Abstract
Recent research using Drosophila melanogaster has seen a resurgence in studies of metabolism and physiology. This review focuses on major methods used to conduct this work. These include protocols for dietary interventions, measurements of triglycerides, cholesterol, glucose, trehalose, and glycogen, stains for lipid detection, and the use of gas chromatography/mass spectrometry (GC/MS) to detect major polar metabolites. It is our hope that this will provide a useful framework for both new and current researchers in the field.
1. Introduction
Metabolism and physiology provided a major focus for Drosophila research in the middle of the last century [1]. This work included fundamental studies of biochemical genetics, starting with classic work on eye pigment biosynthesis and the one gene-one enzyme hypothesis [2], as well as detailed characterization of basic energy physiology [e.g. ref. 3, 4, 5]. Although the advent of recombinant DNA technology shifted the Drosophila field toward studies of developmental biology, recent efforts have refocused on metabolism, with a particular emphasis on the mechanisms that maintain energy homeostasis and the use of Drosophila as a model for studies of diabetes and obesity [6–10]. In this review, we highlight methods that have been developed to conduct current genetic studies of metabolism in Drosophila, with a focus on energy homeostasis and physiology. We begin with an overall discussion of the challenges facing researchers in the field, given the contributions of both environmental and genetic factors to metabolic control. We then move on to review protocols for dietary intervention in Drosophila, followed by widely used assays to quantify basic metabolites in the animal. We end with protocols for metabolomic profiling by gas chromatography-mass spectrometry (GC/MS). Our goal is not to be comprehensive in our coverage of metabolic protocols, since that is beyond the scope of one article, but rather to focus on some major methods currently being used, with the hope that this will provide a useful framework for both new and current researchers in the field.
2. Experimental design
The profound interplay between environment and genetics requires that metabolic studies be undertaken with careful attention to genetic background, diet, stock maintenance, and statistical analysis of data. Particular concern should be directed toward the selection of control strains and the establishment of an appropriately matched genetic background for the mutant being studied. Outcrossing mutants to the control strain provides a good way to achieve this goal, as does confirming results with more than one control line. Matching the genetic background becomes more difficult, however, when GAL4 drivers and UAS transgenes are included in the genotype, although a uniform white mutant background and controls with the driver and/or responder alone can be included. Tissue-specific RNAi provides an invaluable and widely used tool for functional studies of metabolism. In addition to the standard concerns with off-target effects, however, RNAi can have nonspecific effects on physiology [e.g. ref. 11]. Finally, some care should be taken when using commercial kits for quantifying specific metabolites. Although these kits may be widely used in mammalian research, most have not been validated in Drosophila. As discussed below for quantifying simple metabolites such as glucose, glycogen, and triglycerides, protocols need to be adapted to the system. A good way to achieve this for a new assay is to use known mutants in the pathway being studied, and score for the predicted changes in metabolite levels. Alternatively, methods such as GC/MS can be used to confirm the changes in metabolite levels detected by an enzymatic kit. In general, the more labile the metabolite (such as ATP), the more care that is needed for its quantification.
3. Starvation and dietary paradigms
The metabolic state of an animal is intimately linked with its diet. Accordingly, dietary conditions must be carefully controlled when performing metabolic studies. Often this will require the lab making special batches of media to maintain both breeding and experimental stocks. This point becomes particularly important when considering the recent evidence that parental diet can influence the metabolic state of offspring, as has been well documented in mammals [12, 13]. While carefully controlled dietary conditions are essential, dietary manipulation also provides one of the most useful approaches for studies of metabolism and animal physiology. For example, enrichment or restriction of particular nutrients can often evoke robust metabolic phenotypes in mutants that may otherwise appear normal under ideal dietary conditions [14–17].
Starvation provides an important dietary stress that tests the animal’s ability to mobilize stored nutrients for survival. This can be achieved by transferring larvae or flies to either water or PBS. Although moist filter paper is employed for larvae, studies of adult starvation should use 1% agar as a substrate, as it provides a more uniform moist environment for long-term studies (>12 hours).
Alteration of dietary sugar levels is a useful method for investigating potential defects in carbohydrate homeostasis. This can be easily achieved by using an 8% yeast and 1% agar medium to serve as the dietary foundation for manipulating sugar concentrations. A range from 3–15% dietary sugar (ratio of 2:1 glucose to sucrose, by weight) represents the low-to-high spectrum found within a relatively normal diet for Drosophila (Table 1). In our studies, altering the sugar concentration of this media has proven to be an effective tool for both enhancing and suppressing diabetic phenotypes. For example, several Drosophila mutants that are sensitive to dietary sugar can maintain euglycemia on a 3% sugar diet but display increasingly severe hyperglycemia as dietary sugar is increased (W. Barry, D. Bricker, and R. Somer, unpublished results).
Table 1.
Caloric content of Drosophila diets
| Semi-defined (Bloomington) |
g/liter | Carbs (g) | Protein (g) | Fat (g) | Total kcal |
|---|---|---|---|---|---|
| agar | 10 g | 8.9 | 0 | 0.1 | |
| Yeast | 80 g | 30.8 | 38.6 | 3.12 | |
| Yeast extract | 20 g | 7.7 | 8.6 | 1.72 | |
| Peptone | 20 g | 0.1 | 14.6 | 0 | |
| Sucrose | 30 g | 30 | 0 | 0 | |
| Glucose | 60 g | 60 | 0 | 0 | |
| Total grams | 137.5 | 61.8 | 4.94 | ||
| Total kcal | 550 | 247.2 | 42.29 | 839.5 | |
| Percent total kcal | 65.5 | 29.4 | 5.04 |
| 9% Sugar Diet (normal) |
g/liter | Carbs (g) | Protein (g) | Fat (g) | Total kcal |
|---|---|---|---|---|---|
| agar | 10 g | 8.9 | 0 | 0.1 | |
| Yeast | 80 g | 30.8 | 38.6 | 3.12 | |
| Sucrose | 30 g | 30 | 0 | 0 | |
| Glucose | 60 g | 60 | 0 | 0 | |
| Total grams | 129.7 | 38.6 | 3.22 | ||
| Total kcal | 518.8 | 154.4 | 27.56 | 700.8 | |
| Percent total kcal | 74.0 | 22.0 | 3.93 |
| 3% Sugar Diet (low sugar) |
g/liter | Carbs (g) | Protein (g) | Fat (g) | Total kcal |
|---|---|---|---|---|---|
| agar | 10 g | 8.9 | 0 | 0.1 | |
| Yeast | 80 g | 30.8 | 38.6 | 3.12 | |
| Sucrose | 10 g | 10 | 0 | 0 | |
| Glucose | 20 g | 20 | 0 | 0 | |
| Total grams | 69.7 | 38.6 | 3.22 | ||
| Total kcal | 278.8 | 154.4 | 27.56 | 460.8 | |
| Percent total kcal | 60.5 | 33.5 | 5.98 |
| 15% Sugar Diet (high sugar) |
g/liter | Carbs (g) | Protein (g) | Fat (g) | Total kcal |
|---|---|---|---|---|---|
| agar | 10 g | 8.9 | 0 | 0.1 | |
| Yeast | 80 g | 30.8 | 38.6 | 3.12 | |
| Sucrose | 50 g | 50 | 0 | 0 | |
| Glucose | 100 g | 100 | 0 | 0 | |
| Total grams | 189.7 | 38.6 | 3.22 | ||
| Total kcal | 758.8 | 154.4 | 27.56 | 940.8 | |
| Percent total kcal | 80.7 | 16.4 | 2.93 |
| Diabetic diet | g/liter | Carbs (g) | Protein (g) | Fat (g) | Total kcal |
|---|---|---|---|---|---|
| agar | 10 g | 8.9 | 0 | 0.1 | |
| Yeast | 80 g | 30.8 | 38.6 | 3.12 | |
| Yeast extract | 20 g | 7.7 | 8.6 | 1.72 | |
| Peptone | 20 g | 0.1 | 14.6 | 0 | |
| Sucrose | 342 g | 342 | 0 | 0 | |
| Total grams | 389.5 | 61.8 | 4.94 | ||
| Total kcal | 1558 | 247.2 | 42.29 | 1847.5 |
Diabetic diet from Musselman et al. (2011) Dis. Models & Mech. 4: 842–849.
Very high sugar diets have been developed for studies of type 2 diabetes in Drosophila. These diets consist of either 1% agar, 3.4% yeast, 8.3% cornmeal, and 30% sucrose [15] or 1% agar, 8% yeast, 2% yeast extract, 2% peptone, and 34.2% sucrose [18] (Table 1). These diets lead to hallmarks of type 2 diabetes in wild-type Drosophila, including hyperglycemia, insulin resistance, obesity, and cardiac dysfunction [15, 18, 19]. High lipid diets have also been developed for studies of diet-induced obesity in Drosophila. These can be achieved by supplementation of standard growth media with either soy lipids [20] or coconut oil [21, 22], which has been most widely used for this purpose.
Although the above media allow the researcher to crudely manipulate the overall levels of carbohydrates and lipids in the diet, precise addition or removal of dietary components is often desirable. Fortunately, two labs have recently developed chemically defined diets that support Drosophila development and growth (although at a reduced rate), and are well suited for the study of adult physiology and aging [23, 24]. The advent of these defined diets provides a level of control not possible in past work, and should greatly facilitate nutritional and metabolic studies in Drosophila.
3.1 Measuring feeding rate
It is often important to assess whether changes in feeding rate contribute to an observed metabolic phenotype. The use of dietary dyes can provide a crude yet simple method to visually assess food consumption [25]. This approach is particularly useful when analyzing responses to an acute feeding bout, by ensuring that only animals that have consumed the food are analyzed. Dietary dyes are not well suited for quantitative analysis, however, and cannot be used to accurately assess feeding rate over extended periods of time. In contrast, adding radioactive material to the food provides a more quantitative measurement of consumption based on ingested radiation after a fixed period of time [26]. This can be achieved for individual flies by adding 108 counts/min/ml (final volume) α-32P dCTP (or any other nucleotide) to the diet and permitting the animals to feed for 2 hours. Lower levels of radiation can be used if multiple flies are measured. Labeled animals are transferred to unlabeled food for several hours to allow them to clear radioactivity that is nonspecifically bound to the cuticle. They are then subjected to Cerenkov counting in a scintillation counter to quantify food uptake [27, 28]. Scintillation fluid can be used if the animals are sacrificed and lower levels of radiation are employed in the diet.
The Capillary Feeder (CAFE) assay has been developed as a means to directly quantify food consumption by making use of a capillary feeder containing a liquid medium [29]. While this technique allows for direct and precise analysis of ingested food quantities, some researchers have raised concerns that the diet and its method of delivery might not represent a natural feeding scenario [29, 30]. This led to an alternate technique that measures the number of times an animal extends its proboscis to feed in a fixed time period [30]. A more detailed description of these techniques can be found in a recent review on Drosophila feeding behavior [31].
4. Methods to measure basic metabolites: lipids
Under normal feeding conditions, dietary triglycerides (TAG) and cholesterol esters are broken down into free fatty acids, monoacylglycerols, and free sterols in the intestine. These digested lipids can then be absorbed by the intestinal cells, where TAG is resynthesized and packaged together with cholesterol, cholesterol esters, and carrier proteins to form lipoprotein particles that are trafficked throughout the body. These lipids can be either utilized by cells or deposited in storage tissues. Most lipids reside in the fat body, although some are also present in the gut and oenocytes of both larvae and adults. Two different types of assays can be used to detect lipids – either specific assays to quantify TAG or sterols from whole animals or dissected tissues, or lipophilic dyes to visualize neutral lipids within cells. Taken together, these provide a valuable indication of the lipid distribution within the animal.
4.1 Colorimetric quantification of triglycerides
Overall quantification of TAG is often a first step in a metabolic study, along with measurements of basic carbohydrates and protein. An accurate, although insensitive means of measuring TAG is to use Bligh and Dyer lipid extraction followed by thin layer chromatography (TLC) [32, 33]. This allows fractionation and detection of total fatty acids, diacylglycerol (the major circulating form of lipid in insects), and TAG. The most widely used assay for detecting triglycerides, however, is a coupled colorimetric assay that detects free glycerol levels after cleaving TAG with lipoprotein lipase [32, 34, 35]. Although a few studies have suggested that this approach does not provide an accurate assessment of stored fat in insects [36, 37], both TLC and the colorimetric assay produce similar results [32]. It is, however, worth noting that the colorimetric assay releases glycerol from mono- and diacylglycerides [38] in addition to TAG and, therefore, conclusions regarding any particular form of glycerolipid should be validated by TLC. The presence of eye pigment in adult samples could also interfere with accurate absorbance measurements at certain wavelengths; however, assay kits designed to measure absorbance at 540 nm do not detect eye pigment [32].
4.2 Protocol: Coupled colorimetric assay for triglycerides
Collect samples (25 mid-second instar larvae; 5 adult flies).
Rinse several times with 1 ml cold PBS to remove all traces of food that might be attached to the outside of the animal. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS. Snap freeze animals in liquid nitrogen for later homogenization or add 100 µl of cold PBS + 0.05% Tween 20 (PBST).
Rapidly homogenize animals in PBST with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000). Frozen samples should be kept on dry ice until addition of PBST. If samples are not kept cold, stored glycerolipids will be enzymatically degraded into free glycerol by endogenous enzymes and skew the final analysis.
Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80°C for later analysis.
Heat supernatant for 10 min at 70°C. Do not centrifuge the heat-treated lysate because lipids are partially insoluble in PBST. At this time, the heat-inactivated sample can be frozen and stored at −80°C, if desired.
Prepare standards: Dilute 40 µl of the glycerol standard solution (Sigma 2.5 mg/ml triolein equivalent glycerol standard; G7793) with 60 µl PBST (100 µl final volume) to generate a 1.0 mg/ml triolein equivalent standard. Do two 2-fold serial dilutions into PBST (50 µl 1 mg/ml + 50 µl PBST for 0.5 mg/ml standard, etc.) to generate 0.125, 0.25 and 0.5 mg/ml standards. Glycerol stock solutions can be stored at −20°C or 4°C.
Add 20 µl of the glycerol standards, fly samples, and a PBST blank to each of two 1.5 ml microfuge tubes. Add 20 µl of PBST to one tube (this sample will be used to measure free glycerol). Add 20 µl of triglyceride reagent (Sigma; T2449) to the other tube (the TAG in this sample will be digested by lipase to free the glycerol backbone). Incubate tubes at 37°C for 30–60 minutes.
Centrifuge for 3 min at full speed. Transfer 30 µl of each sample to a clear-bottom 96-well plate.
Add 100 µl of free glycerol reagent (Sigma; F6428) to each sample with a multichannel pipette and mix well. Seal the wells with parafilm to prevent evaporation and incubate the plate for 5 min at 37°C. Centrifuge the plate in an appropriate swing-bucket rotor to clear condensate from the sides of the wells and to remove any air bubbles present in the samples. Use a plate reader to measure absorbance at 540 nm.
Determine the TAG concentration for each sample by subtracting the absorbance for the free glycerol in the untreated samples from the total glycerol concentration in samples that have been incubated with triglyceride reagent. The TAG content in each sample is calculated based on the triolein-equivalent standard curve. This assay is linear from 0–1.0 mg/ml TAG.
As with any metabolic measurement, TAG levels must be considered in the larger context of animal growth and physiology. Mutations that delay developmental progression and alter adult body size could produce proportional changes in TAG in that do not reflect metabolic defects. Similarly, wild-type adult flies do not feed for a prolonged period after eclosion, and need to be aged several days to achieve maturity [39]. Thus to prevent inaccurate results, all samples should be developmentally staged and normalized to an internal parameter such as soluble protein (although the effects of a mutation on protein levels may require another method, such as body weight, for normalization). A simple method for measuring protein concentration is to remove 10 µl of homogenized sample prior to heat treatment, dilute between 1:10 and 1:20 in cold PBS, and use 10 µl of the dilution to conduct a Bradford assay [40]. If protein concentration is not measured immediately, samples should be stored at −80°C.
4.3 Cholesterol quantification
Cholesterol is an essential component of cell membranes and also the precursor for steroid hormone biosynthesis. In addition to free cholesterol, cholesterol esterified to long chain fatty acids is present in circulating lipoprotein particles as well as stored in lipid droplets. A fluorometric assay has been widely used to quantify free cholesterol in Drosophila larvae and adults [14, 41–44]. In addition, by sonicating the extracts to emulsify lipids and including cholesterol esterase in the reaction, cholesterol esters can be digested into free cholesterol, which can then be quantified by the fluorometric assay and compared to the background level of free cholesterol present in the sample. We use the Amplex Red Cholesterol assay (Invitrogen; A12216) to detect either free cholesterol or esterified cholesterol, as described below. This kit uses cholesterol oxidase to convert free cholesterol into cholest-4-en-3-one and hydrogen peroxide. The hydrogen peroxide then reacts with 10-acetyl-3,7-dihydroxyphenoxazine in the presence of horseradish peroxidase to generate resorufin, which can be detected by its fluorescence at ~590 nm. It should be noted that this assay will detect all sterols, including the abundant sterols in Drosophila (ergosterol and dehydrocholesterol), all of which are defined by a hydroxyl group at the 3-position of the A-ring [33].
4.4 Protocol: Fluorescent assay for free cholesterol
This assay does not involve sonicating the samples and thus measures primarily free sterols. Sonication is needed to release and emulsify the cholesterol esters stored in lipid droplets. Although cholesterol esterase is included in this assay, it makes little difference in the final result.
Follow the Amplex Red Cholesterol Assay Kit (Invitrogen; A12216) instructions to prepare aliquots of the 20 mM Amplex Red reagent in DMSO, 200 U/ml HRP in 1× reaction buffer, 200 U/ml cholesterol oxidase in 1× reaction buffer, and 200 U/ml cholesterol esterase in 1× reaction buffer. These can be stored at −20°C for later use.
Collect samples in 1.5 ml microfuge tubes (75 embryos, 30 first instar larvae, 5 adult male flies). Rinse several times with cold PBS to remove all traces of food that might be attached to the outside of the animal. Embryos should be dechorionated following standard procedures and subsequently transferred to a 1.5 ml microfuge tube. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS.
Homogenize animals in 100 µl of 1× reaction buffer with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000).
Centrifuge homogenate for 5 min at 2,300 × g at room temperature. Transfer the supernatant to a fresh 1.5 ml microfuge tube, including the upper “fatty” layer and mix gently. Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80°C for later analysis.
Prepare cholesterol standards in 1× reaction buffer using the 2 mg/ml cholesterol reference standard provided in the kit. We prepare a dilution series, ending up with at least 50 µl each of 0, 2, 4, 6, 8 µg/ml cholesterol.
Add 50 µl of the cholesterol standards and a 1× reaction buffer blank to the first row of a black 96 well plate (Optiplate-96 Perkin Elmer; 6005270). In the next row, add 50 µl of each sample into individual wells.
- Prepare the reaction mix using stored aliquots:
- 15 µl 20 mM Amplex Red reagent in DMSO
- 10 µl 200 U/ml HRP in 1× reaction buffer
- 10 µl 200 U/ml cholesterol oxidase in 1× reaction buffer
- 1 µl 200 U/ml cholesterol esterase in 1× reaction buffer
- + 1494 µl 1× reaction buffer
Add 90 µl of reaction mix to each well with a multichannel pipette. Be sure to keep in the dark and incubate at least 30 min at 37°C.
Measure immediately using a fluorescence plate reader with excitation at 530 nm and emission at 590 nm. Each data point resulting from the assay should represent an average of at least three collections of animals, and the assay should be repeated at least three times.
4.5 Protocol: Fluorescent assay for cholesterol esters
This assay uses sonication of the animal homogenates to release and emulsify the cholesterol esters stored in lipid droplets. Each sample is then divided into two tubes, one of which is treated with cholesterol esterase. The amount of cholesterol esters is determined by subtracting the background level of free cholesterol in the untreated sample from the total amount of esterified and free cholesterol present in the sample treated with cholesterol esterase.
Follow the Amplex Red Cholesterol Assay Kit (Invitrogen; A12216) instructions to prepare aliquots of the 20 mM Amplex Red reagent in DMSO, 200 U/ml HRP in 1× reaction buffer, and 200 U/ml cholesterol oxidase in 1× reaction buffer. These can be stored at −20°C for later use.
Collect samples in 1.5 ml microfuge tubes (30 adult male flies). Rinse animals several times with cold PBS to remove all traces of food. Carefully remove all liquid and homogenize animals in 250 µl of cold PBS + 0.05% Tween 20 (PBST) with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000). Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Protein samples can be frozen and stored at −80°C for later analysis.
Add 650 µl of cold PBST to each sample and sonicate on ice using a tip sonicator, three times for 30 seconds each, at a medium setting to prevent foaming.
Split each sample into two 1.5 ml microfuge tubes and add 10 µl of 200 U/ml cholesterol esterase (either Sigma C1403 or C3766) to one set of tubes.
Incubate both sets of tubes overnight (> 16 hrs) at 37°C with occasional vortexing.
Extract all tubes with 900 µl 2:1 chloroform:methanol by shaking the tubes several times vigorously over a period of about 3 minutes. Centrifuge to separate the phases, remove the lower organic phase, and transfer to a fresh 1.5 ml microfuge tube. Use a centrifugal vacuum concentrator to remove all of the organic solvent, leaving an oily lipid residue (approx. 30 minutes).
Solubilize lipids in 500 µl PBST by vortexing and sonicating as above.
Prepare free cholesterol standards in 1× reaction buffer using the 2 mg/ml cholesterol reference standard provided in the kit. We prepare a dilution series, ending up with at least 50 µl each of 0, 2, 4, 6, 8 µg/ml cholesterol.
Add 50 µl of the cholesterol standards and a 1× reaction buffer blank to the first row of a black 96 well plate (Optiplate-96 Perkin Elmer; 6005270). In the next rows, add a 1:1 dilution of each sample with 1× reaction buffer (25 µl of each sample + 25 µl 1× reaction buffer) to each well.
- Prepare the reaction mix using stored aliquots:
- 15 µl 20 mM Amplex Red reagent in DMSO
- 10 µl 200 U/ml HRP in 1× reaction buffer
- 10 µl 200 U/ml cholesterol oxidase in 1× reaction buffer
- + 1495 µl 1× reaction buffer
Add 90 µl of reaction mix to each well with a multichannel pipette. Be sure to keep in the dark and incubate for at least 30 min at 37°C.
Measure immediately using a fluorescence plate reader with excitation at 530 nm and emission at 590 nm.
Determine the amount of cholesterol esters by subtracting the measurements performed on samples that were not treated with cholesterol esterase from those that were treated with the enzyme. Each data point resulting from the assay should represent an average of at least three collections of animals, and the assay should be repeated at least three times.
4.6 Stains
While quantitative assays provide a means of measuring total stored TAG or cholesterol, staining of these neutral lipids in the intestine, fat body, and oenocytes provide a valuable cell based assay. This is especially powerful when combined with clonal analysis of mutant cells [45–47]. If relative quantification between samples is the goal, however, all samples should be prepared simultaneously and analyzed to ensure that the final results are not influenced by variations in temperature, dye concentration, incubation time, or microscope settings. Ideally, experimental and control slides should be randomized and scored blindly to eliminate confirmation bias. Many dyes are available for lipid detection, with Oil Red O, Sudan Black, and Nile Red being the most commonly used.
4.6a Oil Red O
The dye Oil Red O provides a reliable means to visually assess neutral lipids in fixed tissues [48–51]. Animals are dissected and fixed using 4% formaldehyde in PBS for 30 minutes, washed twice in PBS and twice in 100% propylene glycol. Fixed tissues are then stained in a solution of 0.5% Oil Red O (Sigma; O0625) dissolved in propylene glycol, which has been filtered through Whatman #1 filter paper and preheated to 60°C. Allow samples to incubate for 1 hour at 60°C, then wash twice with 85% propylene glycol and twice with PBS at room temperature. Stained samples are mounted on microscope slides using glycerol. The use of this dye in Drosophila is well documented and provides a sensitive means to assay neutral lipids in the oenocytes and intestine. The density of TAG in the fat body, however, results in an Oil Red O stain that is too intense for identifying modest defects in fat storage.
4.6b Sudan Black
Similar to Oil Red O, the dye Sudan Black allows for an accurate assessment of neutral lipid stores in fixed tissues [49, 52]. Tissues that have been fixed with 4% formaldehyde are rinsed twice with PBS, once with 50% ethanol, and then stained for 2 minutes at room temperature with prefiltered 0.5% Sudan Black (Sigma; 199664) dissolved in 75% ethanol. Once staining is completed, samples are sequentially rinsed with 50% ethanol, 25% ethanol, and PBS before mounting on a microscope slide with glycerol. In general, Sudan Black results in less intense tissue stains than Oil Red O and thus is better suited for visualizing the high levels of TAG in the fat body.
4.6c Nile Red
Unlike Oil Red O and Sudan Black, Nile Red can be used to visualize neutral lipids in unfixed tissues [53]. Animals should be quickly dissected in a solution of 0.00002% Nile Red (Sigma; 19123) and 75% glycerol, and visualized using confocal microscopy. Although relatively straightforward, the use of Nile Red is somewhat limited and can be employed improperly [54–56]. While this dye allows for the qualitative assessment of lipid droplet size and shape, it should not be used to quantitatively measure fat reserves.
5. Methods to measure basic metabolites: carbohydrates
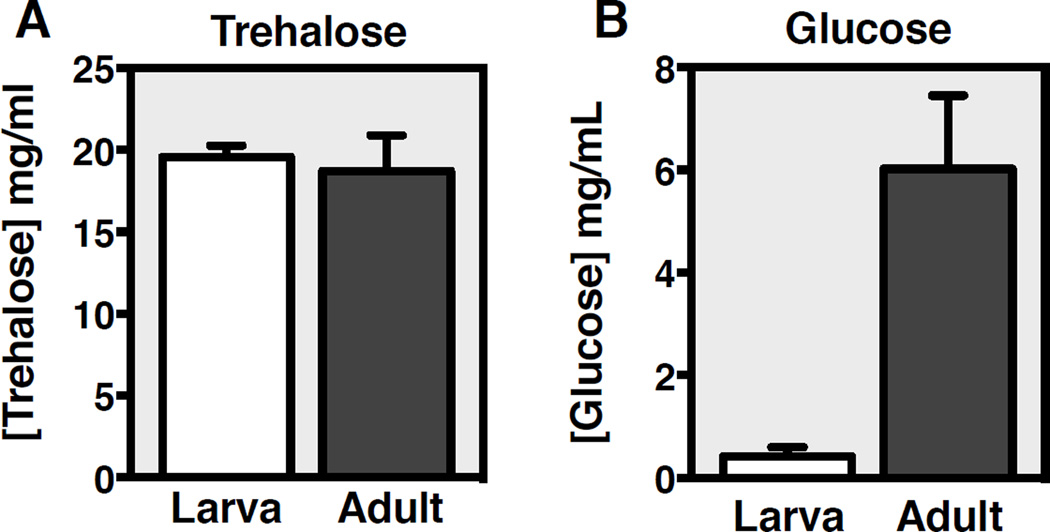
Proper regulation of carbohydrate homeostasis is critical for maintaining normal physiology. The two primary forms of circulating carbohydrates in Drosophila are glucose and trehalose (a disaccharide of glucose). While trehalose is abundant in larval hemolymph, circulating glucose and trehalose can be readily detected in the adult fly (Figure 1). These sugars serve a variety of purposes, including providing an essential energy source through glycolysis, substrates for biosynthetic reactions in growing animals, and energy storage in the form of glycogen.
Figure 1. Circulating trehalose and glucose levels in larvae and adults.
(A) Circulating trehalose is approximately equivalent and abundant during larval and adult stages. (B) In contrast, circulating glucose is low in larvae and is elevated during adulthood. Hemolymph was collected from w1118 mid-third instar larvae and five day old adults maintained on a standard cornmeal-based lab medium by puncturing the cuticle with a tungsten needle followed by centrifugation through glass wool into a collection tube. Circulating trehalose and glucose concentrations were determined using the HK assay kit.
5.1 Enzymatic Assays
A number of enzymatic assays are commercially available to quantify levels of specific sugars and related metabolic intermediates. Although initially developed for use in mammalian systems, many of these assays have been successfully co-opted for use in Drosophila. Care should be taken, however, to properly validate each enzymatic assay for a particular experimental system and/or developmental stage.
5.1a Glucose
Two enzymatic-based methods have been widely used to measure free glucose levels in flies. The first of these utilizes the enzyme Hexokinase (HK) to phosphorylate glucose, producing glucose-6-phosphate [57]. Subsequent oxidation of glucose-6-phosphate by glucose-6-phosphate dehydrogenase produces 6-phosphogluconate and NADH. Spectrophotometric readings at 340 nm can then be used to assess the amount of NADH produced, as it naturally absorbs ultraviolet light at this wavelength. With this method, NADH production is directly proportional to the starting glucose concentration. Alternatively, free glucose levels can be measured through a colorimetric-based enzymatic assay. This method employs Glucose Oxidase (GO) activity to catalyze the oxidation of glucose to hydrogen peroxide and gluconic acid [58]. Subsequently, the hydrogen peroxide and an added compound, o-dianisidine, react to produce an oxidized form of o-dianisidine (orange color) in the presence of peroxidase. Sulfuric acid is then added to stop the reaction and convert the color to a pink hue, which can be quantified at an absorbance of 540 nm.
In our hands, the GO assay cannot be used to accurately quantify carbohydrates in pupae; rather, the HK protocol should be followed when studying this stage in development (W. Barry, unpublished results). This finding emphasizes the need to carefully validate the use of metabolite assays in new experimental contexts prior to their use. In addition, when using either the GO or HK protocol, it is important to keep the samples of homogenized tissue cold and to move rapidly prior to heat inactivation, which is essential to prevent the enzymatic breakdown of glycogen and trehalose into glucose by endogenous enzymes in the extract. Similar to the TAG assay described above, 10 µl of homogenate should be removed prior to heat inactivation and reserved for Bradford quantification of protein levels, providing a means to internally normalize glucose levels (although, as discussed above, effects of a mutation on protein levels may require another method for normalization).
5.1b Protocol: Glucose Quantification
-
1)
Collect samples (25 mid-second instar larvae; 5 adult flies).
-
2)
Rinse several times with 1 ml cold PBS to remove all traces of food that might be attached to the outside of the animal. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS. Snap freeze animals in liquid nitrogen for later homogenization or add 100 µl cold PBS.
-
3)
Rapidly homogenize animals in PBS with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000). Frozen samples should be kept on dry ice until addition of PBS. If samples are not kept cold, glycogen and trehalose will be enzymatically degraded into free glucose by endogenous enzymes and skew the final analysis.
-
4)
Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80°C for later analysis.
-
5)
Heat supernatant for 10 min at 70°C, then centrifuge for 3 min at maximum speed in a refrigerated tabletop centrifuge that has been prechilled to 4°C.
-
6)
Pipette the resulting supernatant into a new 1.5 ml microfuge tube. At this time, the heat-inactivated sample can be frozen and stored at −80°C, if desired.
-
7)
Prepare glucose standards: Dilute 16 µl of 1 mg/ml glucose with 84 µl PBS (100 µl final volume) for 0.16 mg/ml standard. Do four 2-fold serial dilutions into PBS (50 µl 0.16 mg/ml + 50 µl PBS for 0.08 mg/ml standard, etc.) to generate 0.01, 0.02, 0.04, 0.08, and 0.16 mg/ml glucose standards. These assays are linear from 0.01 to 0.16 mg/ml glucose. Glucose stock solutions can be stored at −20°C or 4°C.
-
8)
Add 30 µl of glucose standards and a PBS blank to the first row of a clear-bottom 96 well plate. In the next row, add 30 µl of each sample into an individual well (after diluting them 1:4–1:8 with PBS).
-
9a)
For GO assay:
Add 100 µl of GO reagent (Sigma; GAGO-20) to each well with a multichannel pipette. Seal the wells with parafilm to prevent evaporation and incubate the plate at 37°C for 30–60 min. Stop the reaction by adding 100 µl of 12 N H2SO4 (the samples should visibly change color from yellow/orange to pink). Centrifuge the plate in an appropriate swing-bucket rotor to clear condensate from the sides of the wells and to remove any air bubbles present in the samples. Use a plate reader to measure absorbance at 540 nm. Determine the free glucose concentration by comparing the free glucose measurements for each sample to the glucose standard curve.
-
9b)
For HK assay:
Add 100 µl of HK reagent (Sigma; GAHK20) to each well with a multichannel pipette. Seal the wells with parafilm to prevent evaporation and incubate the plate at room temperature for 15 min. Centrifuge the plate in an appropriate swing-bucket rotor to clear condensate from the sides of the wells and to remove any air bubbles present in the samples. Use a plate reader to measure absorbance at 340 nm. Determine the free glucose concentration by comparing the free glucose measurements for each sample to the glucose standard curve.
Glucose transporters are bidirectional, and thus free glucose will travel freely between intracellular and extracellular compartments. As a result, most free glucose is found in the circulating fluid rather than within cells. Measurements of free glucose from whole animal homogenates therefore provide an approximate assessment of circulating glucose levels. As always, however, accurate measurements of circulating trehalose or glucose can only be made by using hemolymph samples (see below).
5.1c Trehalose
Trehalose represents the major circulating sugar in Drosophila and, together with glucose, provides essential information about the metabolic state of an animal. Trehalose is quantified by an assay that is based on the protocol for measuring glucose levels, described above [59, 60]. The difference is that trehalase is added to the extract to digest the trehalose into free glucose, which can then be quantified by standard assays and compared to the background level of glucose present in the original sample.
5.1d Protocol: Trehalose Quantification
Collect samples (25 mid-second instar larvae; 5 adult flies).
Rinse several times with 1 ml cold PBS to remove all traces of food that might be attached to the outside of the animal. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS. Snap freeze animals in liquid nitrogen for later homogenization or add 100 µl cold Trehalase Buffer (TB) (5 mM Tris pH 6.6, 137 mM NaCl, 2.7 mM KCl).
Rapidly homogenize animals in TB with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000). Frozen samples should be kept on dry ice until addition of TB. If samples are not kept cold, glycogen and trehalose will be enzymatically degraded into free glucose by endogenous enzymes and skew the final analysis.
Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80ºC for later analysis.
Heat supernatant for 10 min at 70°C, then centrifuge for 3 min at maximum speed in a refrigerated tabletop centrifuge that has been prechilled to 4°C.
Pipette the resulting supernatant into a new 1.5 ml microfuge tube. At this time, the heat-inactivated sample can be frozen and stored at −80°C, if desired.
Prepare the trehalase stock (TS) by diluting 3 µl porcine trehalase (Sigma; T8778-1UN) in 1 ml TB.
-
Generate glucose and trehalose standards for standard curves:
For glucose standards, dilute 16 µl of 1 mg/ml glucose stock solution with 84 µl TB (100 µl final volume) to generate a 0.16 mg/ml standard. Conduct a series of 2-fold serial dilutions of the 0.16 mg/ml standard using TB (50 µl 0.16 mg/ml + 50 µl TB for 0.08 mg/ml standard) to generate 0.01, 0.02, 0.04, 0.08, and 0.16 mg/ml glucose standards. Aliquots of glucose stock solution can be stored at −20°C or 4°C.
For trehalose standards, dilute 16 µl of 1 mg/ml trehalose (Sigma; 90208) with 50 µl TS and 34 µl TB (100 µl final volume) for 0.16 mg/ml standard. Conduct a series of 2-fold serial dilutions of the 0.16 mg/ml standard using a 1:1 mix of TB and TS (e.g., 50 µl 0.16 mg/ml + 50 µl TB+TS for 0.08 mg/ml standard) to generate 0.01, 0.02, 0.04, 0.08, and 0.16 mg/ml trehalose standards. Aliquots of trehalose stock solution can be stored at −20°C or 4°C.
Add 30 µl of TB blank or glucose standards to 30 µl TB in 1.5 ml microfuge tubes.
Add 30 µl of TB blank or trehalose standards to two sets of 1.5 ml microfuge tubes. In one set, add 30 µl of TB to determine free glucose. In other set, add 30 µl TS to digest trehalose to glucose.
If necessary, dilute fly samples between 1:2 to 1:4 in TB. Larval samples do not need diluting at this step unless analyzing a mutant that possesses particularly high trehalose levels.
Add 30 µl of each fly sample to two 1.5 ml microfuge tubes. In the first tube, add 30 µl of TB to determine the background level of free glucose. In the second tube, add 30 µl of TS to digest trehalose into free glucose.
Incubate all standards and samples at 37°C for 18–24 hours.
Centrifuge at maximum speed in a tabletop centrifuge for 3 minutes. Transfer 30 µl of each sample into an individual well of a 96-well plate for analysis and use either the GO or HK assay to measure free glucose.
Free glucose concentration in each sample is calculated based on the glucose standard curve. For trehalose measurements, first subtract the absorbance measured for free glucose in the untreated samples from the absorbance of the samples that have been digested with trehalase. The trehalose content in each sample is then calculated based on the trehalose standard curve. This method is linear from 0.01 to 0.16 mg/ml trehalose.
5.1e Hemolymph
While analysis of whole animal homogenates provides a relatively easy and high throughput approach for quantifying glucose and trehalose, this method does not distinguish between circulating and stored sugars, and can be complicated by the presence of food within the gut. Therefore, hemolymph samples are used to directly assay circulating sugars. Several alternate methods for hemolymph collection have been described for both larvae and adults [61–64]. Protocols based on filtration by centrifugation and capillary-based collection of hemolymph are most widely used. The centrifugation approach requires approximately 30–50 adult females (adult males contain less hemolymph per fly and thus require larger numbers) carefully punctured in the thorax using a tungsten needle. Punctured flies are placed in a 0.5 ml microfuge tube that contains a hole at the bottom of the tube, which is packed with glass wool. This tube is then placed within a 1.5 ml collection tube and centrifuged at 9,000 × g for 5 minutes at 4°C, yielding approximately 1.5 µl of hemolymph. Longer centrifugation times, higher speeds, and more flies can increase yield, but may also increase cellular or intestinal contamination. One way to detect possible contamination from intestinal contents is to add food coloring to the medium and check the collected hemolymph for the absence of the dye. Additionally, hemolymph can be analyzed under a dissection scope for the absence of cellular debris.
Hemolymph can also be collected by immobilizing an adult fly wing-side down on a piece of double-sided tape and carefully puncturing the head cuticle with a tungsten needle [64]. Gentle pressure is applied to the thorax with a blunt object, and a capillary tube or pipette tip is used to collect the small drop of hemolymph that is forced out of the puncture site. While this method is labor intensive and not well suited for the collection of large numbers of samples, it minimizes potential hemolymph contamination.
Regardless of which method is used, a 1 µl aliquot of the collected hemolymph is diluted in 99 µl of trehalase buffer (1:100), followed by heat treatment for 5 minutes at 70°C to inactivate endogenous trehalase. After heat treatment, the sample is split into two 50 µl aliquots, one of which is diluted further with an equal volume of trehalase buffer alone, and the other with trehalase buffer plus trehalase [3 µl of porcine trehalase (Sigma; T8778-1UN) per 1 ml of buffer]. The resulting sample will have a final dilution of 1:200. These diluted samples are then placed at 37°C for 18–24 hours to allow for breakdown trehalose into free glucose. A 30 µl aliquot of each sample (+/− trehalase) is then loaded onto a 96 well plate to measure the concentration of hemolymph trehalose and glucose using the enzymatic assays described above. As previously mentioned, the ideal dilution will depend on dietary conditions, genetic background and developmental stage.
5.1f Glycogen
Most carbohydrate in the fly is stored in the form of glycogen, which provides a large and accessible energy source during times of fasting and intense activity. Most glycogen in the adult fly is stored in the fat body, flight muscle, halteres, and gut, while larval glycogen is primarily located in the body wall muscle [3, 65]. Although periodic acid/Schiff staining can be used to visually assess glycogen deposits, the easiest and most quantitative method uses an enzymatic assay that breaks down glycogen into molecules of free glucose, which is then quantified using the GO or HK assays described above [66]. As with the other assays above, homogenized tissue samples must be kept cold and moved rapidly to the heat treatment step to prevent the breakdown of glycogen into free glucose by endogenous enzymes present in the extract. Due to the high level of glycogen in the samples, the resulting supernatant should be appropriately diluted to ensure that the concentrations of all samples are within the linear range of the assay. If the concentration of any sample exceeds that of the 0.16 mg/ml glycogen standard, then the assay should be repeated using more dilute samples. Although either the GO or HK assays can be used to measure glycogen, the HK assay is preferred for glycogen measurements in pupae, when the GO kit can be unreliable.
5.1g Protocol: Glycogen Quantification using the GO kit
Collect samples (25 mid-second instar larvae; 5 adult flies).
Rinse several times with 1 ml cold PBS to remove all traces of food that might be attached to the outside of the animal. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS. Snap freeze animals in liquid nitrogen for later homogenization or add 100 µl cold PBS.
Rapidly homogenize animals in PBS with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000). Frozen samples should be kept on dry ice until addition of PBS. If samples are not kept cold, glycogen and trehalose will be enzymatically degraded into free glucose by endogenous enzymes and skew the final analysis.
Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80°C for later analysis.
Heat supernatant for 10 min at 70°C, then centrifuge for 3 min at maximum speed in a refrigerated tabletop centrifuge that has been prechilled to 4°C.
Pipette the resulting supernatant into a new 1.5 ml microfuge tube. At this time, the heat-inactivated sample can be frozen and stored at −80°C, if desired.
Set up glucose and glycogen standards. Using 1 mg/ml glucose or glycogen stock solutions, make a dilution series of glucose or glycogen standards in PBS, both in the range of 0 – 0.16 mg/ml, as outlined in glucose assay protocol. Glycogen stock solutions can be stored at −20°C or 4°C.
Add 30 µl of each glycogen standard (including PBS blank) to the top row of a clear-bottom 96 well plate. Similarly, add 30 µl of each glucose standard in the next row down.
Dilute heat-treated fly samples for glycogen measurement 1:5 in PBS (the required dilution may range from 1:5 to 1:20 depending on experimental conditions) and load 30 µl into each well. Samples should be loaded in duplicate rows such that one row will be used to measure glycogen + glucose (treated with amyloglucosidase) and the adjacent row will be used to measure glucose alone (no amyloglucosidase).
Prepare the GO reagent (see glucose assay protocol) for your glycogen measurements by adding 1 µl amyloglucosidase (Sigma A1602; 25mg) per 1 ml of GO reagent.
Using a multichannel pipette, add 100 µl of GO reagent + amyloglucosidase to the glycogen standards and the first row of each set of duplicate samples. Then add 100 µl of GO reagent alone (without amyloglucosidase) to the glucose standards and the remaining samples.
Seal the wells with parafilm to prevent evaporation and incubate the plate at 37°C for 30–60 minutes. Briefly centrifuge the plate in an appropriate swing-bucket rotor to clear condensate from the sides of the wells and remove any air bubbles present in the samples. Then add 100 µl of 12 N sulfuric acid and measure absorbance at 540 nm using a plate reader.
Free glucose concentration in each sample is calculated based on the glucose standard curve. For glycogen measurements, first subtract the absorbance measured for free glucose in the untreated samples from the absorbance of the samples that have been digested with amyloglucosidase. The glycogen content in each sample is then calculated based on the glycogen standard curve. This method is linear from 0.01 to 0.16 mg/ml glycogen.
5.1h Protocol: Glycogen Quantification using the HK kit
Collect samples (25 mid-second instar larvae; 5 adult flies).
Rinse several times with 1 ml cold PBS to remove all traces of food that might be attached to the outside of the animal. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS. Snap freeze animals in liquid nitrogen for later homogenization or add 100 µl cold PBS.
Rapidly homogenize animals in PBS with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000). Frozen samples should be kept on dry ice until addition of PBS. If samples are not kept cold, glycogen and trehalose will be enzymatically degraded into free glucose by endogenous enzymes and skew the final analysis.
Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80°C for later analysis.
Heat supernatant for 10 min at 70°C, then centrifuge for 3 min at maximum speed in a refrigerated tabletop centrifuge that has been prechilled to 4°C.
Pipette the resulting supernatant into a new 1.5 ml microfuge tube. At this time, the heat-inactivated sample can be frozen and stored at −80°C, if desired.
Prepare amyloglucosidase stock (AS) solution by adding 1.5 µl amyloglucosidase (Sigma A1602; 25mg) to 1 ml of PBS.
Set up glycogen standards by diluting 16 µl of glycogen stock (1 mg/ml) with 50 µl of AS solution and 34 µl of PBS (100 µl final volume) for 0.16 mg/ml standard. Do four 2-fold serial dilutions into 1:1 mix of AS and 1×PBS (50 µl 0.16 mg/ml + 50 µl AS+PBS for 0.08 mg/ml standard, etc.) to generate 0.01, 0.02, 0.04, 0.08, and 0.16 mg/ml glycogen standards. Glucose standards should be diluted as outlined in the glucose assay protocol (no amyloglucosidase added).
In 1.5 ml microfuge tubes, dilute the heat-treated fly samples 1:3 in PBS (this dilution can range from 1:2 to 1:5 depending on experimental conditions) by combining 20 µl of sample with 40 µl PBS. Aliquot 20 µl of these diluted samples into two separate tubes, one containing 20 µl of AS solution, and the other containing 20 µl of PBS (final sample dilution will equal 1:6).
Add 30 µl of each glycogen standard (including PBS blank) to one row at the top of the plate. Add 30 µl of the glucose standards (and PBS blank) to the next row.
Load 30 µl of each diluted sample to the plate. Samples should be loaded in duplicate rows such that one row will be used to measure glycogen + glucose (amyloglucosidase treated) and the adjacent row will be used to measure glucose alone (no amyloglucosidase).
Seal the wells with parafilm and incubate the plate at 37°C for 60 minutes to digest the glycogen. Spin briefly to clear the condensate from the sides of the wells.
Add 100 µl of HK reagent to each well with a multichannel pipette and incubate at room temperature for 15 minutes. Measure absorbance at 340 nm using a plate reader.
Free glucose concentration in each sample is calculated based on the glucose standard curve. For glycogen measurements, first subtract the absorbance measured for free glucose in the untreated samples from the absorbance of the samples that have been digested with amyloglucosidase. The glycogen content in each sample is then calculated based on the glycogen standard curve. This method is linear from 0.01 to 0.16 mg/ml glycogen.
6. ATP
ATP measurements represent a direct readout of cellular energy levels and thus can provide important insights into metabolic phenotypes. This analysis can be conducted with a luciferase-based assay kit that uses endogenous ATP to generate light [67]. Because ATP is so unstable, a chaotropic buffer is used to preserve as much of the intact metabolite as possible.
6.1 Protocol: Fluorescence Assay for ATP Quantification
Collect samples (25 mid-second instar larvae; 5 adult flies).
Rinse several times with 1 ml cold PBS to remove all traces of food that might be attached to the outside of the animal. Larvae can be washed in a 1.5 ml microfuge tube, but adults should be rinsed in a 9-well glass plate. Transfer adult flies to a 1.5 ml microfuge tube. Carefully remove all liquid. For larvae, centrifuge at 3,000 × g and remove all remnants of PBS.
Prior to homogenizing the samples, prepare the ATP reaction mix (Molecular Probes ATP kit; A22066) by mixing 3.56 ml ddH20, 200 µl 20× reaction buffer, 40 µl 0.1 M DTT, 200 µl 10 mM D-luciferin, and 1 µl firefly luciferase. The resulting mix is sufficient for 40 reactions, and should be kept on ice and protected from light.
Animals are rapidly homogenized in 100 µl of homogenization buffer [6 M guanidine HCL, 100 mM Tris (pH 7.8), 4 mM EDTA] with a pellet pestle (Kontes; 749521-1500) on ice. A motor can be used to facilitate homogenization (Kontes; 749540-0000).
Remove 10 µl of homogenized sample to measure protein content with a Bradford assay. Keep samples on ice and do not heat-treat. Protein samples can be frozen and stored at −80°C for later analysis.
The remaining samples are boiled for 5 minutes and centrifuged for 3 min at maximum speed in a refrigerated tabletop centrifuge that has been prechilled to 4°C.
Transfer 10 µl of the supernatant into a 1.5 ml microfuge tube and dilute 1:10 with 90 µl dilution buffer [25 mM Tris (pH 7.8), 100 µM EDTA], then transfer 10 µl of the diluted supernatant to a second 1.5 ml tube and dilute 1:75 by adding 740 µl of dilution buffer (final dilution of 1:750). The diluted homogenate is centrifuged at 20,000 × g, and 10 µl of the supernatant is transferred to individual wells of a white, opaque 96 well plate (Corning; 3362).
Prepare a series of ATP standards by diluting the 5 mM ATP stock solution provided with the assay kit with ddH2O (0, 0.01, 0.05, 0.1, 0.5, 1 µM). Add 10 µl of each ATP standard solution to the first row of the plate to provide a standard curve. ATP standards can be stored at −20°C for several weeks.
Start the assay by adding 100 µl of the luciferase reaction mix with a multichannel pipette and immediately begin measuring luminescence with a plate reader. A minimum of three sequential measurements should be made for the entire plate, and the results should be averaged.
Determine the ATP concentration by comparing the luminescence measurements for each sample to the ATP standard curve.
7. Metabolomics
The emerging field of metabolomics provides an unprecedented opportunity to simultaneously measure hundreds of metabolic compounds in animal extracts. This technique has proven to be a powerful tool for conducting studies in the fly [17, 68–71]. In addition, when combined with classical genetic analysis, metabolomics can be used to precisely identify metabolic reactions that are affected by a mutation, providing key insights into gene function. The field of metabolomics is too large to summarize in a single review because a variety of methods have been developed to assay several classes of metabolites, ranging from small, charged molecules to lipids. Rather, we provide here a relatively simple protocol that allows users to quantify ~100 small, polar molecules, which includes the basic amino acids, sugars, and intermediates in glycolysis and the TCA cycle. This range of compounds is sufficient to provide insights into the metabolic state of the corresponding animal.
7.1 Genetic background
Metabolomic analysis is particularly sensitive to genetic background effects. An example of this is the use of rosy mutant strains as a control for transgenic lines that were established by scoring for rescue of the eye phenotype. ry mutations render animals unable to synthesize uric acid, which not only results in elevated levels of purine-related intermediates [72], but also induces significant changes in a diverse group of metabolites, including tryptophan, kynurenine, and related compounds [73]. Studies of transgenic lines in which the ry background is not controlled would therefore inaccurately identify widespread metabolic defects. Similarly, yellow mutants exhibit defects in lysine metabolism [74], and other widely-used mutant lines, such as white or vermillion, may produce similar metabolomic artifacts. Therefore, the same genetic background should be used for all samples, ideally using multiple control genotypes for internal confirmation.
7.2 Sample Collection
The amount of material required for a successful metabolomics experiment varies depending on the developmental stage and type of analysis. For a basic survey of small, polar metabolites using GC/MS, samples should contain at least 300 embryos, 25 second instar larvae, and 20 mature adult males. All samples, except for larvae, are collected in screw-cap tubes that contain 1.4 mm ceramic beads (MoBio; 13113-50) and flash frozen in liquid nitrogen. These tubes are designed for the Omni Bead Ruptor, described below. Embryos do not need to be dechorionated prior to collection, but should be washed gently with a paintbrush on a piece of Whatman filter paper soaked in PBS. Larvae are collected in a 1.5 ml tube, repeatedly washed with ice-cold PBS, and flash frozen in liquid nitrogen. The frozen pellet is then dislodged by gently flicking the tube and transferred into a pre-chilled screw cap tube with ceramic beads.
7.3 Sample Processing for GC-MS
All samples should be stored at −80°C until processing, at which point they are transferred to an enzyme-type carrier caddy (Nunc Lab-Top cooler) that has been chilled to −20°C. Add 800 µl of prechilled 90% methanol containing 1.25 µg/ml succinic-d4 acid (Sigma-Aldrich; 293075) and 6.25 µg/ml U-13C, U-15N amino acid mix (Cambridge Isotope; CDNLM-6784) to each tube with a positive displacement pipette. These stable-isotope labeled internal standards provide a means to normalize samples, provide quality control, and allow for the monitoring of instrument efficiency across batches. Additionally, negative controls that contain no fly tissue should be prepared to detect chemical contamination and false-positive peaks during the subsequent GC/MS analysis.
The hard cuticle of the fly is difficult to homogenize and requires a strong physical disruption to efficiency release metabolites. We have found that the Omni Bead Ruptor 24 homogenizer (Omni International) is ideally suited to rapidly and efficiently disrupt fly tissue, with samples homogenized for 30 seconds at 6.45 m/sec. While a variety of bead-filled tubes can be used for this purpose, screw-cap tubes containing 1.4 mm ceramic beads are optimal for removing extraction solvent after processing. Homogenized samples are incubated at −20°C for one hour to enhance protein precipitation and centrifuged at 20,000 × g for 5 minutes at 4°C to remove the resulting precipitate. The supernatant is transferred to a 1.5 ml microfuge tube and the solvent removed with a Speed-Vac (Genevac).
7.4 GC-MS analysis
We perform GC-MS analysis with a Waters GCT Premier mass spectrometer fitted with an Agilent 6890 gas chromatograph and a Gerstel MPS2 autosampler. Dried samples are suspended in 40 µl of 40 mg/ml O-methoxylamine hydrochloride (MOX) (MP Biomedicals; 155405) in pyridine solution (EMD Millipore; PX2012-7) and incubated for one hour at 30°C. Samples are then centrifuged for 5 minutes at 20,000 × g to remove particulate matter, and 25 µl of the supernatant is placed in an autosampler vial (Agilent; 8010-0172 and 5181-1215) with a 250 µl deactivated glass microvolume liner (Agilent; 5183-2086). Forty microliters of N-methyl-N-trimethylsilyltrifluoracetamide containing 1% TMCS (MSTFA) (Thermo Scientific; TS-48915) is added automatically via a Gerstal autosampler and the samples are incubated for 60 minutes at 37°C with shaking. Following incubation, 3 µl of a fatty acid methyl ester standard solution (FAMES; Table 2) is added via the autosampler and 1 µl of the prepared sample is injected to the gas chromatograph inlet at a 10:1 split ratio with the inlet temperature held at 250°C. Fatty acid methyl esters do not occur naturally, but will elute in a highly reproducible manner across the entire chromatogram. The retention time curve of FAMES solution, therefore, allows for the building of reliable retention time libraries of metabolites. Furthermore, this solution is used to assess column quality because a degraded column will exhibit peak tailings of the FAMES standards.
Table 2.
Components of FAMES solution (0.1 mg/ml for each compound)
| Compound | Sigma Aldrich Catalog # |
|---|---|
| methyl heptanoate | 75218 |
| methyl caprylate | 21719 |
| methyl caprate | 21479 |
| methyl laurate | 61689 |
| methyl myristate | 70129 |
| methyl palmitate | 76159 |
| methyl stearate | 85769 |
| methyl arachidate | 10941 |
| methyl behenate | 11940 |
| methyl tetracosanoate | 87115 |
The gas chromatograph is set to an initial temperature of 95°C for one minute followed by a 40°C/min ramp to 110°C and a hold time of 2 minutes. This is followed by a 5°C/min ramp to 250°C and a third ramp to 330°C with a final hold time of 3 minutes. A 30 meter Phenomex ZB5-5 MSi column with a 5 meter long guard column is employed for chromatographic separation. For each analysis, instrument performance is assessed by analyzing the negative control samples, which only contain the internal standard. Experimental samples are only processed if the instrument passes a preset sensitivity and peak shape criteria. Finally, samples are run in a randomized order, except for quality control samples, which are analyzed every nine samples.
7.5 Data Analysis
An initial dataset is prepared using a targeted approach to identify known metabolites. Chromatograms are analyzed using the MassLynx utility QuanLynx, and metabolites are identified based on known retention times and mass fragmentation patterns. The peak area for each metabolite is recorded, and the data is exported to Excel. For our analyses, metabolite identity in QuanLynx is experimentally established using pure, purchased standards, and in limited cases by the commercially available NIST library (National Institute of Standards and Technology; version 11). MarkerLynx is used for peak identification during a second, nontargeted analysis of the chromatograms, and the formatted data is transferred to SIMCA-P+ for principle component analysis (PCA) and partial least squares-discriminate (PLS-DA) analysis. If the PCA analysis identifies significant separation between the experimental groups, PLS-DA analysis is employed to detect significantly altered metabolites. The peaks of unknown metabolites are quantified and the resulting data is exported to Excel. Statistical analysis can be performed with any number of software packages that are available for uni- and multivariate analysis, including JMP and Statistica.
Acknowledgements
We thank L.P. Musselman, A.-F. Ruaud, and M. Sieber for their contributions to these protocols, and D. Bricker, M. Horner, S. Marxreiter, and R. Somer for helpful comments on the manuscript. This work was supported by an NIH K99/R00 Pathway to Independence Award to J.M.T. (K99GM101341), an NIH Developmental Biology Training Grant to W.B. (5T32 HD07491), and NIH R01 DK075607 (C.S.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Brien S, MacIntyre R. In: The Genetics and Biology of Drosophila. Ashburner M, Wright T, editors. New York: Academic Press; 1978. pp. 396–551. [Google Scholar]
- 2.Beadle G, Tatum E. Amer. Nat. 1941;75:107–116. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wigglesworth V. J. Expt. Biol. 1949;26:150–163. doi: 10.1242/jeb.26.2.150. [DOI] [PubMed] [Google Scholar]
- 4.Clare MR. Biological Bulletin. 1925;49:440–460. [Google Scholar]
- 5.Bodine JH, Orr PR. Biological Bulletin. 1925;48:1–14. [Google Scholar]
- 6.Baker KD, Thummel CS. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birse RT, Bodmer R. Crit Rev Biochem Mol Biol. 2011;46:376–385. doi: 10.3109/10409238.2011.599830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhnlein RP. Results Probl Cell Differ. 2010;52:159–173. doi: 10.1007/978-3-642-14426-4_13. [DOI] [PubMed] [Google Scholar]
- 9.Leopold P, Perrimon N. Nature. 2007;450:186–188. doi: 10.1038/nature06286. [DOI] [PubMed] [Google Scholar]
- 10.Teleman AA. Biochem J. 2010;425:13–26. doi: 10.1042/BJ20091181. [DOI] [PubMed] [Google Scholar]
- 11.Alic N, Hoddinott MP, Foley A, Slack C, Piper MD, Partridge L. PLoS One. 2012;7:e45367. doi: 10.1371/journal.pone.0045367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buescher JL, Musselman LP, Wilson CA, Lang T, Keleher M, Baranski TJ, Duncan JG. Dis Model Mech. 2013;6:1123–1132. doi: 10.1242/dmm.011924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matzkin LM, Johnson S, Paight C, Markow TA. PLoS One. 2013;8:e59530. doi: 10.1371/journal.pone.0059530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. Genes & Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasco MY, Leopold P. PLoS One. 2012;7:e36583. doi: 10.1371/journal.pone.0036583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havula E, Teesalu M, Hyotylainen T, Seppala H, Hasygar K, Auvinen P, Oresic M, Sandmann T, Hietakangas V. PLoS Genet. 2013;9:e1003438. doi: 10.1371/journal.pgen.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, Redin C, Boudina S, Gygi SP, Brivet M, Thummel CS, Rutter J. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ. Dis Model Mech. 2011;4:842–849. doi: 10.1242/dmm.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, Cagan R. PLoS Genet. 2013;9:e1003175. doi: 10.1371/journal.pgen.1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunte AS, Matthews KA, Rawson RB. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, Ocorr K, Bodmer R, Oldham S. Cell Metab. 2010;12:533–544. doi: 10.1016/j.cmet.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed LK, Williams S, Springston M, Brown J, Freeman K, DesRoches CE, Sokolowski MB, Gibson G. Genetics. 2010;185:1009–1019. doi: 10.1534/genetics.109.113571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee WC, Micchelli CA. PLoS One. 2013;8:e67308. doi: 10.1371/journal.pone.0067308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piper MD, Blanc E, Leitao-Goncalves R, Yang M, He X, Linford NJ, Hoddinott MP, Hopfen C, Soultoukis GA, Niemeyer C, Kerr F, Pletcher SD, Ribeiro C, Partridge L. Nat Methods. 2013;11:100–105. doi: 10.1038/nmeth.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgecomb RS, Harth CE, Schneiderman AM. The Journal of experimental biology. 1994;197:215–235. doi: 10.1242/jeb.197.1.215. [DOI] [PubMed] [Google Scholar]
- 26.King RC, Wilson LP. J. Exp. Zool. 1955;130:71–82. [Google Scholar]
- 27.Carvalho GB, Kapahi P, Benzer S. Nat Methods. 2005;2:813–815. doi: 10.1038/nmeth798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seay DJ, Thummel CS. J Biol Rhythms. 2011;26:497–506. doi: 10.1177/0748730411420080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Proc Natl Acad Sci USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong R, Piper MD, Wertheim B, Partridge L. PLoS One. 2009;4:e6063. doi: 10.1371/journal.pone.0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itskov PM, Ribeiro C. Front Neurosci. 2013;7:12. doi: 10.3389/fnins.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hildebrandt A, Bickmeyer I, Kuhnlein RP. PLoS One. 2011;6:e23796. doi: 10.1371/journal.pone.0023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietveld A, Neutz S, Simons K, Eaton S. J Biol Chem. 1999;274:12049–12054. doi: 10.1074/jbc.274.17.12049. [DOI] [PubMed] [Google Scholar]
- 34.Clark AG, Gellman W. Drosophila Information Service. 1985;61:190. [Google Scholar]
- 35.Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Current Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- 36.Al-Anzi B, Zinn K. PLoS One. 2010;5:e12353. doi: 10.1371/journal.pone.0012353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams CM, Thomas RH, MacMillan HA, Marshall KE, Sinclair BJ. J. Ins. Phys. 2011;57:1602–1613. doi: 10.1016/j.jinsphys.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Van Veldhoven PP, Swinnen JV, Esquenet M, Verhoeven G. Lipids. 1997;32:1297–1300. doi: 10.1007/s11745-006-0166-1. [DOI] [PubMed] [Google Scholar]
- 39.Chiang H. Amer. Midland Naturalist. 1963;70:329–338. [Google Scholar]
- 40.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Carvalho M, Schwudke D, Sampaio JL, Palm W, Riezman I, Dey G, Gupta GD, Mayor S, Riezman H, Shevchenko A, Kurzchalia TV, Eaton S. Development. 2010;137:3675–3685. doi: 10.1242/dev.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X, Warren JT, Buchanan J, Gilbert LI, Scott MP. Development. 2007;134:3733–3742. doi: 10.1242/dev.004572. [DOI] [PubMed] [Google Scholar]
- 43.Sieber MH, Thummel CS. Cell Metab. 2012;15:122–127. doi: 10.1016/j.cmet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voght SP, Fluegel ML, Andrews LA, Pallanck LJ. Cell Metab. 2007;5:195–205. doi: 10.1016/j.cmet.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Reis T, Van Gilst MR, Hariharan IK. PLoS Genet. 2010;6:e1001206. doi: 10.1371/journal.pgen.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parvy JP, Napal L, Rubin T, Poidevin M, Perrin L, Wicker-Thomas C, Montagne J. PLoS Genet. 2012;8:e1002925. doi: 10.1371/journal.pgen.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiefenbock SK, Baltzer C, Egli NA, Frei C. EMBO J. 2010;29:171–183. doi: 10.1038/emboj.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seecof RL, Dewhurst S. Cell Diff. 1974;3:63–70. doi: 10.1016/0045-6039(74)90041-4. [DOI] [PubMed] [Google Scholar]
- 49.Butterworth FM, Bodenstein D, King RC. The J. Exp. Zool. 1965;158:141–153. doi: 10.1002/jez.1401580203. [DOI] [PubMed] [Google Scholar]
- 50.Palanker L, Tennessen JM, Lam G, Thummel CS. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutierrez E, Wiggins D, Fielding B, Gould AP. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 52.Zinke I, Kirchner C, Chao LC, Tetzlaff MT, Pankratz MJ. Development. 1999;126:5275–5284. doi: 10.1242/dev.126.23.5275. [DOI] [PubMed] [Google Scholar]
- 53.Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Brooks KK, Liang B, Watts JL. PLoS One. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, Watts JL, Lawrenson AL, Hermann GJ. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kunst A, Draeger B, Ziegenhorn J. Methods of Enzymatic Analysis. 3rd ed. New York: Academic Press; 1984. [Google Scholar]
- 58.Bergmeyer HU, Bernt E. Methods of Enzymatic Analysis. 2nd ed. New York: Academic Press; 1974. [Google Scholar]
- 59.Teleman AA, Chen YW, Cohen SM. Dev. Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Chen Q, Ma E, Behar KL, Xu T, Haddad GG. J Biol Chem. 2002;277:3274–3279. doi: 10.1074/jbc.M109479200. [DOI] [PubMed] [Google Scholar]
- 61.Lee G, Park JH. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rulifson EJ, Kim SK, Nusse R. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 63.Piyankarage SC, Featherstone DE, Shippy SA. Anal. Chem. 2012;84:4460–4466. doi: 10.1021/ac3002319. [DOI] [PubMed] [Google Scholar]
- 64.Haselton AT, Fridell YW. J Vis Exp. 2011 doi: 10.3791/2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruaud AF, Lam G, Thummel CS. Mol Endocrinol. 2011;25:83–91. doi: 10.1210/me.2010-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clark AG, Keith LE. Bioch. Genet. 1989;27:263–277. doi: 10.1007/BF00554162. [DOI] [PubMed] [Google Scholar]
- 67.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 68.Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. Cell Metab. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Padmanabha D, Gentile LB, Dumur CI, Beckstead RB, Baker KD. PLoS Genet. 2013;9:e1003230. doi: 10.1371/journal.pgen.1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chintapalli VR, Al Bratty M, Korzekwa D, Watson DG, Dow JA. PLoS One. 2013;8:e78066. doi: 10.1371/journal.pone.0078066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knee JM, Rzezniczak TZ, Barsch A, Guo KZ, Merritt TJ, Chromatogr J. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2013;936:63–73. doi: 10.1016/j.jchromb.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 72.Hilliker AJ, Duyf B, Evans D, Phillips JP. Proc Natl Acad Sci USA. 1992;89:4343–4347. doi: 10.1073/pnas.89.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamleh MA, Hobani Y, Dow JA, Watson DG. FEBS letters. 2008;582:2916–2922. doi: 10.1016/j.febslet.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 74.Bratty MA, Chintapalli VR, Dow JA, Zhang T, Watson DG. FEBS Open Bio. 2012;2:217–221. doi: 10.1016/j.fob.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]