Abstract
While it is well established that clinically ill livestock represent a reservoir of Salmonella, the importance of subclinical shedders as sources of human salmonellosis is less well defined. The aims of this study were to assess the subtype diversity of Salmonella in healthy dairy cattle and associated farm environments and to compare the subtypes isolated from these sources with the Salmonella subtypes associated with clinical human cases in the same geographic area. A total of 1,349 Salmonella isolates from subclinical dairy cattle and farm environments (46 farms) were initially characterized by traditional or molecular serotyping and tested for antimicrobial susceptibility. A set of 381 representative isolates was selected for further characterization by pulsed-field gel electrophoresis (PFGE); these isolates represented unique combinations of sampling date, serovar, antimicrobial resistance pattern, farm of origin, and source, to avoid overrepresentation of subtypes that were re-isolated from a given source. These 381 isolates represented 26 Salmonella serovars; the most common serovars were Cerro [(38.8%, 148/381) isolated from 21 farms], Kentucky [16.3%; 10 farms], Typhimurium [9.4%; 7 farms], Newport [7.6%; 8 farms], and Anatum [6.3%; 6 farms]. Among the 381 isolates, 90 (23.6%) were resistant to between 1 and 11 antimicrobial agents, representing 50 different antimicrobial resistance patterns. Overall, 61 XbaI-PFGE types were detected among these 381 isolates, indicating considerable Salmonella diversity on dairy farms without evidence of clinical salmonellosis. Fourteen PFGE types, representing 12 serovars, exactly matched PFGE types from human isolates, suggesting that subclinically infected dairy cattle could be sources of human disease-associated Salmonella.
Keywords: Salmonella, cattle, subclinical, dairy, subtyping, PFGE
Introduction
Salmonella is a zoonotic foodborne pathogen and the etiologic agent of salmonellosis. Salmonellosis is a major health concern as it is one of the leading causes of foodborne illness in the United States. It has been estimated that approximately 1.0 million nontyphoidal Salmonella illnesses, 20,000 hospitalizations and 370 deaths occur through foodborne transmission in the U.S. each year (Scallan et al., 2011). Salmonella is comprised of two species, Salmonella bongori and Salmonella enterica, and more than 2,600 recognized serovars (Guibourdenche et al., 2010). However, most human salmonellosis cases are caused by relatively few serovars within S. enterica (Jones et al., 2008). Salmonella serovars have different hosts and reservoirs ranging from cold-blooded (e.g., reptiles) to warm-blooded animals (e.g., mammals) (Hoelzer et al., 2011b). Furthermore, Salmonella can survive in farm and other environments for prolonged periods of time (Holley et al., 2006; Cummings et al., 2009a). Some previous studies have established that certain serovars may be overrepresented among specific hosts and/or associated with specific hosts; for example, while serovar Dublin has been isolated from both bovine and human hosts, it is most commonly isolated from cattle and rarely found in other non-primate hosts and is thus typically considered “bovine associated”.
Salmonella is transmitted to animals and humans through the fecal-oral route. Animals can become infected after ingestion of feed and water contaminated with Salmonella. Similarly, humans can become infected by foodborne transmission or after direct or indirect contact with infected animals (Hoelzer et al., 2011b). In livestock, clinical signs typically appear 6 to 24 hours after exposure and include profuse diarrhea, fever, dehydration, inappetence, foul-smelling feces, and mucus or blood in feces (Cummings et al., 2010a). Disease manifestations in people include diarrhea, fever, abdominal cramps and septicemia in severe cases, appearing 12 to 72 hours after ingestion. Salmonella can also be carried subclinically by both humans and animals (Murase et al., 2000; Perron et al., 2008; USDA-NAHMS, 2011). The purpose of this study was to investigate the phenotypic and genotypic diversity and distribution of Salmonella serovarsisolated from subclinically infected cattle and associated farm environments within New York dairy farms.
2. Materials and Methods
2.1. Study design
Bovine fecal and environmental samples were collected from dairy farms in New York between October 2007 and August 2009 as described by Cummings et al. (2010b); 44 of the farms detailed in that study (Cummings et al., 2010b) as well as 2 additional New York dairy herds included here yielded Salmonella-positive samples, for a total of 46 farms with Salmonella-positive samples. Briefly, Salmonella surveillance included both environmental screening and disease monitoring within each herd for a period of at least 12 months. A positive Salmonella culture result arising from either surveillance method would trigger subsequent visits for cattle sampling, and 50–70 fecal samples were collected from apparently healthy cattle at each visit depending on herd size. Overall, 8,948 samples (1,420 environmental and 7,528 fecal) were collected from the 46 dairy herds.
All Salmonella isolates obtained from the 46 farms were used here for phenotypic characterization (i.e., serotyping and antimicrobial susceptibility testing, as detailed below), totaling 1,349 isolates obtained from environmental (n = 402) and fecal samples (n = 947) (see Supplemental Table 1 for a list of all isolates). Isolates were obtained over multiple sample collections at the same farm, with a median number of 4 sample collection dates with Salmonella positive samples per farm (range: 1–18); the wide range reflects that some farms might have only had a single sample collection that yielded Salmonella, while others might have had a large number of sample collections that yielded Salmonella (for details see Cummings et al., 2010b).
2.2. Salmonella isolation from environmental and bovine fecal samples
Salmonella isolation procedures have previously been reported (Cummings et al., 2009a, 2010a, and 2010b). Briefly, environmental samples were taken from four different locations on each farm (i.e., calf housing, cow housing, sick pen, and manure storage area), using sterile 4×4 gauze swabs saturated in double-strength skim milk. Gauze swabs were placed into a sterile flip98 top container, and samples were stored at 4°C and brought to the laboratory for Salmonella isolation. Fecal samples from cattle without clinical signs of salmonellosis (i.e., animals that do not show diarrhea, fever, etc.) were collected via rectal retrieval using a new sleeve for each sample. Ten grams of fecal matter were placed into a Para-Pak bottle, sealed, and sent to the laboratory for culture and further analyses. Cultural testing of these environmental and fecal samples yielded 1,349 Salmonella isolates that were included in the study reported here.
2.3. Serotyping
Traditional serotyping was performed by agglutination (as described by Edwards and Ewing, 1986) at the National Veterinary Services Laboratories (NVSL), a division of the United State Department of Agriculture (USDA) Animal and Plant Health Inspection Service (APHIS, Ames, Iowa).
2.4. Molecular serotyping
Molecular serotyping was performed on selected isolates if (i) the isolates were not serotyped by traditional serotyping, (ii) the isolates were classified as untypeable by traditional serotyping, or (iii) serovars predicted by PFGE did not match the serovars reported based on traditional serotyping (Supplemental Table 2). Molecular serotyping was performed, as reported by Ranieri et al. (2013), using a combination of (i) PCR-based characterization of O antigens and (ii) sequencing-based characterization of the genes encoding the H1 and H2 antigen (i.e., fliCand fljB, respectively). Briefly, PCR-based characterization of O antigens used a multiplex PCR targeting genes specific for serogroups B, C1, C2-C3, D1, and E1 (Herrera-León et al., 2007) as well as separate PCRs detecting genes specific for serogroups G and K (Fitzgerald et al., 2007, 2006). fliC and fljB sequences obtained were aligned and compared to 236 sequences representing 131 Salmonella serovars (Ranieri et al., 2013) in order to predict H1 and H2 antigens. O antigen PCR and H1 and H2 antigen sequencing results were combined to assign serovar designation consistent with the White-Kauffman-Leminor scheme designations.
2.5. Antimicrobial susceptibility testing
Susceptibility testing was performed at Cornell University’s Animal Health Diagnostic Center using the Sensititre® system (Trek Diagnostic Systems Ltd., Cleveland, OH). All 1,349 isolates were examined for susceptibility to 15 antimicrobial agents included in the National Antimicrobial Resistance Monitoring System (NARMS) Gram-negative panel: amikacin, amoxicillin/clavulanic acid, ampicillin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, tetracycline, and trimethoprim/sulfamethoxazole. Clinical and Laboratory Standards Institute guidelines were used for the interpretation of MIC values when available (CLSI, 2008). Otherwise, MIC values were interpreted using NARMS breakpoints (FDA, 2012).
2.6. Pulsed field gel electrophoresis (PFGE)
PFGE typing was performed on 381 representative Salmonella isolates. We defined representative isolates as isolates with a unique combination of sampling date, serovar, antimicrobial resistance pattern, farm of origin, and source (i.e., environmental or bovine fecal). If two or more isolates shared a unique combination of the aforementioned criteria, then only one was randomly selected for PFGE typing (using www.random.org) to avoid overrepresentation of subtypes that were re-isolated on the same farm. If a serovar and its variants (e.g., S. Typhimurium and S. Typhimurium var. O 5 –) were isolated from the same farm on the same date, one representative of each was selected for PFGE typing. PFGE typing was performed using the standard CDC PulseNet protocol for Salmonella (Ribot et al., 2006). Bacterial cultures were embedded in 1% agarose plugs (Lonza SeaKem Gold Agarose, Rockland, ME) and digested with 50 U/plug of XbaI (Roche Applied Science, Indianapolis, IN) at 37°C. A subset of Salmonella Cerro isolates (n = 10) were also digested with 40 U/plug of NotI at 37°C, to improve the resolution (Zheng et al., 2007) of the subtyping results by XbaI. The restriction fragments were separated by agarose PFGE using the Chef Mapper® XA or the CHEF-DR II® electrophoresis systems (Bio-Rad, Hercules, California). The initial switch time was 2.16 s and the final switch time was 63.8 s for XbaI, and 2 s and 20 s for NotI. The gel images were captured using Gel Doc equipment (Bio-Rad, Hercules, California). The tiff images were analyzed using the BioNumerics software program version 5.1 (Applied Maths, Sint-Martens- Latem, Belgium). XbaI-PFGE type numbers were assigned after a comparison against 5,828 PFGE types in the BioNumerics database of the Food Safety Laboratory. Clustering analyses were performed using the unweighted pair group method with arithmetic mean algorithm (UPGMA) based on the DICE similarity coefficient with 1.5% position tolerance.
3. Results
3.1. Initial phenotypic subtyping data suggest frequent re-isolation of isolates characterized by a combination of identical serovar and antimicrobial resistance patterns on a given farm
The initial isolate set included 1,349 Salmonella isolated from 46 New York dairy farms; 70.2% and 29.8% of isolates were obtained from subclinical dairy cattle and farm environments, respectively (Supplemental Table 1). The median herd size for all 46 positive farms was 978 female dairy cattle (range: 245–7,412). Traditional serotyping was performed for 1,344 isolates; ten of these isolates were classified as untypeable by the NVSL (Supplemental Table 1). These Salmonella isolates were used to initially select 403 representative isolates for PFGE typing; these isolates represented a unique combination of sampling date, serovar, antimicrobial resistance pattern, farm of origin, and source. For example, on farm 1, a total of 8 pansusceptible serovar Meleagridis isolates were obtained from bovine fecal samples on 12/11/2007; one isolate was randomly selected to be included in the set of representative isolates (Supplemental Table 1).
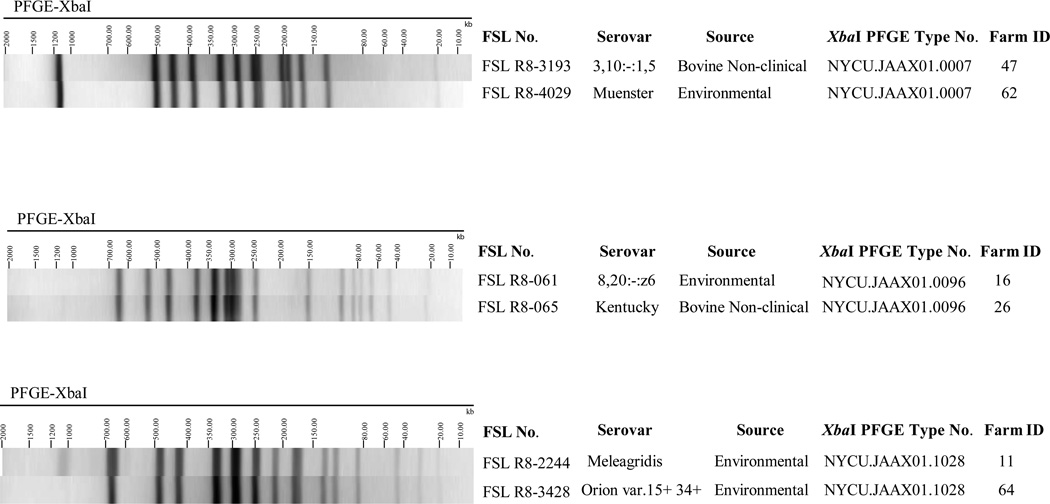
Initial PFGE analysis of the 403 representative isolates identified 47 isolates where the PFGE type for a given isolate matched one or more isolates that were classified into a different serovar (Supplemental Table 2). For 16 of these 47 isolates, additional isolates with a matching PFGE type were isolated from the same farm as a given isolate. These 16 isolates thus were reclassified as the serovar they were predicted to have, based on PFGE, since the other isolates with the matching PFGE pattern also showed this PFGE predicted serovar. For example, isolate FSL R8-346 from farm 1 was predicted by PFGE to represent serovar Meleagridis but was initially classified as serovar Kentucky by traditional serotyping; as 14 other isolates with the same PFGE isolated from the same farm had been classified as serovar Meleagridis (by classical serotyping), isolate FSL R8-346 was reclassified as Meleagridis (Supplemental Table 1). For the other 31 of these 47 isolates, molecular serotyping was performed (see Supplemental Table 2 for details). In addition, 10 isolates classified as untypeable and 5 isolates that had not been serotyped were characterized by molecular serotyping (see Supplemental Table 2 for details). After completion of molecular serotyping and PFGE analysis, a total of 381 isolates were identified as representative; these isolates were used for the subsequent analyses described below; some of the 403 isolates that were initially identified as representative were removed from the set of representative isolates as the additional characterization described here revealed that the initial serovar represented a misclassification and that after reclassification this given isolate matched another isolate also included among the isolate set that represented unique subtypes. Interestingly, as part of this analysis we also identified a number of isolates where agiven PFGE type represented two different serovars, including (i) PFGE type NYCU.JAAX01.1028, representing serovars Orion 15+ 34+ and Meleagridis; (ii) PFGE type NYCU.JAAX01.0096, representing Kentucky and 8,20:–:z6; and (iii) PFGE type NYCU.JAAX01.0007, representing Muenster and 3,10:–:1,5 (see Figure 1 for examples).
Figure 1.
Three instances where Salmonella isolates with different serovars shared the same XbaI-PFGE type.
3.2. More than 50% of representative environmental and subclinical isolates represented serovars Cerro and Kentucky
Among the 381 representative Salmonella isolates, 61.6% (235/381) and 38.3% (146/381) were obtained from the farm environment and subclinical dairy cattle, respectively. These isolates represented 26 serovars (Table 1). The 5 most common serovars were Cerro (38.8%; 148 isolates obtained from 21 farms); Kentucky (16.3%; 62 isolates from 10 farms); Typhimurium, including variant O 5– (9.4%; 36 isolates from 7 farms); Newport (7.6%; 29 isolates from 8 farms); and Anatum, including variant 15+ (6.3%, 24 isolates from 6 farms). The remaining serovars accounted for 21.5% (82/381). The number of Salmonella serovars isolated per farm ranged from 1 to 6 (Table 2); for example, farm 19 yielded isolates representing six different serovars, including Typhimurium (including Typhimurium var. O 5–), Kentucky, Newport, Oranienburg, Anatum, and 6,7:–:1,5.
Table 1.
Serovar diversity (by source and farm) among the 381 representative Salmonella isolates obtained from dairy cattle and farm environments in NY state.
| Serovara | Number of environmental isolates |
Number of bovine non- clinical isolates |
Total Number of isolates |
Total Number of farms |
Farm ID |
|---|---|---|---|---|---|
| Agona | 3 | — | 3 | 3 | 10, 28, 46 |
| Anatum | 13 | 7 | 20 | 6 | 4,19,21,39,52,56 |
| Anatum var. 15+ | 2 | 2 | 4 | 1 | 39 |
| Cerro | 92 | 56 | 148 | 21 | 15,17,18,26,27,28,30,45,47,49,50,52, 53,54,55,57,59,60,61,62,65 |
| Heidelberg | 1 | — | 1 | 1 | 46 |
| Infantis | 4 | — | 4 | 4 | 10,18,36,55 |
| Kentucky | 36 | 26 | 62 | 10 | 14,15,16,17,18,19,21,26,42,53 |
| Mbandaka | 6 | — | 6 | 4 | 10,40,48,52 |
| Meleagridis | 13 | 6 | 19 | 2 | 1,11 |
| Minnesota | 1 | 3 | 4 | 2 | 49,62 |
| Montevideo | 5 | 1 | 6 | 2 | 41,60 |
| Muenster | 5 | 3 | 8 | 5 | 1,3,23,26,62 |
| Newport | 13 | 16 | 29 | 8 | 14,17,18,19,29,35,60,62 |
| Oranienburg | 4 | — | 4 | 2 | 10,19 |
| Orion var. 15+, 34+ | 2 | — | 2 | 1 | 64 |
| Paratyphi B var. L-tartrate+ | 1 | — | 1 | 1 | 20 |
| Senftenberg | — | 1 | 1 | 1 | 46 |
| Typhimurium | 13 | 14 | 27 | 7 | 1,17,19,22,25,51 |
| Typhimurium var. O 5– (Copenhagen) | 6 | 3 | 9 | 4 | 1,19,25,60 |
| Tennessee | 1 | — | 1 | 1 | 46 |
| Thompson | 1 | — | 1 | 1 | 57 |
| 3,10:–:1,5 | 2 | 2 | 4 | 1 | 47 |
| 3,10:–:l,w | 1 | — | 1 | 1 | 1 |
| 3,10:e,h:– | 1 | — | 1 | 1 | 11 |
| 4,5,12:i:– | 1 | 1 | 2 | 2 | 26,52 |
| 6,7:–:1,5 | 1 | — | 1 | 1 | 19 |
| 8,20:–:z6 | 1 | — | 1 | 1 | 16 |
| Untypeableb | 5 | 5 | 10 | 8 | 1,15,16,19,39,42,57,65 |
| Total | 235 | 146 | 381 | — |
A total of 26 serovars were found in the 46 farms in this study. Serovar variants (e.g., S. Typhimurium and S. Typhimurium var. O 5 –) were not considered as individual serovars.
All untypeable isolates were characterized by molecular serotyping, and then classified as Anatum (n=2), Cerro (n=2), Kentucky (n=4), Dublin (n=1), and Meleagridis (n=1) (See Supplemental Table 2).
Table 2.
Sample collection dates when Salmonella positive samples were obtained from each of the 46 farms.
| Farm ID | No. of serovars per farm (no. of untypeable isolates) |
No. of sample collection dates |
Sample collection dates |
|---|---|---|---|
| 1 | 4 (1) | 11 | 10/2/2007–10/22/2008 |
| 3 | 1 | 1 | 02/07/2008 |
| 4 | 1 | 2 | 07/15/2008–08/18/2008 |
| 10 | 4 | 4 | 11/27/2007–09/01/2008 |
| 11 | 2 | 5 | 10/31/2007–10/30/2008 |
| 14 | 2 | 5 | 10/11/2007–08/30/2008 |
| 15 | 2 (1) | 6 | 10/11/2007–12/09/2008 |
| 16 | 2 (2) | 6 | 12/04/2007–11/11/2008 |
| 17 | 4 | 9 | 10/11/2007–09/10/2008 |
| 18 | 4 | 7 | 10/18/2007–09/08/2008 |
| 19 | 6 (1) | 7 | 10/18/2007–10/02/2008 |
| 20 | 1 | 1 | 06/10/2008 |
| 21 | 2 | 4 | 10/19/2007–06/16/2008 |
| 22 | 1 | 3 | 12/17/2007–03/14/2008 |
| 23 | 1 | 1 | 10/19/2007 |
| 25 | 1 | 9 | 10/19/2007–09/03/2008 |
| 26 | 4 | 9 | 10/24/2007–10/27/2008 |
| 27 | 1 | 2 | 10/24/2007–01/25/2008 |
| 28 | 2 | 1 | 10/24/2007 |
| 29 | 1 | 1 | 10/24/2007 |
| 30 | 1 | 18 | 10/24/2007–10/15/2008 |
| 35 | 1 | 4 | 10/23/2008–12/09/2008 |
| 36 | 1 | 1 | 11/13/2007 |
| 39 | 1 (1) | 5 | 11/13/2007–12/03/2008 |
| 40 | 1 | 2 | 11/14/2007–1/10/2008 |
| 41 | 1 | 2 | 08/04/2008–10/03/2008 |
| 42 | 1 (1) | 7 | 11/15/2007–09/17/2008 |
| 45 | 1 | 1 | 11/29/2007 |
| 46 | 4 | 3 | 11/29/2007–09/25/2008 |
| 47 | 2 | 3 | 09/11/2008–01/06/2009 |
| 48 | 1 | 1 | 03/2008 |
| 49 | 2 | 16 | 01/31/2008–02/23/2009 |
| 50 | 1 | 4 | 11/01/2008–02/09/2009 |
| 51 | 1 | 1 | 05/09/2008 |
| 52 | 4 | 6 | 04/26/2008–02/14/2009 |
| 53 | 2 | 4 | 11/06/2008–03/03/2009 |
| 54 | 1 | 1 | 11/26/2008 |
| 55 | 2 | 3 | 09/29/2008–03/12/2009 |
| 56 | 1 | 5 | 02/22/2008–03/25/2009 |
| 57 | 2 (2) | 12 | 03/19/2008–03/24/2009 |
| 59 | 1 | 1 | 09/2008 |
| 60 | 4 | 7 | 06/17/2008–04/13/2009 |
| 61 | 1 | 6 | 06/17/2008–04/12/2009 |
| 62 | 4 | 10 | 06/22/2008–01/22/2009 |
| 64 | 1 | 2 | 09/27/2008–02/17/2009 |
| 65 | 1 (1) | 10 | 09/12/2008–08/11/2009 |
3.3. About 25% of environmental and subclinical isolates were resistant to one or more antimicrobial agents, with Salmonella Typhimurium representing the most common drug resistant serovar
While 76.4% of the 381 isolates designated as representative were pansusceptible, 23.6% (90 isolates) showed resistance to 1 to 11 antimicrobial agents (see Supplemental Table 1). These isolates represented 50 different antimicrobial resistance patterns (see Supplemental Table 3). All isolates were susceptible to amikacin and ciprofloxacin (Table 3). Of the 90 resistant isolates, 42were isolated from subclinical cattle and 48 were isolated from the farm environment. Among these 90 isolates, the most commonly observed resistances were to ampicillin (found in 72% of these 90 isolates), tetracycline (63% of isolates), and amoxicillin/clavulanic acid (58% of isolates) (Table 3). Both pansusceptible as well as resistant Salmonella isolates were obtained from 48% (22/46) of the farms; 46% and 6% of farms yielded only pansusceptible or resistant isolates, respectively. Interestingly, in some cases isolates resistant to antimicrobial agents shared the same serovar-XbaI-PFGE-type combination as pansusceptible isolates suggesting resistance gene acquisition and/or deletion events that did not affect the PFGE banding pattern.
Table 3.
Antimicrobial resistance observed among the 381 representative Salmonella isolated from healthy dairy cattle and associated farm environments in New York.
| Resistant | Intermediate Resistance | |||
|---|---|---|---|---|
| Antimicrobials | Concentration (mg/mL) |
No. of isolates |
Concentration (mg/mL) |
No. of isolates |
| Amikacin | ≥ 64 | 0 | 32 | 0 |
| Amoxicillin/Clavulanic Acid | ≥32/16 | 52 | 16/8 | 5 |
| Ampicillin | ≥ 32 | 65 | 16 | 2 |
| Cefoxitin | ≥ 32 | 43 | 16 | 2 |
| Ceftiofur | ≥ 8 | 48 | 4 | 5 |
| Ceftriaxone | ≥ 64 | 5 | 16–32 | 28 |
| Chloramphenicol | ≥ 32 | 32 | 16 | 13 |
| Ciprofloxacin | ≥ 4 | 0 | 2 | 0 |
| Gentamicin | ≥ 16 | 0 | 8 | 1 |
| Kanamycin | ≥ 64 | 29 | 32 | 0 |
| Nalidixic Acid | ≥ 32 | 1 | - | 0 |
| Streptomycin | ≥ 64 | 41 | - | 0 |
| Sulfisoxazole | ≥ 512 | 48 | - | 0 |
| Tetracycline | ≥ 16 | 57 | 8 | 2 |
| Trimethoprim/Sulfamethoxazole | ≥ 4/76 | 2 | - | 0 |
The 90 isolates that showed resistance to at least one antimicrobial included 14 different serovars, including Typhimurium (25 isolates, representing 20 different resistance patterns), Cerro (13 isolates, representing 12 different resistance patterns), Newport (22 isolates, representing 7 different patterns), Kentucky (8 isolates representing 6 different resistance patterns) (Supplemental Table 3).
3.4. PFGE typing of 381 representative environmental and subclinical isolates revealed presence of PFGE types that exactly match human clinical isolates
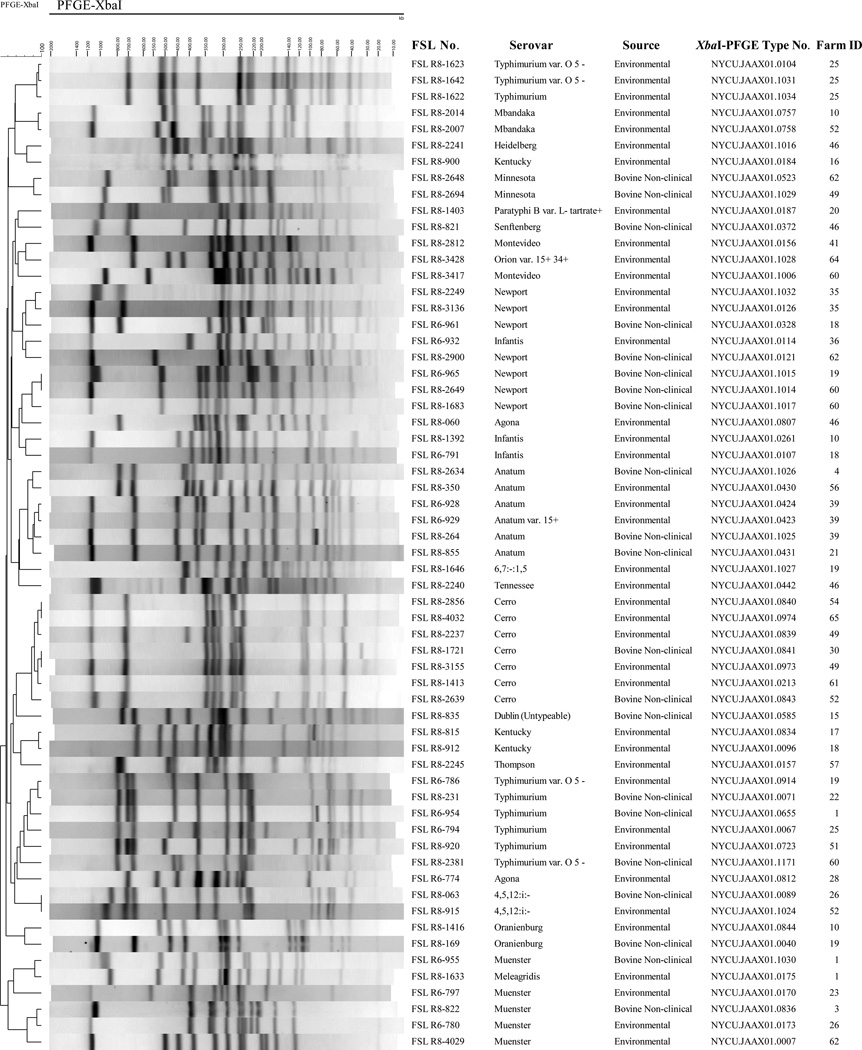
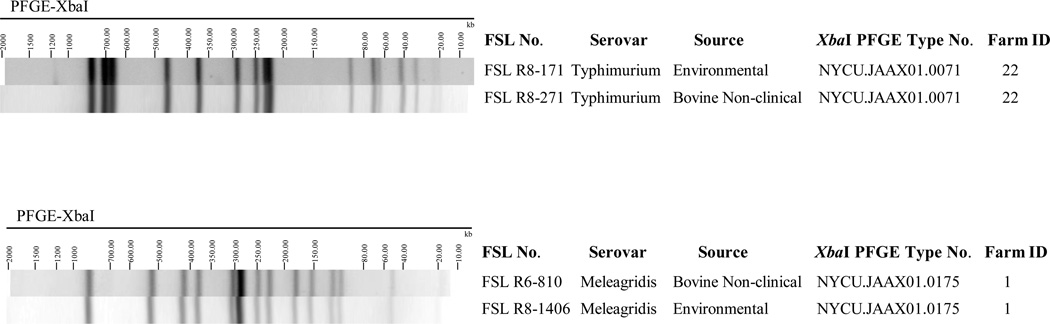
Among the 381 isolates, a total of 61 different XbaI-PFGE types were identified (Figure 2). On 27 farms, we identified isolates from both environmental samples and fecal samples that shared the same PFGE type (see Figure 3 for two examples). Salmonella Cerro XbaI-PFGE type NYCU.JAAX01.0213 was the most widely distributed PFGE pattern; this pattern represented 134 of the 148 S. Cerro isolates (90.5 %), and isolates with this pattern were obtained from 19 farms in 11 different counties in New York. Furthermore, NotI-PFGE analyses of 10 randomly chosen serovar Cerro isolates (representing 4 XbaI-PFGE types, including 7 isolates with XbaI- PFGE type NYCU.JAAX01.0213) yielded two NotI-PFGE types with 9 isolates sharing the same NotI-PFGE type (Supplemental Figure 1).
Figure 2.
The 61 XbaI-PFGE types found among the 381 representative Salmonella isolates characterized here. These isolates were obtained from subclinical dairy cattle and associated dairy farm environments samples collected in 46 New York state dairy farms.
Figure 3.
Examples where Salmonella isolated from different sources within the same farm shared undistinguishable XbaI-PFGE types. Shown here are environmental and fecal isolates from Farms 1 and 22 that shared the same PFGE type; overall (including these two farms), there were 27 farms where isolates from different sources within the same farm shared undistinguishable XbaI-PFGE types.
The 61 XbaI-PFGE types identified here were also compared to PFGE types represented among 1,849 human isolates (provided by the New York state Department of Health) in our database; these human isolates were obtained between January 2001 and December 2010, while the 381 isolates characterized here were obtained between October 2007 and August 2009. Human isolates represented a longer time frame to capture a larger number of PFGE types; the goal of this comparison was not to determine whether the farms sampled here were the specific source of human infections but rather to determine whether the PFGE types found among healthy cattle and associated farm environments and human isolates represent overlapping populations. Overall, 14 of the PFGE types represented among the 381 isolates from dairy farms exactly matched PFGE types found among human isolates. These PFGE patterns represent 12 different serovars (Table 4). A total of 143 human isolates (7.7% of the 1,849 human isolates) were represented by PFGE types that were identical to PFGE types found among Salmonella isolates from dairy farms (Table 4).
Table 4.
PFGE types found among both, isolates from subclinical dairy cattle and farm environments, (collected between October 2007 and August 2009) and of isolates from human clinical cases (reported to the New York State Department of Health between January 2001 and December 2010).
| PFGE type | Serovar | No. of human isolates | No. of isolates obtained from farm environments and subclinical animals |
|---|---|---|---|
| NYCU.JAAX01.0040 | Oranienburg | 13 | 1 |
| NYCU.JAAX01.0071 | Typhimurium | 31 | 14 |
| NYCU.JAAX01.0089 | 4,5,12:i:– | 31 | 1 |
| NYCU.JAAX01.0096 | Kentucky | 1 | 66 |
| NYCU.JAAX01.0114 | Infantis | 1 | 1 |
| NYCU.JAAX01.0121 | Newport | 4 | 15 |
| NYCU.JAAX01.0126 | Newport | 8 | 8 |
| NYCU.JAAX01.0156 | Montevideo | 11 | 3 |
| NYCU.JAAX01.0157 | Thompson | 31 | 1 |
| NYCU.JAAX01.0213 | Cerro | 3 | 136 |
| NYCU.JAAX01.0442 | Tennessee | 5 | 1 |
| NYCU.JAAX01.0757 | Mbandaka | 2 | 1 |
| NYCU.JAAX01.0914 | Typhimurium | 1 | 2 |
| NYCU.JAAX01.1016 | Heidelberg | 1 | 1 |
4. Discussion
4.1. Salmonella serovars Cerro and Kentucky were the most common serovars isolated from subclinical dairy cattle and dairy farm environments in New York
Salmonella serovars Cerro and Kentucky were the two most common serovars isolated from subclinical dairy cattle and dairy farm environments. While farms that yielded Salmonella isolates on multiple sample collection dates could drive serovar and subtype frequencies (as isolates with a given serovar may have been included for multiple sampling dates from a given farm), re-isolation of a serovar also makes exposure and dispersal of this serovar more likely. Our data are consistent with the NAHMS Dairy 2007 study, which found serovars Cerro and Kentucky to be the most common serovars isolated from healthy cattle on U.S. dairy operations; in this study, 121 dairy farms were sampled in 17 states that host 79.5% of U.S. dairy herds and82.5% of U.S. dairy cows (USDA-NAHMS, 2011). Salmonella Cerro was also the most common serovar found in bulk tank milk and milk filters tested as part of the NAHMS study (van Kessel et al., 2011). van Kessel et al. (2007) previously suggested that S. Cerro might behave as a commensal organism in dairy cattle, based on a protracted outbreak of subclinical S. Cerro infection. However, S. Cerro has been described as an emerging pathogen among dairy cattle in the northeastern U.S. over the last few years, as evidenced by a sharp rise in S. Cerro isolations from bovine clinical cases (Cummings et al., 2010b; Hoelzer et al., 2011a; Tewari et al., 2012). These studies have also reported that, based on PFGE analysis, U.S. Salmonella Cerro isolates represent a highly clonal population, consistent with our PFGE data reported here. While our data support frequent isolation of S. Cerro from cattle that are subclinically infected, experimental infection studies will be needed to further characterize this serovar’s behavior as a commensal vs. pathogenic organism.
In addition to serovars Cerro and Kentucky, a number of other serovars commonly isolated from the New York dairy farms in our study, were also reported by the 2007 NAHMS study (USDA-NAHMS, 2011) as commonly isolated from dairy operations and healthy cattle around the U.S. (e.g., Muenster, Meleagridis, and Mbandaka). Interestingly, none of these serovars (i.e., Cerro, Kentucky, Muenster, Meleagridis, and Mbandaka) are among the 20 most common human disease-associated serovars reported by CDC (CDC, 2011). These findings suggest that a number of Salmonella serovars that are common among subclinically infected cattle are rare among human clinical cases. Future phenotypic and genotypic studies on these serovars may be warranted to identify possible mechanisms that may explain an association of these serovars with bovine hosts.
4.2. Serovars resulting from traditional serotyping could be confirmed using a combination of PFGE typing and molecular serotyping
PFGE typing has previously been demonstrated to be useful in serovar prediction (Zou et al., 2010), particularly if large databases are available to facilitate serovar prediction (Ranieri et al., 2013). We thus used the PFGE data for our isolates to determine whether serovar identification by classical serotyping matched the serovar predicted based on a comparison of PFGE types to a database of previously characterized isolates with both PFGE and classical serovar data. Here, this approach successfully identified a number of isolates that did not have the correct serovar assigned to them. We also identified a number of instances where different serovars with similar antigenic formula were found to have indistinguishable PFGE types, such as Kentucky (8,20:i:z6) and 8,20:–:z6, among others. This is consistent with previous reports (e.g., Soyer et al., 2010; Torpdahl et al., 2005) that also identified isolates, with similar antigenic formula, that shared indistinguishable PFGE types. These findings support that prediction of serovar using banding pattern-based subtyping methods is likely to yield at least some inconsistent results. In contrast, use of a molecular serotyping approach based on PCR amplification of O-antigen specific genes and sequencing of fliC and fljB (Ranieri et al., 2013) provided good differentiation of isolates and clarification of ambiguous traditional serotyping results (e.g., where traditional serotyping data and PFGE-based prediction of serovars did not match), as well as for improved classification of untypeable isolates. These findings suggest that the previously described (Ranieri et al., 2013) molecular serotyping approach used here provides a suitable tool for the assessment of Salmonella serovars, particularly as PCR reactions that identify additional O antigens are developed.
4.3. Some Salmonella serovars and subtypes frequently recovered from human clinical cases were also regularly found among healthy cattle and dairy farm environments
In addition to Salmonella serovars that were frequently isolated from healthy cattle and dairy farm environments but rare among human clinical cases, we also identified a number of serovars commonly associated with human clinical cases. For example, the 3rd and 4th most commonly isolated serovars in this study were Typhimurium and Newport, which are among the top 3 Salmonella serovars isolated from laboratory-confirmed human cases in the U.S. (CDC, 2011). Furthermore, 10 of the 26 serovars isolated here were reported among the 20 most commonly isolated serovars from human clinical cases in 2009 (CDC, 2011). When the PFGE types for 381 isolates from subclinical cattle and farm environments were compared to 1,849 PFGE types from human isolates collected in New York, a total of 14 PFGE types representing 12 serovars were indistinguishable from PFGE types from human isolates. Even though Salmonella Cerro is rarely associated with human illness, three human isolates matched Cerro isolates from healthy cattle and dairy farm environments by PFGE, indicating that Cerro may, in rare cases, cause human illnesses, consistent with a few reports of Salmonella Cerro-associated human salmonellosis (CDC, 1985; Mammina et al., 2000; Tewari et al., 2012). While our data do not specifically show that the farms sampled here were sources of human cases, our data support that subclinically infected cattle as well as farm environments may represent reservoirs or sources of Salmonella serovars and PFGE types commonly associated with human disease. Similarly, other reports have shown that a considerable proportion of Salmonella isolates from dairy cattle with clinical signs can match human clinical isolates by PFGE (Gupta et al., 2003).
4.4. Subclinical cattle and dairy farm environments are sources of drug-resistant Salmonella
Our data also showed that a number of Salmonella isolates from healthy cattle and farm environments are resistant to antimicrobial drugs, including some isolates resistant to 8 or more drugs. Identification of a number of MDR S. Newport is consistent with previous studies that found a considerable number of MDR S. Newport among cattle with clinical salmonellosis (Cummings et al., 2009b; Rankin et al., 2002). Previous studies have also found antimicrobial drug-resistant Salmonella, including MDR isolates, in healthy cattle and dairy farms in Thailand and the U.S. (Chuanchuen et al., 2010; Cobbold et al., 2006). In another study, Perron et al. (2008) showed that Salmonella from subclinically infected livestock representing species otherthan cattle (i.e., swine) can also be drug-resistant. Overall, these data suggest that not only clinically affected animals, but also healthy animals and farm environments not associated with animal salmonellosis cases or outbreaks, can be possible sources of MDR Salmonella.
5. Conclusion
Our data indicate that healthy livestock and farm environments may represent a potentially important reservoir of Salmonella serovars and subtypes associated with human infections, particularly considering that nearly 25% of Salmonella isolates were resistant to multiple antimicrobial agents. Subclinical shedding of Salmonella by dairy cattle thus represents a potential public health issue, particularly because fecal shedding results in widespread environmental contamination and an increased risk of within-herd transmission, both of which can promote zoonotic transmission through foodborne exposure or direct contact (e.g., for farm visitors). In addition, shedding animals cannot be recognized through clinical signs, which reduces the likelihood of adequate biosecurity efforts and quarantine efforts for these non- clinical shedders. While some studies have suggested that direct contact with cattle or other livestock is a risk factor for acquiring human salmonellosis (Cummings et al., 2012), further studies will be needed to quantitatively define the risk of transmission from healthy dairy cattle or cattle with clinical signs of salmonellosis and dairy-associated environments to humans.
Supplementary Material
List of the 1,349 Salmonella isolates obtained from dairy cattle and dairy farm environments from 46 NY state farms.
Salmonella isolates for which additional serovar analyses were performed.
Antimicrobial resistance types among 90 Salmonella isolates with resistance to at least one antimicrobial.
NotI-PFGE types for 10 Salmonella Cerro isolates represent 4 different XbaI-PFGE types. These isolates were differentiated into 2 NotI-PFGE types.
Acknowledgements
This project was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number N01-AI-30054.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Centers for Disease Control and Prevention (CDC) Epidemiologic notes and reports salmonellosis associated with carne seca-New Mexico, MMWR. Morbidity and Mortality Weekly Report. 1985 [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Salmonella Surveillance: Annual Summary, 2009. US Department of Health and Human Services; 2011. [Google Scholar]
- Chuanchuen R, Ajariyakhajorn K, Koowatananukul C, Wannaprasat W, Khemtong S, Samngamnim S. Antimicrobial resistance and virulence genes in Salmonella enterica isolates from dairy cows. Foodborne Pathog. Dis. 2010;7:63–69. doi: 10.1089/fpd.2009.0341. [DOI] [PubMed] [Google Scholar]
- Clinical Laboratory Standard Institute (CLSI) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; Approved standard, 3rd Edition. CLSI document M31-A3. Wayne, Pennsylvania: 2008. [Google Scholar]
- Cobbold RN, Rice DH, Davis MA, Besser TE, Hancock DD. Long-term persistence of multi–drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 2006;228:585–591. doi: 10.2460/javma.228.4.585. [DOI] [PubMed] [Google Scholar]
- Cummings KJ, Warnick LD, Alexander KA, Cripps CJ, Gröhn YT, James KL, McDonough PL, Reed KE. The duration of fecal Salmonella shedding following clinical disease among dairy cattle in the northeastern USA. Prev. Vet. Med. 2009a;92:134–139. doi: 10.1016/j.prevetmed.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KJ, Warnick LD, Alexander KA, Cripps CJ, Gröhn YT, McDonough PL, Nydam DV, Reed KE. The incidence of salmonellosis among dairy herds in the northeastern United States. J. Dairy Sci. 2009b;92:3766–3774. doi: 10.3168/jds.2009-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KJ, Warnick LD, Elton M, Gröhn YT, McDonough PL, Siler JD. The effect of clinical outbreaks of salmonellosis on the prevalence of fecal Salmonella shedding among dairy cattle in New York. Foodborne Pathog. Dis. 2010a;7:815–823. doi: 10.1089/fpd.2009.0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KJ, Warnick LD, Elton M, Rodriguez-Rivera LD, Siler JD, Wright EM, Gröhn YT, Wiedmann M. Salmonella enterica serotype Cerro among dairy cattle in New York: An Emerging Pathogen? Foodborne Pathog. Dis. 2010b;7:659–665. doi: 10.1089/fpd.2009.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KJ, Warnick LD, Davis MA, Eckmann K, Gröhn YT, Hoelzer K, MacDonald K, Root TP, Siler JD, McGuire SM, Wiedmann M, Wright EM, Zansky SM, Besser TE. Farm animal contact as risk factor for transmission of bovine-associated Salmonella subtypes. Emerg. Infect. Dis. 2012;18:1929–1936. doi: 10.3201/eid1812.110831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PR, Ewing WH. Edwards and Ewing’s identification of Enterobacteriaceae. 4th ed. Elsevier, University of Michigan; 1986. [Google Scholar]
- Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 2007;45:3323–3334. doi: 10.1128/JCM.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald C, Gheesling L, Collins M, Fields PI. Sequence analysis of the rfb loci, encoding proteins involved in the biosynthesis of the Salmonella enterica O17 and O18 antigens: serogroup-specific identification by PCR. Appl. Environ. Microbiol. 2006;72:7949–7953. doi: 10.1128/AEM.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration (FDA) National Antimicrobial Resistance Monitoring System – enteric bacteria (NARMS): 2010 Executive Report. Rockville, MD: U.S. Department of Health and Human Services, Food and Drug Administration; 2012. [Google Scholar]
- Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, Weill F-X. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2010;161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Gupta A, Fontana J, Crowe C, Bolstorff B, Stout A, Van Duyne S, Hoekstra MP, Whichard JM, Barrett TJ, Angulo FJ. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 2003;188:1707–1716. doi: 10.1086/379668. [DOI] [PubMed] [Google Scholar]
- Herrera-León S, Ramiro R, Arroyo M, Díez R, Usera MA, Echeita MA. Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res. Microbiol. 2007;158:122–127. doi: 10.1016/j.resmic.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Hoelzer K, Cummings KJ, Wright EM, Rodriguez-Rivera LD, Roof SE, Moreno Switt AI, Dumas N, Root T, Schoonmaker-Bopp DJ, Gröhn YT, Siler JD, Warnick LD, Hancock DD, Davis MA, Wiedmann M. Salmonella Cerro isolated over the past twenty years from various sources in the US represent a single predominant pulsed field gel electrophoresis type. Vet. Microbiol. 2011a;150:389–393. doi: 10.1016/j.vetmic.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzer K, Moreno Switt AI, Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Vet. Res. 2011b;42:34. doi: 10.1186/1297-9716-42-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley RA, Arrus KM, Ominski KH, Tenuta M, Blank G. Salmonella survival in manure-treated soils during simulated seasonal temperature exposure. J. Environ. Qual. 2006;35:1170–1180. doi: 10.2134/jeq2005.0449. [DOI] [PubMed] [Google Scholar]
- Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D’Angelo M, Hurd S, Medus C, Cronquist A, Angulo FJ. Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 2008;198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- Mammina C, Cannova L, Carfi Pavia S, Natasi A. Endemic presence of Salmonella enterica serotype Cerro in southern Italy. Euro Surveill. 2000;5:84–86. doi: 10.2807/esm.05.07.00028-en. [DOI] [PubMed] [Google Scholar]
- Murase T, Yamada M, Muto T, Matsushima A, Yamai S. Fecal excretion of Salmonella enterica serovar Typhimurium following a food-borne outbreak. J. Clin. Microbiol. 2000;38:3495–3497. doi: 10.1128/jcm.38.9.3495-3497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron GG, Quessy S, Bell G. A reservoir of drug-resistant pathogenic bacteria in asymptomatic hosts. PLoS One. 2008;3:e3749. doi: 10.1371/journal.pone.0003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri ML, Shi C, Moreno Switt AI, den Bakker HC, Wiedmann M. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J. Clin. Microbiol. 2013;51:1786–1797. doi: 10.1128/JCM.03201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SC, Aceto H, Cassidy J, Holt J, Young S, Love B, Tewari D, Munro DS, Benson CE. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. Appl. Environ. Microbiol. 2002;40:4679–4684. doi: 10.1128/JCM.40.12.4679-4684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer Y, Alcaine SD, Schoonmaker-bopp DJ, Root TP, Warnick LD, Mcdonough PL, Dumas NB, Gröhn YT. Pulsed-field gel electrophoresis diversity of human and bovine clinical Salmonella isolates. Foodborne Pathog. Dis. 2010;7:707–717. doi: 10.1089/fpd.2009.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari D, Sandt CH, Miller DM, Jayarao BM, M’ikanatha NM. Prevalence of Salmonella Cerro in laboratory-based submissions of cattle and comparison with human infections in Pennsylvania, 2005–2010. Foodborne Pathog. Dis. 2012;9:928–933. doi: 10.1089/fpd.2012.1142. [DOI] [PubMed] [Google Scholar]
- Torpdahl M, Skov MN, Sandvang D, Baggesen DL. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J. Microbiol. Meth. 2005;63:173–184. doi: 10.1016/j.mimet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- USDA-NAHMS. SalmonellaListeria and Campylobacter on US Dairy Operations 1996–2007. Fort Collins, CO: 2011. Dairy 2007. [Google Scholar]
- Van Kessel JS, Karns JS, Lombard JE, Kopral CA. Prevalence of Salmonella enterica Listeria monocytogenes and Escherichia coli virulence factors in bulk tank milk and in-line filters from U.S. dairies. J. Food Prot. 2011;74:759–768. doi: 10.4315/0362-028X.JFP-10-423. [DOI] [PubMed] [Google Scholar]
- Van Kessel JS, Karns JS, Wolfgang DR, Hovingh E, Schukken YH. Longitudinal study of a clonal, subclinical outbreak of Salmonella enterica subsp enterica serovar Cerro in a U.S. dairy herd. Foodborne Pathog. Dis. 2007;4:449–461. doi: 10.1089/fpd.2007.0033. [DOI] [PubMed] [Google Scholar]
- Zheng J, Keys CE, Zhao S, Meng J, Brown EW. Enhanced subtyping scheme for Salmonella enteritidis. Emerg. Infect. Dis. 2007;13:1932–1935. doi: 10.3201/eid1312.070185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Lin W-J, Foley SL, Chen C-H, Nayak R, Chen JJ. Evaluation of pulsed field gel electrophoresis profiles for identification of Salmonella serotypes. J. Clin. Microbiol. 2010;48:3122–3126. doi: 10.1128/JCM.00645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the 1,349 Salmonella isolates obtained from dairy cattle and dairy farm environments from 46 NY state farms.
Salmonella isolates for which additional serovar analyses were performed.
Antimicrobial resistance types among 90 Salmonella isolates with resistance to at least one antimicrobial.
NotI-PFGE types for 10 Salmonella Cerro isolates represent 4 different XbaI-PFGE types. These isolates were differentiated into 2 NotI-PFGE types.