold ischemic delay on global gene expression is rather limited affecting only a small subset of the transcripts (Table 3). To assess the number of transcripts affected by cold ischemic delay, we compared the baseline specimen in a pair‐wise differential analysis with each of the delayed specimens within each tumor sample. The average number of genes with significantly reduced expression (log2‐fold > 3) ranged from 83 (baseline vs. 20 min delay) to 129 (baseline vs. 180 min delay) (Table 3), with the mean expression level of these genes ranging from 6.8 to 8.3, well above noise levels. Therefore, globally, the effect of cold ischemic delay is limited.
Table 3.
Global transcriptional effects of prolonged cold ischemic stress.
| Condition | Concordance correlation coefficient | Differentially expressed Genesa | ||
|---|---|---|---|---|
| Average | Range | Average | Range | |
| Baseline vs. 20 min | 0.916 | 0.868–0.955 | 83.2 | 13–232 |
| vs. 40 min | 0.911 | 0.874–0.942 | 88.3 | 24–239 |
| vs. 60 min | 0.901 | 0.828–0.957 | 129.9 | 15–535 |
| vs. 120 min | 0.889 | 0.774–0.953 | 161.0 | 19–762 |
| vs. 180 min | 0.899 | 0.844–0.938 | 128.8 | 35–377 |
Log2‐fold (Baseline vs. Time Delay) > 3.
3.3. Cold ischemia induces bidirectional changes in specific genes
We identified RNA transcripts that are most responsive to cold ischemia stress across the 11 breast tumor specimens based on univariate mixed‐effects linear regression analyses of the expression level of probe sets with increasing of cold ischemic time accounting for between tumor variation. Of the 16,588 pre‐filtered probe sets, 41 were found to have a significant cold ischemic effect (slope) at FDR < 0.01. Prolonged cold ischemia significantly increased the expression of 18 transcripts and reduced the expression of 23 transcripts (Figure 3). Expression trends of these probe sets with increasing cold ischemic time are shown in Supplementary Figure S2 and annotation details are provided in Supplementary Table S3 and S4. Among the induced transcripts were stress response genes (HYOU1, PRPH2, and FOS), apoptotic response or negative regulators of cell proliferation (DPF1, MCC, GDF9), and transcription factors (NOTCH4, BATF). Among downregulated genes, 12 are involved in metabolism, mRNA processing or protein processing, and three (BUB3, RBL2, MYH10) in cell cycle regulation, providing a clear picture of reduced cell proliferation in response to ischemic stress. A broader list of transcripts with significant cold ischemic effects at FDR <0.05 (140 induced and 183 reduced) is provided for reference in Supplementary Tables S5 and S6.
Figure 3.
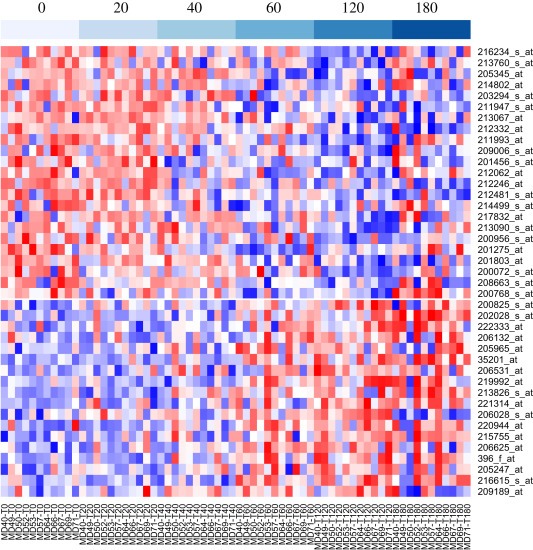
Heatmap showing the expression levels of the 41 probe sets that were significantly affected by prolonged cold ischemic stress. Expression data were standardized to the same mean and standard deviation within each sample. Blue indicates low expression and red high expression. Probe sets were ordered from those most strongly reduced (top) to those most strongly induced (bottom) with increasing cold ischemic delay. Samples (columns) were ordered by cold ischemic delay, as shown at the top, and then by sample number.
3.4. Gene index for sensitivity to cold ischemia stress
We combined the expression levels of the 41 probe sets identified to significantly respond to cold ischemic stress to derive a quantitative index that could be an indicator for the extent of cold ischemic stress in a tumor specimen, as described in the Materials and Methods. The relationship between the gene‐based ischemic stress index (ISI) and the actual cold ischemic time is shown in Figure 4A. The linear relationship is consistent across samples and throughout the entire range of cold ischemic time evaluated in this study. Mixed effects regression provided an estimate for the common slope of 2.63 (P < 0.001), a null intercept (P = 0.83), and an intraclass correlation coefficient of 0.74 confirming a consistent model fit across samples.
Figure 4.
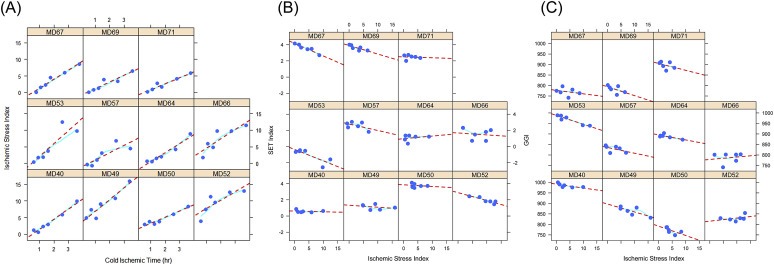
Ischemic Stress Index (ISI) assessed from the expression levels of 41 mRNA transcripts that were identified as being particularly sensitive to cold ischemic stress. The plots show that within each tumor sample, the ISI increases linearly with the actual cold ischemic delay at room temperature imposed on the tumor specimens before stabilizing them in RNAlater (A). Also shown is the effect of increasing cold ischemic stress assessed through the Ischemic Stress Index (ISI) on two genomic signatures, the Endocrine Sensitivity Index — SET (B) and the Genomic Grade Index — GGI (C). Circles represent the ISI measured from the gene expression profile obtained from a tissue aliquot preserved at the indicated time, dotted lines represent the linear regression trend, and solid lines represent a locally weighted polynomial regression (LOESS smoother).
To evaluate the consistency of the linear relationship between ISI and cold ischemic time and the ability of the ISI to estimate the length of cold ischemia in unknown samples, we performed complete random cross‐validation as described in the methods. The linear relationship between ISI and cold ischemic time was consistent under cross‐validation (Supplementary Figure S3), with an average R 2 of 0.8 (standard deviation SD 0.11; Supplementary Table S7 and Figure S4). The average prediction root mean squared error was about twice as large than the average residual regression error (Supplementary Table S7 and Figure S4). Yet, a specimen's length of cold ischemia was predicted accurately on average for 86% (SD 12%) of the held out specimens (Supplementary Table S7 and Figure S4). These results suggest that the linear relationship between cold ischemic time and ISI is robust and that this relationship can be used to reliably estimate the length of cold ischemia from the expression levels of the most sensitive transcripts.
We calculated the ISI and two multi‐gene indices, the endocrine sensitivity (SET) index (Symmans et al., 2010) and the genomic grade index (GGI) (Sotiriou et al., 2006), for the specimens in this study in order to elucidate the potential utility of the ISI as an indicator of cold ischemic stress in breast cancer samples because such information may not be available at the time of sample procurement. For many of the tumor samples, there was a linear reduction of the value of the index with increasing cold ischemic stress as assessed by the ISI (Figure 4B and C). To assess potential cold ischemic effects in published datasets, we computed the ISI in several gene expression datasets published in the GEO. As Figure 5 shows, the median ISI for most datasets was less than 5, suggesting mild cold ischemic effects (equivalent <2 h ischemic delay; Figure 4), although the ISI distributions were relatively broad with some of the samples having ISI values twice as high as the median. Tissue specimen type or tumor subtype appear to have a small effect, if any, on the ISI. However, contrary to what was expected, FNAs appear to be associated with relatively higher ISI in TNBC but not in the other subtypes (Figure 5).
Figure 5.
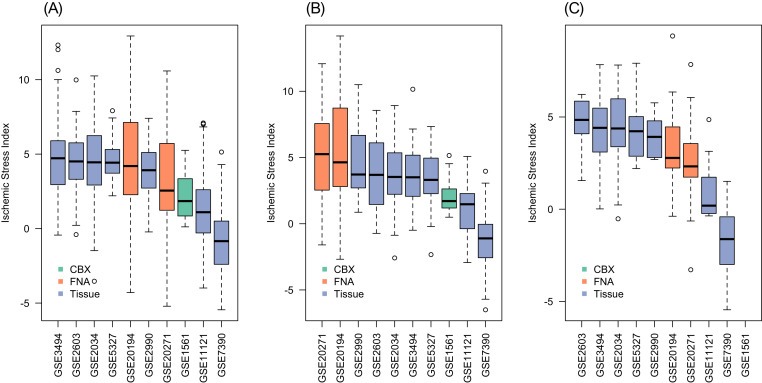
Boxplots showing the distribution of Ischemic Stress Index (ISI) evaluated for gene expression profiles from different datasets in the GEO repository. All transcriptional profiles were generated using the Affymetrix HGU133A microarrays and were normalized by MAS5: (A) non‐triple negative breast cancer (ER+/HER2‐) datasets involving a total of 1096 profiles (range of size per dataset 19–223); (B) TNBC datasets involving a total of 391 profiles (range of size per dataset 12–63); (C) HER2‐positive datasets involving a total of 118 profiles (range of size per dataset 7–26). The color of the bars reflects the source of the tissue (CBX – core biopsy; FNA – fine needle aspiration biopsy; Tissue – surgically resected tissue). Generally, the median ISI for all datasets is 5 or lower, which corresponds to about 2 h of cold‐ischemic delay (see Figure 4), although some samples appear to have ISI as high as 10 or greater. Further details on the datasets and sources are provided in Supplemental Table S8.
4. Discussion
In our previous study, we systematically evaluated the effects of tissue preservation and prolonged cold ischemia on RNA yield and quality and on the expression of ESR1, ERBB2, MKI67 and of two multi‐gene indices, the SET index and the GGI (Hatzis et al., 2011). Here, we extend the analysis to characterize global gene expression effects using 11 breast cancer tissue samples preserved in RNAlater or by snap freezing at different times after surgery.
Sample handling is known to alter the expression levels of a large number of genes, with reported effects ranging from 88 genes (1.5%) following surgical manipulation in prostate cancer (Lin et al., 2006) to 461 genes (3%) in one breast cancer study (De Cecco et al., 2009). An ischemic delay of 40 min had marginal effects on overall gene expression levels, which is in agreement with previous studies (Line et al., 2006). Yet, the effect was smaller in specimens preserved in RNAlater (average CCC = 0.92) compared to snap frozen samples (average CCC = 0.88), indicating better overall preservation consistency with RNAlater. Intriguingly, neither preservation method appears to be optimal for all transcripts, since each apparently preserves a different subset of transcripts. Overall, the levels of 481 transcripts (3%) were significantly different in aliquots from the same tumor preserved in RNAlater vs. snap freezing, with about half of them being significantly higher in RNAlater aliquots and the other half in the snap frozen aliquots. The RNA of genes involved in protein binding (non‐covalent interactions with other proteins or protein complexes) and in metabolic processes appear to be particularly sensitive to the preservation method, with about half of them been better preserved in RNAlater and the other half by snap freezing (Supplementary Figure 1). However, gene set enrichment analysis showed that different metabolic pathways are significantly overrepresented in the two sets of differentially preserved transcripts. RNAlater appears to be better in preserving transcripts involved in primary metabolism, cell cycle control, and signaling pathways important to cancer, whereas snap freezing seems to be more optimal for preserving transcripts that are involved in protein synthesis and drug metabolism (Table 2). Furthermore, it appears that the pool of mRNA transcripts from highly expressed genes might be better preserved by snap freezing the tissue (Figure 2B). However, preservation in RNAlater might be suboptimal for other types of molecular analyses as it cannot preserve tissue morphology (Staff et al., 2013), and although it preserves DNA, it can potentially interfere with DNA extraction (Michaud and Foran, 2011).
Despite best efforts at standardizing tissue collection, extended time delays in preservation of the resected tissue can occur, and the effect of prolonged cold ischemic delays on gene expression has not been well characterized. We previously showed that prolonged cold ischemic delays had a minimal effect on breast cancer related single genes and multi‐gene signatures (Hatzis et al., 2011). Globally, the overall concordance in gene expression between aliquots from the same sample preserved at baseline or after 3 h delay at room temperature was 0.90 (Table 3). This appears comparable to the concordance between duplicate aliquots preserved in RNAlater (preliminary data not shown here, pending on analysis of a larger study designed specifically to characterize technical sources of variation). Although the global effect of prolonged cold ischemia is rather marginal, there were some transcripts that appeared to be particularly sensitive to cold ischemic stress. We identified 41 such transcripts at a false discovery rate of ≤1%, of which 23 showed reduced expression while 18 showed increased expression with prolonged ischemia (Figure 3).
Increased transcription levels of stress response genes (HYOU1, PRPH2, and FOS) imply that these genes are induced in response to cold ischemic stress. HYOU1 has cyto‐protective properties and is known to be upregulated in response to hypoxic stress (Bando et al., 2004), but its overexpression in breast cancer is also associated with lymphovascular invasion, regional nodal involvement and generally with poor prognosis (Stojadinovic et al., 2007). Hypoxia can also induce FOS, a transcription factor with a role in cell proliferation, differentiation and endocrine resistance (Gee et al., 1999; Nathaniel et al., 2012; Wong et al., 2008). In a recent meta‐analysis, higher FOS levels were associated with better relapse free survival after tamoxifen treatment in breast cancer (Mihaly et al., 2013). It is therefore evident how hypoxic stress caused by prolonged cold ischemia could confound the assessment of prognostic gene‐expression based biomarkers.
NOTCH signaling plays a key role in cell proliferation, apoptosis and vascularity (Ma et al., 2011). Aberrant NOTCH4 activity can induce mammary gland carcinoma and vessel perfusion of mammary tumors (Costa et al., 2013; Han et al., 2011). Higher cancer cell proliferation would require greater vascular nourishment and may induce hypoxic conditions for tumor growth. Knowing that NOTCH4 is primarily expressed in vascular endothelial cells (Costa et al., 2013), the observed higher expression of NOTCH4 with prolonged ischemia in our study is consistent with previous reports (Han et al., 2011; Hiyama et al., 2011; Murphy et al., 2012; Reedijk, 2012; Speiser et al., 2012). High NOTCH expression has also been shown to correlate with poor outcome in breast cancer (Reedijk, 2012; Speiser et al., 2012), with the majority of triple negative breast cancers (TNBC) overexpressing both NOTCH1 and NOTCH4 receptors (Speiser et al., 2012). We also observed ischemia‐induced expression of MCC, a known colorectal tumor suppressor gene that is thought to negatively regulate cell cycle proliferation (Fukuyama et al., 2008; Kohonen‐Corish et al., 2007; Pangon et al., 2010). Such effects have not been reported previously. Again, induced gene expression in response to prolonged ischemia could potentially confound interpretation and use of these biomarkers in breast cancer.
On the other hand, transcripts negatively affected by prolonged ischemia were from genes involved in metabolism, mRNA or protein processing, and in cell cycle regulation (BUB3, RBL2, MYH10). Lower expression of these genes may be related either to response to ischemic stress or to transcript loss due to mRNA degradation. Among these genes, BARD1 is a tumor suppressor gene and since high levels of this tumor suppressor gene are typically associated with poor differentiation and poor prognosis in breast cancer (Wu et al., 2006), susceptibility to cold ischemic stress could result in reduced reported marker levels and erroneous prognosis assessment from mishandled samples.
Although global gene expression changes on individual transcripts were overall marginal, the effects can be additive for multi‐gene indices. The ischemic stress index (ISI) described herein appears to estimate consistently the length of cold ischemia in breast cancer specimens and could be useful for assessing gene expression profiles as a means of pre‐analytical quality assessment in addition to the aforementioned standardization of tissue handling practices for transcriptional marker assessment. Our assessment of more than 1500 breast cancer profiles from public repositories showed similar ISI distributions across 10 different datasets, irrespective of molecular subtype or tumor specimen type. One dataset (GSE7390) had noticeably lower ISI values. This was the only study that used Trizol for RNA isolation (Supplemental Table S8), suggesting potential bias by tissue processing protocols. Although ISI levels from FNA biopsies were generally low among ER+/HER2− and HER2+ specimens as expected due to the typically reduced pre‐analytical delays associated with such biopsies, they were the highest among triple negative (TNBC) specimens. The pathologic features of TNBCs are more likely to contain rapid growth with zonal or central necrosis and central regions of fibrous scarring (Reis‐Filho and Tutt, 2008), particularly when such tumors are locally advanced. Such locally advanced tumors are amenable to FNA and are more likely to be represented in cohorts of patients who received neoadjuvant chemotherapy (intended to convert the tumor to operability). The high ISI in these specimens might reflect the hypoxic conditions in the central necrotic zones that are generally associated with such carcinomas.
This pilot study however has a number of limitations. First, RNA decay is a dynamic process that can be affected by many factors including genetic sequence variants, transcription factors, enzymatic and environmental factors (Duan et al., 2013), and therefore this study may be too small to account for such effects. Secondly, cold ischemic delay is only one of the aspects of pre‐analytical tissue‐handling after surgical excision that could potentially influence tissue quality for biomarker assessment. Other factors with potentially confounding effects are warm ischemic time, tissue storage length and storage conditions. Finally, this study was based on only one tissue type (breast cancer) and the effect of ischemia on global gene expression may be different in other tumor types (Webster, 2006).
4.1. Funding
This work was supported in whole or in part with Federal Funds form the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E to WFS and CH.
4.2. Notes
The sponsors reviewed and approved the design of the study and monitored its conduct by quarterly teleconferences led by Dr. Helen Moore, Director, Biospecimen Research Network, National Cancer Institute. The sponsors did not have a role in the collection of samples, analysis, interpretation of the data, writing of the article or decision to submit the article. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.02.002
Aktas Bilge, Sun Hongxia, Yao Hui, Shi Weiwei, Hubbard Rebekah, Zhang Ya, Jiang Tingting, Ononye Sophia N., Wali Vikram B., Pusztai Lajos, Symmans W. Fraser and Hatzis Christos, (2014), Global gene expression changes induced by prolonged cold ischemic stress and preservation method of breast cancer tissue, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.02.002.
References
- Abdullah-Sayani, A. , Bueno-de-Mesquita, J.M. , van de Vijver, M.J. , 2006. Technology Insight: tuning into the genetic orchestra using microarrays–limitations of DNA microarrays in clinical practice. Nat. Clin. Pract. Oncol.. 3, 501–516. [DOI] [PubMed] [Google Scholar]
- Bando, Y. , Tsukamoto, Y. , Katayama, T. , Ozawa, K. , Kitao, Y. , Hori, O. , Stern, D.M. , Yamauchi, A. , Ogawa, S. , 2004. ORP150/HSP12A protects renal tubular epithelium from ischemia-induced cell death. FASEB J.. 18, 1401–1403. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. , Hochberg, Y. , 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 57, 289–300. [Google Scholar]
- Borgan, E. , Navon, R. , Vollan, H.K. , Schlichting, E. , Sauer, T. , Yakhini, Z. , Lingjaerde, O.C. , Sorlie, T. , Borresen-Dale, A.L. , 2011. Ischemia caused by time to freezing induces systematic microRNA and mRNA responses in cancer tissue. Mol. Oncol.. 5, 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M.J. , Wu, X. , Cuervo, H. , Srinivasan, R. , Bechis, S.K. , Cheang, E. , Marjanovic, O. , Gridley, T. , Cvetic, C.A. , Wang, R.A. , 2013. Notch4 is required for tumor onset and perfusion. Vasc. Cell. 5, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco, L. , Musella, V. , Veneroni, S. , Cappelletti, V. , Bongarzone, I. , Callari, M. , Valeri, B. , Pierotti, M.A. , Daidone, M.G. , 2009. Impact of biospecimens handling on biomarker research in breast cancer. BMC Cancer. 9, 409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, J. , Shi, J. , Ge, X. , Dolken, L. , Moy, W. , He, D. , Shi, S. , Sanders, A.R. , Ross, J. , Gejman, P.V. , 2013. Genome-wide survey of interindividual differences of RNA stability in human lymphoblastoid cell lines. Sci. Rep.. 3, 1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige, S. , Pfaffl, M.W. , 2006. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med.. 27, 126–139. [DOI] [PubMed] [Google Scholar]
- Fukuyama, R. , Niculaita, R. , Ng, K.P. , Obusez, E. , Sanchez, J. , Kalady, M. , Aung, P.P. , Casey, G. , Sizemore, N. , 2008. Mutated in colorectal cancer, a putative tumor suppressor for serrated colorectal cancer, selectively represses beta-catenin-dependent transcription. Oncogene. 27, 6044–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee, J.M. , Willsher, P.C. , Kenny, F.S. , Robertson, J.F. , Pinder, S.E. , Ellis, I.O. , Nicholson, R.I. , 1999. Endocrine response and resistance in breast cancer: a role for the transcription factor Fos. Int. J. Cancer. 84, 54–61. [DOI] [PubMed] [Google Scholar]
- Gentleman, R.C. , Carey, V.J. , Bates, D.M. , Bolstad, B. , Dettling, M. , Dudoit, S. , Ellis, B. , Gautier, L. , Ge, Y. , Gentry, J. , Hornik, K. , Hothorn, T. , Huber, W. , Iacus, S. , Irizarry, R. , Leisch, F. , Li, C. , Maechler, M. , Rossini, A.J. , Sawitzki, G. , Smith, C. , Smyth, G. , Tierney, L. , Yang, J.Y. , Zhang, J. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol.. 5, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, M.E. , Hayes, D.F. , Dowsett, M. , Allred, D.C. , Hagerty, K.L. , Badve, S. , Fitzgibbons, P.L. , Francis, G. , Goldstein, N.S. , Hayes, M. , Hicks, D.G. , Lester, S. , Love, R. , Mangu, P.B. , McShane, L. , Miller, K. , Osborne, C.K. , Paik, S. , Perlmutter, J. , Rhodes, A. , Sasano, H. , Schwartz, J.N. , Sweep, F.C. , Taube, S. , Torlakovic, E.E. , Valenstein, P. , Viale, G. , Visscher, D. , Wheeler, T. , Williams, R.B. , Wittliff, J.L. , Wolff, A.C. , 2010. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol.. 28, 2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J. , Hendzel, M.J. , Allalunis-Turner, J. , 2011. Notch signaling as a therapeutic target for breast cancer treatment?. Breast Cancer Res.. 13, 210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis, C. , Sun, H. , Yao, H. , Hubbard, R.E. , Meric-Bernstam, F. , Babiera, G.V. , Wu, Y. , Pusztai, L. , Symmans, W.F. , 2011. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J. Natl. Cancer Inst.. 103, 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama, A. , Skubutyte, R. , Markova, D. , Anderson, D.G. , Yadla, S. , Sakai, D. , Mochida, J. , Albert, T.J. , Shapiro, I.M. , Risbud, M.V. , 2011. Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: implications in degenerative disc disease. Arthritis Rheum.. 63, 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis, J.P. , 2007. Is molecular profiling ready for use in clinical decision making?. Oncologist. 12, 301–311. [DOI] [PubMed] [Google Scholar]
- Kohonen-Corish, M.R. , Sigglekow, N.D. , Susanto, J. , Chapuis, P.H. , Bokey, E.L. , Dent, O.F. , Chan, C. , Lin, B.P. , Seng, T.J. , Laird, P.W. , Young, J. , Leggett, B.A. , Jass, J.R. , Sutherland, R.L. , 2007. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 26, 4435–4441. [DOI] [PubMed] [Google Scholar]
- Li, X. , Deavers, M.T. , Guo, M. , Liu, P. , Gong, Y. , Albarracin, C.T. , Middleton, L.P. , Huo, L. , 2013. The effect of prolonged cold ischemia time on estrogen receptor immunohistochemistry in breast cancer. Mod. Pathol.. 26, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.W. , Coleman, I.M. , Hawley, S. , Huang, C.Y. , Dumpit, R. , Gifford, D. , Kezele, P. , Hung, H. , Knudsen, B.S. , Kristal, A.R. , Nelson, P.S. , 2006. Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J. Clin. Oncol.. 24, 3763–3770. [DOI] [PubMed] [Google Scholar]
- Luo, W. , Friedman, M.S. , Shedden, K. , Hankenson, K.D. , Woolf, P.J. , 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinform.. 10, 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D. , Dong, X. , Zang, S. , Ma, R. , Zhao, P. , Guo, D. , Dai, J. , Chen, F. , Ye, J. , Ji, C. , 2011. Aberrant expression and clinical correlation of Notch signaling molecules in breast cancer of Chinese population. Asia Pac. J. Clin. Oncol.. 7, 385–391. [DOI] [PubMed] [Google Scholar]
- Michaud, C.L. , Foran, D.R. , 2011. Simplified field preservation of tissues for subsequent DNA analyses. J. Forensic Sci.. 56, 846–852. [DOI] [PubMed] [Google Scholar]
- Mihaly, Z. , Kormos, M. , Lanczky, A. , Dank, M. , Budczies, J. , Szasz, M.A. , Gyorffy, B. , 2013. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res. Treat. 140, 219–232. [DOI] [PubMed] [Google Scholar]
- Murphy, P.A. , Kim, T.N. , Lu, G. , Bollen, A.W. , Schaffer, C.B. , Wang, R.A. , 2012. Notch4 normalization reduces blood vessel size in arteriovenous malformations. Sci. Transl Med.. 4, 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musella, V. , Verderio, P. , Reid, J.F. , Pizzamiglio, S. , Gariboldi, M. , Callari, M. , Milione, M. , De Cecco, L. , Veneroni, S. , Pierotti, M.A. , Daidone, M.G. , 2013. Effects of warm ischemic time on gene expression profiling in colorectal cancer tissues and normal mucosa. PLoS One. 8, e53406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathaniel, T.I. , Otukonyong, E. , Abdellatif, A. , Soyinka, J.O. , 2012. Effect of hypoxia on metabolic rate, core body temperature, and c-fos expression in the naked mole rat. Int. J. Dev. Neurosci.. 30, 539–544. [DOI] [PubMed] [Google Scholar]
- Neumeister, V.M. , Anagnostou, V. , Siddiqui, S. , England, A.M. , Zarrella, E.R. , Vassilakopoulou, M. , Parisi, F. , Kluger, Y. , Hicks, D.G. , Rimm, D.L. , 2012. Quantitative assessment of effect of preanalytic cold ischemic time on protein expression in breast cancer tissues. J. Natl. Cancer Inst.. 104, 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangon, L. , Sigglekow, N.D. , Larance, M. , Al-Sohaily, S. , Mladenova, D.N. , Selinger, C.I. , Musgrove, E.A. , Kohonen-Corish, M.R. , 2010. The “mutated in colorectal Cancer” protein is a novel target of the UV-induced DNA damage checkpoint. Genes Cancer. 1, 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounds, S. , Cheng, C. , 2004. Improving false discovery rate estimation. Bioinformatics. 20, 1737–1745. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2013. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing; Vienna, Austria: URL: http://www.R-project.org/ [Google Scholar]
- Reedijk, M. , 2012. Notch signaling and breast cancer. Adv. Exp. Med. Biol.. 727, 241–257. [DOI] [PubMed] [Google Scholar]
- Reis-Filho, J.S. , Tutt, A.N. , 2008. Triple negative tumours: a critical review. Histopathology. 52, 108–118. [DOI] [PubMed] [Google Scholar]
- Schroeder, A. , Mueller, O. , Stocker, S. , Salowsky, R. , Leiber, M. , Gassmann, M. , Lightfoot, S. , Menzel, W. , Granzow, M. , Ragg, T. , 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol.. 7, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. , 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol.. 3, Article3 [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. , Michaud, J. , Scott, H.S. , 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 21, 2067–2075. [DOI] [PubMed] [Google Scholar]
- Sotiriou, C. , Wirapati, P. , Loi, S. , Harris, A. , Fox, S. , Smeds, J. , Nordgren, H. , Farmer, P. , Praz, V. , Haibe-Kains, B. , Desmedt, C. , Larsimont, D. , Cardoso, F. , Peterse, H. , Nuyten, D. , Buyse, M. , Van de Vijver, M.J. , Bergh, J. , Piccart, M. , Delorenzi, M. , 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst.. 98, 262–272. [DOI] [PubMed] [Google Scholar]
- Sparano, J.A. , Solin, L.J. , 2010. Defining the clinical utility of gene expression assays in breast cancer: the intersection of science and art in clinical decision making. J. Clin. Oncol.. 28, 1625–1627. [DOI] [PubMed] [Google Scholar]
- Speiser, J. , Foreman, K. , Drinka, E. , Godellas, C. , Perez, C. , Salhadar, A. , Ersahin, C. , Rajan, P. , 2012. Notch-1 and Notch-4 biomarker expression in triple-negative breast cancer. Int. J. Surg. Pathol.. 20, 139–145. [DOI] [PubMed] [Google Scholar]
- Staff, S. , Kujala, P. , Karhu, R. , Rokman, A. , Ilvesaro, J. , Kares, S. , Isola, J. , 2013. Preservation of nucleic acids and tissue morphology in paraffin-embedded clinical samples: comparison of five molecular fixatives. J. Clin. Pathol.. 66, 807–810. [DOI] [PubMed] [Google Scholar]
- Stojadinovic, A. , Hooke, J.A. , Shriver, C.D. , Nissan, A. , Kovatich, A.J. , Kao, T.C. , Ponniah, S. , Peoples, G.E. , Moroni, M. , 2007. HYOU1/Orp150 expression in breast cancer. Med. Sci. Monit.. 13, BR231–239. [PubMed] [Google Scholar]
- Symmans, F.W. , Hatzis, C. , Sotiriou, C. , Andre, F. , Peintinger, F. , Regitnig, P. , Daxenbichler, G. , Desmedt, C. , Domont, J. , Marth, C. , Delaloge, S. , Bauernhofer, T. , Valero, V. , Booser, D. , Hortobagyi, G.N. , Pusztai, L. , 2010. Genomic index of sensitivity to endoccrine therapy for breast cancer. J. Clin. Oncol.. 28, 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, M.J. , 2006. Tissue preparation and banking. Prog. Brain Res.. 158, 3–14. [DOI] [PubMed] [Google Scholar]
- Wolff, A.C. , Hammond, M.E. , Schwartz, J.N. , Hagerty, K.L. , Allred, D.C. , Cote, R.J. , Dowsett, M. , Fitzgibbons, P.L. , Hanna, W.M. , Langer, A. , McShane, L.M. , Paik, S. , Pegram, M.D. , Perez, E.A. , Press, M.F. , Rhodes, A. , Sturgeon, C. , Taube, S.E. , Tubbs, R. , Vance, G.H. , van de Vijver, M. , Wheeler, T.M. , Hayes, D.F. , American Society of Clinical, O., College of American, P, 2007. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol.. 25, 118–145. [DOI] [PubMed] [Google Scholar]
- Wong, V. , Wang, D.Y. , Warren, K. , Kulkarni, S. , Boerner, S. , Done, S.J. , Leong, W.L. , 2008. The effects of timing of fine needle aspiration biopsies on gene expression profiles in breast cancers. BMC Cancer. 8, 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.Y. , Vlastos, A.T. , Pelte, M.F. , Caligo, M.A. , Bianco, A. , Krause, K.H. , Laurent, G.J. , Irminger-Finger, I. , 2006. Aberrant expression of BARD1 in breast and ovarian cancers with poor prognosis. Int. J. Cancer. 118, 1215–1226. [DOI] [PubMed] [Google Scholar]
- Yildiz-Aktas, I.Z. , Dabbs, D.J. , Bhargava, R. , 2012. The effect of cold ischemic time on the immunohistochemical evaluation of estrogen receptor, progesterone receptor, and HER2 expression in invasive breast carcinoma. Mod. Pathol.. 25, 1098–1105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


