Abstract
Drosophila oogenesis is an excellent system for the study of developmental cell biology. Active areas of research include stem cell maintenance, gamete development, pattern formation, cytoskeletal regulation, intercellular communication, intercellular transport, cell polarity, cell migration, cell death, morphogenesis, cell cycle control, and many more. The large size and relatively simple organization of egg chambers make them ideally suited for microscopy of both living and fixed whole mount tissue. A wide range of tools is available for oogenesis research. Newly available shRNA transgenic lines provide an alternative to classic loss-of-function F2 screens and clonal screens. Gene expression can be specifically controlled in either germline or somatic cells using the Gal4/UAS system. Protein trap lines provide fluorescent tags of proteins expressed at endogenous levels for live imaging and screening backgrounds. This review provides information on many available reagents and key methods for research in oogenesis.
Keywords: Drosophila, Oogenesis, Live-cell imaging, Genetics, RNAi
1. Introduction
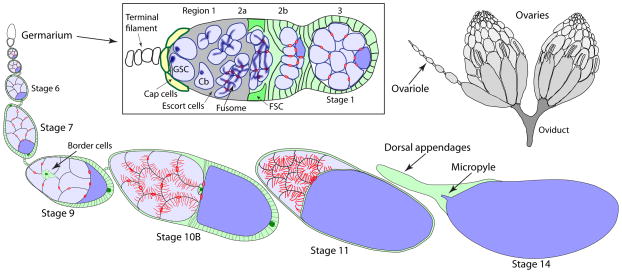
Oogenesis in Drosophila [1,2] supports an impressively high level of fecundity. Ovaries in adult females consist of ~16 parallel tubes called ovarioles that contain developing egg chambers arranged a linear array of progressive developmental stages (Fig. 1). Movement of egg chambers is facilitated by peristaltic contractions of circular muscles surrounding each ovariole and a muscle mesh surrounding the whole ovary [3–5]. The ovarioles of each ovary converge at a lateral oviduct, which connects to a common oviduct and then the uterus. Mature stage 14 eggs move into the uterus where they are fertilized by a single sperm and then laid. The development of each egg takes about eight days: roughly half of this time is spent in the germarium for egg chamber formation, and the remaining four days are required for egg chamber development. Each of the ovarioles produces approximately two eggs per day, or over 60 eggs from a young, well-fed female [6]. This high output of eggs depends on an abundant source of food, major contributions from support cells in egg chambers and a wide range of cellular interactions. An overview of oogenesis and examples of how its study has contributed to our understanding of developmental biology are summarized in this section.
Figure 1.
Overview of oogenesis. Drosophila females have a pair of ovaries (top right), each of which consists of ~15 ovaioles. Oogenesis begins in the germarium (center box), where germline and somatic stem cells (GSC and FSC, respectively) divide continuously to support the formation of new egg chambers. See text for further details.
1.1. The germarium and early oogenesis
The formation of egg chambers (also called follicles) takes place during the first four days of oogenesis in the germarium. Germline stem cells at the apical end of germaria are maintained by signaling from adjacent niche cells called cap cells. Stem cell daughters called cystoblasts leave the niche and undergo four mitotic divisions to produce a cyst of 16 cells. Incomplete cytokinesis in these divisions leaves intercellular bridges that are later stabilized by the accumulation of filamentous actin to form ring canals that persist until the end of oogenesis. Somatic escort cells encase the dividing cysts. After completing mitosis, escort cells are exchanged for follicle cells to complete stage 1 egg chamber assembly. The follicle cells are generated by two follicle-cell stem cells located between germarium regions 2a and 2b. Thus, the germarium contains two types of stem cells maintained in separate niches whose division rates must be coordinated to produce cells necessary for egg chamber production. Only one of the 16 germline-derived cells develops into an oocyte while the remaining 15 differentiate into nurse cells with polyploid nuclei. By the time an egg chamber emerges from the germarium, the oocyte is already positioned at the posterior as a result of a signaling cascade emanating from the next most mature egg chamber. Thus, the first four days of oogenesis produce an oocyte already endowed with anterior/posterior axis information and accompanied by a suite of nurse cells and follicle cells poised to support oocyte development. Examination of egg chamber assembly carried out in many laboratories has yielded a huge amount of information on stem cell and stem cell niche biology [7,8] and the origins of polarity [9]. This research takes advantage of powerful genetic and cytological tools developed over many years.
1.2. Previtellogenic development, stages 2–8
The second half of oogenesis (~3.5 days) takes place in stage 2–14 egg chambers as they move within ovarioles toward the oviducts. Most of this time is needed for previtellogenic egg chamber development during stages 2–8, when oocyte growth is mediated entirely by intercellular movement of cytoplasm from nurse cells to the oocyte through ring canals. The diameter of ring canals slowly expands as egg chambers grow, mediated by active actin filament polymerization in the ring canal rim [10]. The polarization of oocytes is established during these stages as a consequence of the posterior location of the oocyte nucleus. The gurken mRNA accumulates at the posterior with the oocyte nucleus, and produces a ligand for EGFR activation in posterior follicle cells [11]. EGFR activation transmits a signal back to the germline that causes microtubule reorganization in the anterior/posterior axis. In addition, many maternal mRNAs and RNA-binding proteins accumulate specifically in oocytes through a combination of microtubule-based directed transport and trapping within the oocyte [12].
Follicle cells also make crucial contributions to previtellogenic oogenesis. Egg chambers change shape from spherical to elongated starting during stage 5. Interestingly, this shape change is driven by egg chamber rotations that occur between stages 5 and 9. Follicle cells drive the rotations as they migrate perpendicular to the axis of the ovariole, laying down a girdle of polarized extracellular matrix as they go. As a result, expansion of egg chambers takes place anisotropically toward the poles [13]. A major change in cell cycle also takes place in follicle cells. Mitotic divisions cease at the end of stage six, followed by three endoreplication cycles during stages 7–10A [14]. The Notch-Delta signaling pathway controls this transition [15,16], providing an excellent opportunity for studying cell cycle changes under developmental control. Like the germline cysts, the follicle cells are also syncytial, as they remain interconnected with a number of sibling cells by small (~200 nm diameter) ring canals that result from incomplete cytokinesis [17–19]. These ring canals do not grow in size during oogenesis, but they are able to support intercellular movement of protein between cells, raising the interesting possibility that they serve an important function in oogenesis [20,21].
Stage 8 egg chambers do not progress into vitellogenesis (yolk uptake) if egg chambers have severe patterning defects or if environmental conditions are unlikely to support the survival of progeny. Limiting the availability of protein in the diet of females causes egg chamber apoptosis at the end of stage 8, thus avoiding the metabolic cost of completing egg development and depleting the female’s energy stores. During egg chamber apoptosis, follicle cells lose their epithelial organization and become phagocytic, engulfing the cytoplasm of germline cells [22,23]. If egg chambers are sound and protein is restored to the food, oogenesis resumes and stage 14 eggs can develop from surviving stage 8 egg chambers within one day. Thus, stage 8 serves as a metabolic checkpoint that triggers egg chamber destruction while preserving less mature egg chambers poised to resume development when conditions improve.
1.3. Completing oogenesis, stages 9–14
The final day of oogenesis produces a huge increase in oocyte volume due to yolk uptake from hemolymph and the complete transfer of nurse cell cytoplasm to the oocyte. Yolk uptake beginning at the end of stage 8 causes the rate oocyte growth to overtake nurse cell growth so that the oocyte takes up half the volume of egg chambers by stage 10A. During these stages, several key patterning molecules are localized within the oocyte: oskar mRNA at the posterior, bicoid mRNA at the anterior, and gurken mRNA at the dorsal anterior domain [24]. The movement and anchoring of these maternal mRNAs are active areas of research that benefit from the ability to do live-cell imaging to reveal conserved mechanisms of mRNA localization [25,26]. The final phase of oocyte growth happens during stage 11 when nurse cells contract and squeeze (or ‘dump’) their remaining cytoplasm into the oocyte in about 30 minutes, accompanied by robust microtubule-mediated mixing of the oocyte cytoplasm. Nurse cell death after dumping has some of the hallmarks of apoptosis, although it is a caspase-independent process [23,27]. In preparation for this nurse cell dumping, stage 10B egg chambers produce cables of unipolar actin filaments that grow from the nurse cell membranes inward until they reach the nuclear envelope [28,29]. The actin cables prevent nurse cell nuclei from being squeezed into ring canals where they would block the flow of cytoplasm to the oocyte. Prostaglandin signaling is involved with triggering the formation of nurse cell actin cables [30].
Follicle cells form a secretory epithelium in egg chambers with apical/basal polarity and extensive microvilli on their apical surfaces facing the germline cells. They secrete yolk protein during vitellogenesis, vitelline membrane proteins during stage 9–11, and finally chorion proteins beginning in stage 11. In preparation for chorion protein production, stage 9 follicle cells switch from endoreplication to synchronous amplification of the two chorion-gene clusters through repeated firings of replication origins in the clusters [31,32]. The endocycle to amplification switch is mediated by down-regulation of Notch and activation of the Ecdysone receptor [33].
In addition to driving egg chamber rotation, follicle cell migrations dramatically change egg chamber organization. During stage 9, 6–10 border cells delaminate from the anterior follicle cell epithelium and migrate between nurse cells to the anterior of the oocyte carrying two polar cells as passengers. The migration of these cells is guided by the PVR growth factor produced in the oocyte [34], and propelled by acto-myosin dynamics [35]. As border cell migrate between nurse cells, other follicle cells move around nurse cells toward the oocyte where they form a columnar epithelium. During stage 10A, follicle cells at the anterior circumference of the oocyte migrate centripetally, separating the oocyte from nurse cells. Nurse cell dumping is completed just as centripetal follicle cells completely cover the oocyte anterior at the end of stage 11. The border cells participate in forming the micropyle through which a sperm reaches the oocyte. Two groups of 65–80 follicle cells in the anterior, dorsal domain form specialized eggshell structures called dorsal appendages that facilitate gas exchange during embryogenesis. The specification and morphogenesis of dorsal appendages are mediated by patterning cues in specific follicle cell groups [36].
2. Methods to manipulate gene expression during oogenesis
2.1. Gal4/UAS
Extensive Gal4/UAS tools are available for manipulating gene expression in germline and somatic cells of egg chambers. However, using the right UAS vector is critical. The original UASt vectors include P element ends for transformation and work very well in the somatic cells of the ovary; however, UASt-mediated expression is extremely poor in germline cells. Pernille Rørth [37] determined that the basal promoter from the hsp70 gene used in UASt is not expressed in germline cells. She produced new UASp vectors that instead use the basal promoter from the P element transposase gene. UASp vectors are included in the T. Murphy Collection of Gateway vectors at the Bloomington DGRC. A version of UASp that includes the attB sequence for phiC31-mediated integration was reported recently [38]. The UASt vector is available at the DGRC in Bloomington, and versions of UASt with the attB recombination sequence are available at FlyC31. New UAS, attB vectors made in the Rubin lab at Janelia Farm are optimized for expression in the nervous system [39]; however, they work very well in follicle cells and also support expression in germline cells. Rubin lab vectors are listed on their website and are available at Addgene.
A multitude of Gal4 lines is available from stock centers and labs that are useful for driving expression in specific cells of the ovary. The best source for comprehensive general information is the Gal4 page of the Bloomington Drosophila Stock Center (BDSC), although this page has limited information on expression. Many of these lines are derived from Gal4 enhancer trap screens aimed at identifying lines expressed in during oogenesis and early embryogenesis [40,41]. Commonly used drivers for oogenesis research are listed in Table 1, including lines for expression in the stem cell niche (germarium), germline cells and follicle cells. Gal4 expression from the nanos promoter is in germline stem cells, then recedes in young egg chambers and resumes later in oogenesis. The otu promoter produces more uniform expression in the germarium and egg chambers, but tapers off toward the end of oogenesis. A line called Maternal Triple Driver (MTD)-Gal4 has three Gal4 constructs (P{otu-GAL4::VP16.R}1, w*; P{GAL4-nos.NGT}40; P{GAL4::VP16-nos.UTR}MVD1), providing robust germline expression throughout oogenesis [42]. For germline expression outside the germarium (starting in stage 2), the Matα-TubGal4 (MAT) line is ideal. Most of the lines driving expression in somatic follicle cells are also expressed in other tissues of the fly. An exception is Vm26a-Gal4, which has the promoter from the vitelline membrane 26Aa gene and is specific for follicle cells in stage 10–14 egg chambers [36].
Table 1.
Commonly used Gal4 lines.
| Name | Symbol | Chr. | BDSC # | Kyoto DGRC # | Expression | Reference | |
|---|---|---|---|---|---|---|---|
| Stem cell niche | c587-Gal4 | P{GAL4}C587 | 1 | Escort cells | [118] | ||
| bab1-Gal4 | P{GawB}bab1[A gal4-5] | 3 | 6802 | Terminal filament and cap cells | [119] | ||
| 109-30-Gal4 | P{GawB}109-30 | 2 | 7023 | Follicle stem cells and germarium follicle cells | [120] | ||
| 13C06-Gal4 | P{GMR13C06-GAL4}attP2 | 3 | 47860 | Posterior escort cells, follicle stem cells, prefollicle cells at region 2a/2b border | [121,122] | ||
| Germline | bam-Gal4 | P{bamP-Gal4:VP16} | 2 | Cystoblasts and late stages | [123]; | ||
| Matα-TubGal4 (aka, MAT) | P{matα4-GAL-VP16}V37 | 3 | 7063 | Starting in stage 2 | [124] | ||
| Matα-TubGal4 (aka, MAT) | P{matα4-GAL-VP16}V2H | 2 | 7062 | Starting in stage 2 | [124] | ||
| MTD (Maternal Triple Driver)-Gal4 | P{otu-GAL4::VP16.R}1, w*; P{GAL4-nos.NGT}40; P{GAL4::VP16-nos.UTR} | 1; 2; 3 | 31777 | All germline cells, starting with stem cells | [42] | ||
| NGT40 | P{GAL4-nos.NGT}40 | 2 | 4442 | 107748 | Germline cells | [125] | |
| nos-Gal4-VP16 | P{GAL4::VP16-nos.UTR}MVD2 | 1 | 7303 | Stem cells, low in young egg chambers, high starting in stage ~5 | [126] | ||
| nos-Gal4-VP16 | P{GAL4::VP16-nos.UTR}CG632 5[MVD1] | 3 | 4937 | 107955 | Stem cells, low in young egg chambers, high starting in stage ~5 | [126] | |
| pCOG-Gal4 | P{otu-GAL4::VP16.R}1 | 1 | Included in 31777 | Germarium and young egg chambers | [37] | ||
| tub-Gal4 | P{tubP-GAL4}LL7 | 3 | 5138 | 108069 | All cells (moderate level) | [69] | |
| Follicle cell | CY2-Gal4 | P{GawB}CY2 | 2 | Starting in stage 8 | [127] | ||
| cb16-Gal4 | P{GawB}cb16 | 2 | 6722 | Starting in germarium | [41] | ||
| C587-Gal4 | P{GAL4}C587 | X | Somatic cells in the germarium | [46,128] | |||
| e22c-Gal4 | P{en2.4-GAL4}e22c | 2 | 1973 | 106609 | All cells, including stem cells | [129] | |
| tub-Gal4 | P{tubP-GAL4}LL7 | 3 | 5138 | 108069 | All cells | [69] | |
| TJ-Gal4 | P{GawB}NP1624 | 2 | 104055 | All cells (including follicle stem cells and escort cells) | [130] | ||
| T80-Gal4 | P{GawB}T80 | 2 | 1878 | 106551 | All cells | [131] | |
| GR1-Gal4 | P{GawB}GR1 | 3 | 36287 | All cells | [132] | ||
| actin-Gal4 | P{Act5C-GAL4}25FO1 | 2 | 4414 | 107727 | All cells (low in germline) | [133] | |
| slbo-Gal4 | P{GAL4-slbo.2.6}1206 | 2 | 6458 | Border cells | [134] | ||
| c306-Gal4 | P{GawB}c306 | 1 | 3743 | 107215 | Anterior and border cells | [40] | |
| Vm26Aa-GAL4 | 1, 2 or 3 | Stage 10–14 (specific) | [36] | ||||
| Rho-Gal4 | 3 | Dorsal anterior (floor cells) | [135] |
A curious aspect of Gal4/UAS expression in egg chambers is frequent mosaicism. A group of follicle cells with one level of reporter expression is often adjacent to another group with a very different level. Nurse cells can also display different levels of expression [43]. The explanation for follicle cell mosaicism may involve epigenetic marks inherited by progeny during mitotic proliferation [44], though the level of variation in expression is somewhat masked by intercellular movement of proteins between follicle cells through ring canals [20]. The probability of mosaic expression levels should be taken into consideration when interpreting results of an experiment involving Gal4-mediated expression.
A method to impose an additional layer of temporal control on Gal4 driven gene expression is the TARGET system [45]. TARGET relies on ubiquitous expression of Gal80ts, a specific, temperature sensitive repressor of Gal4, to inhibit Gal4 driven transgene expression at 18° C, the permissive temperature for Gal80ts. The Gal80ts-mediated repression is relieved by shifting flies to the restrictive temperature for Gal80ts (~29° C), which allows Gal4 to activate transcription. This method can works follicle cells [46], but in our experience Gal80ts is not able to repress strong Gal4 expression from Matα-TubGal4 in the germline. Repression of weaker germline Gal4 drivers may be possible; however, but this needs further testing.
2.2. Promoter fusions
For rescue experiments or screening backgrounds, it can be desirable to have gene expression under the control of a promoter directly fused to a cDNA rather than using the bipartite Gal4/UAS system. Several vectors are available with promoters for germline-specific expression, or for ubiquitous expression in egg chambers (Table 2). Vectors using the promoter from the α-Tubulin at 67C (αTub67C) gene direct strong, uniform expression in germline cells beginning in stage 2 egg chambers. The ovarian tumor (otu) promoter produces low to moderate levels of germline expression throughout oogenesis, and the hsp26 promoter produces moderate levels of expression starting in stage 8 egg chambers. The nanos (nos) or vasa (vas) promoter directs expression in primordial germ cells in developing animals, and in the germline cells of the adult germarium. Expression is low in early egg chambers, and high in late stages. For ubiquitous expression, the Ubiquitin-63E (Ubi-p63E) or spaghetti-squash promoter is a good choice. The α-Tubulin at 84B (αTub84B) promoter also provides expression in both germline and follicle cells, although expression is lower in the germline.
Table 2.
Promoters useful for driving gene expression in oogenesis.
| Expression | Promoter | Vectors | Reference |
|---|---|---|---|
| Off or low in germarium, very strong and uniform expression in vitellarium. | α-Tubulin at 67C | pCaTub67CMatpolyA | [136] |
| D277 pCaTub67CMatpolyA | |||
| D277Matg | [137] | ||
| Low to moderate levels throughout oogenesis; may taper in later stages. | ovarian tumor | pCOG | [138] |
| On in primordial germ cells, on in germarium, low in early stages, then high in late stages. | nanos | [139] | |
| On in primordial germ cells. In adults, moderate and uniform expression throughout oogenesis. | vasa | [140] | |
| Moderate expression in germline (stage 8 and later) | Heat shock protein 26 | pGerm80 | [141] |
| pGerm90 | |||
| Ubiquitous. | α-Tubulin at 84B | [69] | |
| Ubiquitous. | Ubiquitin-63E | pWUM2 | [142] |
| pUp2-RHX poly-A | [143] | ||
| pCasPeR-Ubi | [144] | ||
| Ubiquitous | spaghetti squash | [145] [146] [147] |
3. Genetic methods
The greatest strength of Drosophila as an experimental system is the ability to use genetic analysis to understand cellular and developmental processes. Work from many labs during the past decades has resulted in remarkable innovations in the genetic tools available to study the genes and developmental mechanisms controlling oogenesis. We present here the basis for classical genetic approaches, and also summarize more recently developed techniques that rely on transgenic manipulations.
3.1. Classical genetics: female-sterile mutations
Initial genetic approaches to study oogenesis focused on screens for recessive loss-of-function mutations affecting female fertility. The two broad classes of mutations affecting female fertility are female-sterile (fs) mutations and maternal-effect lethal mutations (mel). By definition, female-sterile mutations cause a defect during oogenesis such that normal eggs are not produced. In contrast, females bearing maternal-effect lethal mutations are able to produce normal eggs, indicating that oogenesis is able to proceed normally, but these eggs fail to produce viable offspring due to the lack of a gene product essential for embryonic development that is normally provided to the egg during oogenesis. Maternal-effect lethal mutations have been invaluable in understanding the mechanisms of early embryonic development.
Screening for recessive female-sterile and/or maternal-effect lethal mutations requires an F3 screen, where mutagenized chromosomes are balanced, made homozygous, and tested for effects on fertility. This task therefore requires substantial effort, and several large-scale of this type were conducted in 1970s and 1980s. As a result of these efforts, most genes that can be mutated to specifically affect female fertility have likely been identified [47–53].
3.2. Mosaic analysis
While female-sterile screens succeeded in isolating many genes that are critical for female fertility, the majority of genes that function in oogenesis are essential genes for which loss of function mutations do not produce a female-sterile phenotype. The study of these genes requires a means to inactivate or knock down gene function specifically in the ovary without compromising essential functions elsewhere in the fly. Historically, three experimental approaches have been developed to engineer genetically mosaic ovaries: pole cell transplantation, mitotic recombination, and tissue-specific RNAi. The latter two approaches are far easier to carry out and are more commonly used, but transplantation experiments can be useful in certain circumstances.
3.2.1. Pole cell transplantation
Primordial germ cells form early in embryonic development and are the first embryonic cells to undergo cellularization. This allows them to be unambiguously identified in the embryo, and they can be transplanted from one embryo to another using micromanipulation. If the recipient embryo is derived from a strain lacking a functional germline (usually by carrying the dominant female-sterile gene ovoD1), then the resulting germline will be formed from the transplanted germ cells. Transplanted pole cells give rise to only germline tissue [54,55], so this technique can be used to determine whether a gene affecting oogenesis functions in the germ cells or the associated somatic cells. Experiments using such approaches led to important conclusions regarding sex determination and intercellular communication in Drosophila. A recent publication [56] describes an updated protocol on the technique that was originally developed in the 1970s [57,58].
3.2.2. Mitotic recombination
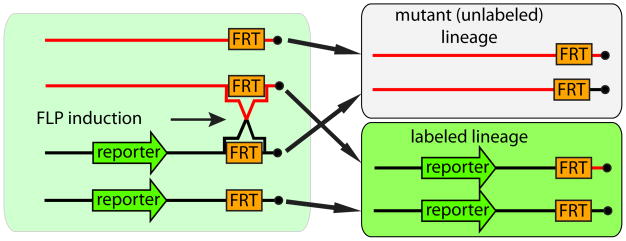
A versatile and widely used system to generate genetic mosaics in oogenesis is mitotic recombination. In this system, flies heterozygous for a mutation of interest are induced to undergo recombination during the G2 phase of the cell cycle so that inter-homolog exchange distal to a centromere will give rise to genetically distinct daughter cells. The wild-type chromosome typically includes a marker or reporter gene to allow for identification of recombined cells (Figure 2).
Figure 2.
Mitotic recombination induced during G2 by expressing the FLP enzyme in heterozygous cells. Homologous chromosomes have aligned FRT sequences located near centromeres (black dots). Subsequent segregation of chromosomes results in two genetically distinct lineages. This scheme shows depicts a negative marking strategy, where mutant cells are marked by loss of a ubiquitous reporter gene.
When mitotic recombination was first used to generate genetic mosaics, recombination was induced by exposing flies to ionizing radiation [59]. This method is relatively inefficient, however, and the introduction of the yeast FLP/FRT site-specific recombination system into flies vastly improved the efficiency of genetic mosaic analysis [60]. In this system, the yeast FLP enzyme acts on two FLP recombination targets (FRTs) and catalyzes DNA strand exchange. Transgenes carrying FRT sequences have been recovered in centrosome-proximal positions on all five chromosome arms, enabling efficient mitotic recombination schemes to generate homozygous mutant clones for essentially all Drosophila genes [61,62]. Identifying mutant germline mosaics can be done genetically using Dominant Female Sterile (DFS) method or visually using reporters.
FLP-DFS system
The most efficient method to identify germline clones for later-staged egg chambers is the FLP-DFS technique [63–65]. This method uses a genetic selection to specifically mark germline mosaics resulting from mitotic recombination. The selection relies on the dominant female-sterile mutation ovoD1, which when heterozygous results in an early arrest of germline development. In FLP-DFS, the experiment is designed so that ovoD1 and the mutation are on FRT-bearing chromosomes that are trans-heterozygous with each other. When recombination is induced in germ line stem cells or their progenitors, germ cells will be generated that are homozygous for the mutation of interest and, importantly, lack the ovoD1 mutation. The only egg chambers that survive to later stages are the non-ovoD1 egg chambers that are homozygous for the mutation of interest. Screens using elegant versions of this approach identified several genes involved in oocyte specification and development in [66–68].
Other marking systems
While the FLP-DFS system is an efficient means of generating late-stage germline clones, it is not useful for identifying germline clones during early oogenesis or for identifying clones in somatic cells. Instead, mosaics in these cell types can be identified using a variety of cytological markers. The simplest and most commonly used marking system for genetic mosaic analysis is negative marking, where mutant cells are marked by loss of a marker. Among these markers, nuclear-localized GFP expressed from the ubiquitin promoter is most commonly used, but other markers, including ubiquitously expressed β-galactosidase and the myc epitope fused to either nuclear or membrane localization domains can also be useful when GFP fluorescence is not desirable [62]. Chromosomes with these markers recombined onto the commonly used FRTs are available at the Bloomington stock center.
It is important to note that the syncytial nature of ovarian cells can have important consequences for negative marking strategies. As mentioned in Section 1.1, follicle cells remain interconnected in syncytial nests of varying size, and small proteins can readily diffuse between connected follicle cells [20]. Thus it is probable that in when homozygous mutant cells are marked by loss of GFP, some GFP may diffuse from wild-type GFP producing cells to the neighboring mutant cells that lack the GFP transgene. Similarly, wild-type gene product may (or may not, depending on its ability to diffuse) move from a genetically wild-type cell to a homozygous mutant cell. The extent to which this is a concern in clonal analysis of follicle cells depends on the ability of the marker and the wild-type gene product to diffuse (for a more complete discussion of these issues, see [21]).
In addition to negative marking, the MARCM system (Mosaic analysis with a repressible clonal marker) allows mutant cells to be positively marked by a UAS reporter transgene [69]. In MARCM, mitotic recombination is induced in a background where both Gal4 and its repressor Gal80 are ubiquitously expressed. Reporter gene expression is repressed by Gal80, which is expressed from a transgene that is heterozygous with the mutant of interest. Mitotic recombination results in progeny cells that are homozygous for the mutant and lack the Gal80 transgene; only these cells will express the UAS reporter. While this system is more complicated to set up, many stocks are available at Bloomington to facilitate MARCM crossing schemes (MARCM stocks). A further advantage of MARCM is that other UAS transgenes in addition to visible markers can be incorporated into MARCM experiments. This has been done in ovarian follicle cells for example, where mutant clones of a vacuolar ATPase were tested for their ability to suppress the effects of UAS-driven constructs that expressed activated forms of the Notch signaling protein [70]. In addition, it is possible to use biologically informative reporters as clonal markers; for example, a probe such as UAS-GFP::tubulin can be used as both a clonal marker and as a label for the microtubule cytoskeleton.
Lineage analysis
Mitotic recombination can also be used to trace cell lineage in a tissue as the cells divide. Lineage tracing experiments have been used to understand how somatic and germline stem cells contribute to ovarian tissue homeostasis (e.g., [71,72]. Several techniques based on FLP/FRT recombination have been developed that allow distinct labeling of mitotic lineages in oogenesis. The “X-15” lineage tracing system relies on mitotic recombination between FRT sequences to fuse the strong, ubiquitous α-tub84B promoter with the E. coli LacZ gene [73]. When recombination is induced during mitosis, one lineage inherits the promoter-LacZ fusion and will express high levels of β-galactosidase, which can be easily detected using either an X-gal staining reaction or immunofluorescence. Two additional lineage tracing systems, the twin spot generator (TSG; [74]) and twin spot MARCM [75] have been recently developed in which both cell lineages resulting from mitotic recombination are labeled. The twin spot generator is similar to the X-15 system in concept, but in this case FRT recombination reconstitutes ubiquitously driven RFP and GFP transgenes in the two lineages. Twin spot MARCM functions similarly to conventional MARCM, except that instead of Gal80-mediated repression, GFP and RFP are repressed by synthetic microRNAs in un-recombined cells. It is also possible to use the twin spot marking systems to compare wild type and mutant lineages, but only if a gene of interest is located distal to the FRT and transgene sites. Currently the twin spot MARCM transgenes are set up on chromosome arm 2L, while twin-spot generator transgenes are available at centromere-proximal locations on chromosome arms 2L, 3L, and 3R [74].
3.2.3. RNA interference
RNAi in Drosophila has become an immensely powerful tool, and at least three genomic-scale projects have been undertaken with the goal of establishing transgenic RNAi lines for essentially all Drosophila genes [76,77] and RNAi FLY, unpublished (available lines are listed at the following sites: (VDRC, TRiP, and RNAi FLY). For many cell types in Drosophila, RNAi is efficiently induced by expressing an inverted-repeat “hairpin” RNA [78]. The Vienna and RNAi FLY transgenic RNAi collection consists of transgenes that express hairpin RNAs [76] under UAS control. However, hairpin RNAs do not effectively silence genes in the female germline, limiting their usefulness in studying oogenesis. More recently, a novel transgenic RNAi system has been developed that allows potent gene silencing in all cell types, including ovarian germ cells [79]. This method was designed to express short RNA hairpins in a microRNA backbone; these RNAs appear to be more efficient in gene knockdown [80]. Experimental tests of this system demonstrated that the short micro-RNAs (shRNAs) produced a potent knockdown of target genes in all cell types examined, including ovarian germ cells [79]. A new large-scale transgenic RNAi collection is being generated using the shRNA strategy, and these lines are also available at TRiP. Effective RNAi silencing in the germline enabled a large-scale screen for genes involved in ovarian stem cell regulation [81]. By testing RNAi lines driven by either MTD-Gal4, which drives UAS-RNAi expression beginning in stem cells and their progenitors, or Matα-TubGal4, which begins driving expression after germline cyst formation is complete, it was possible to identify genes that function in the germarium, outside the germarium, or both.
4. Imaging
The cells that make up the ovary are remarkable from a cell biological perspective. Both somatic and germline cells are unusually large, with the germ line nurse cells up to 100 μ in diameter, and the somatic follicle cells up to nearly 30 μ in height. The cells of the ovary therefore represent a unique opportunity to study a variety of cell biological processes. Preparing fixed ovaries for direct imaging or immunofluorescence is straightforward, and standard formaldehyde fixation/immunofluorescence protocols give excellent results for many purposes. However, some special considerations and protocol variations are needed for preservation of certain cellular structures and antigens; we discuss these below.
A further advantage of studying cell biology in the Drosophila ovary is the large number of proteins and cellular compartments can be labeled using publicly available reagents. Several large-scale screens for proteins traps have resulted in hundreds of genes being tagged with an artificial GFP exon [82–84]. A subset of protein traps with localization patterns that are useful in oogenesis are listed in Table 3. In addition, transgenes have been engineered that express fluorescent protein fusions allowing the localization of many proteins and structures in oogenesis; some of these are listed in Table 4, and more can be found at the Bloomington Drosophila Stock Center. Programed cell death plays several important roles in Drosophila oogenesis [23], and a wealth of reagents and methods to study apoptosis, necrosis, and autophagy have been developed [85,86]. Finally, the Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa is an excellent resource for monoclonal antibodies that recognize 215 Drosophila proteins. These antibodies are available to academic research labs for a nominal fee.
Table 3.
| Category | Where | Name | Gene | Chr. | BDSC | Kyoto DGRC |
|---|---|---|---|---|---|---|
| cytoskeleton | actin cytoskeleton (germline) | YC0059 | Lasp (lethal) | 3 | 50866 | |
| microtubules, polar cells | ZCL2129, ZCL2183 | Jupiter | 3 | 110853, 110858 | ||
| ring canals | CA07004 | Vsg | 3 | 50812 | ||
| ER | ER lumen, fusome | G00198 | Pdi | 3 | 110624 | |
| ER, fusome | G00024 | l(1)G0320 | 1 | 50839 | ||
| ER, fusome | ZCL0488 | Sec61alpha | 2 | 110726 | ||
| smooth ER (germline), fusome | G00071, G00199 | Rtnl1 | 2 | 110579, 110625 | ||
| follicle cell | follicle cells cytoplasm & nuclei | YC0023 | Sm | 2 | 50862 | |
| soma in germarium, polar cells, muscle | G00258 | Fas3 | 2 | 50841 | ||
| germarium | cap cells | CB03410 | Drat | 2 | 50838 | |
| escort cells | YC0102 | Fax | 3 | 50870 | ||
| germline stem cells | CC06238 | Skap | 3 | 51576 | ||
| terminal filament | CB02069 | CG14207 | 1 | 50833 | ||
| membrane | apical membranes | CC01941 | Baz | 1 | 51572 | |
| apical membranes (follicle cells) | YC0005 | Dlg1 | 1 | 50859 | ||
| basement membrane | ZCL1700, ZCL1973 | Trol | 1 | 110807, 110836 | ||
| basement membrane | G00205 | Vkg | 2 | 110626 | ||
| plama membranes (follicle cells) | G00305 | Nrg | 1 | 110658 | ||
| lateral membranes (follicle cells) | YC0017 | Indy (lethal) | 3 | 50860 | ||
| plasma membranes | CA07474 | Picot | 2 | 50822 | ||
| muscle | muscle | ZCL0663 | Zasp66 | 3 | 110740 | |
| muscle | G00189 | Zasp52 | 2 | 110621 | ||
| nuclei | chromosomes | YB0011 | D1 | 3 | 50850 | |
| histones | G00280 | His2Av | 3 | 110647 | ||
| nuclei | G00131 | Dek | 2 | 50840 | ||
| nuclei | YB0077 | Rm62 | 3 | 50852 | ||
| ribosomes | ribosomes | ZCL3170 | RpS2 | 2 | 110957 | |
| RNP | RNP | ZCL1607 | Yps | 3 | 110796 |
Table 4.
Fluorescent protein fusions
| Category | Engineered | Expression | BDSC | reference |
|---|---|---|---|---|
| Actin | UASp-LifeAct | Gal4/UASp | [29] | |
| UASp-Actin-GFP | Gal4/UASp | 7309, 7310, 7311 | [148,150] | |
| UASp-Moesin | Gal4/UASp | [151] | ||
| Moesin | ubiquitous | [146] | ||
| Autophagosomes | UASp-mCherry-DrAtg8a | Gal4/UASp | 37749. 37750 | [152] |
| Germ cells | Bam-GFP | endogenous | [123] | |
| GFP-Nanos | endogenous | [139] | ||
| Cherry-Vasa | endogenous | [153] | ||
| GFP-Vasa | endogenous | [140] | ||
| Endoplasmic reticulum | EYFP-ER | ubiquitous | 7195 | [154] |
| UASp-GgalLYZ.GFP.KDEL | Gal4/UASp | 30903 | [155] | |
| UASp-RFP.KDEL | Gal4/UASp | 30909, 30910 | [155] | |
| Fusome | ShAdd-YFP | germline | [42] | |
| Golgi | EYFP-Golgi | ubiquitous | 7193 | [154] |
| UASp-GFP.Golgi | Gal4/UASp | 30902 | [155] | |
| UASp-RFP.Golgi | Gal4/UASp | 30907, 30908 | ||
| UASp-γCop.mRFP | Gal4/UASp | 29713, 29714 | ||
| UASp-γCop.GFP | Gal4/UASp | 29711 | ||
| Microtubules | EB1-GFP | Gal4/UASp | [156,157] | |
| EB1-GFP | ubiquitious | [158,159] | ||
| UASp-GFP-tubulin | Gal4/UASp | 7373 | [160] | |
| Tau-GFP | germline | [136] | ||
| EYFP-mito | ubiquitous | 7194 | [154] | |
| mRNA | bicoid-ms2 | endogenous | [161] | |
| gurken-ms2 | endogenous | [162] | ||
| oskar-ms2 | endogenous | [163] | ||
| nanos-ms2 | endogenous | [164] | ||
| Nuclei (chromosomes) | His2Av-Cherry | ubiquitous | 23650, 23651 | |
| Plasma Membrane | myr-mCherry | ubiquitous | [147] | |
| Ring canals | Ovhts-GFP | germline | [42] | |
| Tec29-GFP | germline | [165] | ||
| Vesicles | UASp-YFP-Rabs | Gal4/UASp | many | [166] |
4.1. Ovary dissection
The actual removal and dissection of ovaries is not difficult and with practice the task is accomplished easily, as most of the tissue within the abdomen of a well-fed female is ovary. Ovary dissection has been described in detail in several methods papers [87–89], and there was also an excellent video of ovary dissection in preparation for fixation accompanying Zimmerman et al [89]. Ovaries can be dissected into PBS, or more specialized media designed for Drosophila ex vivo culture (e.g., EBR [87], IMADS [90], or Grace’s or Schneider’s Drosophila media [88,91]. Once the ovaries are removed, dissecting forceps are typically used to tease apart the ovary into individual ovarioles. Note that one method for dissociating ovaries that relies on rapid pipetting of the tissue can damage the cells and lead to phenotypic artifacts [92], so this method should be avoided in most instances.
4.2. Fixation
Preservation of many ovarian antigens and cellular structures can be accomplished with a short (5–20 minute) fixation using buffered 4–6% formaldehyde [87,93]. Some investigators prefer a fixation solution made from freshly dissolved paraformaldehyde, though many structures are well preserved using formaldehyde diluted from a commercial reagent-grade 37% stock. Reagent grade formaldehyde contains ~10% methanol, which may interfere with preservation of some antigens, so a good alternative is commercial methanol-free electron microscopy grade formaldehyde sold by electron microscopy vendors (e.g., Polysciences, Electron Microscopy Sciences). Mature stage 14 oocytes/eggs are encased in a hydrophobic vitelline membrane, and protocols for embryo fixation often include heptane in the fixative. Inclusion of heptane in ovary fixatives can aid in the permeabilization of late-stage oocytes, but is not necessary if the focus of an experiment is to examine pre-stage 14 egg chambers.
An occasional problem with formaldehyde fixed tissue is poor penetration of antibodies into the germline. This results in no apparent antibody labeling in the center of mid- to late-stage egg chambers, leaving only a peripheral fluorescent signal. Strategies to alleviate this problem include altering the duration of fixation, as well as inclusion of detergent or DMSO during or after fixation to aid in permeabilization [93,94].
In addition to formaldehyde-based fixatives, other alternative fixations can be used to preserve antigens or cellular structures that are not detectable using aldehyde fixation. Fixation in 100% methanol results in protein denaturation, and some antigens are only detectable using such a method (e.g., Filamin protein on fusomes [95]). Fixation with methanol has several drawbacks, however, as it prevents F-actin labeling with phalloidin, and in many cases Methanol reduces or eliminates signal from fluorescent proteins. Finally, a heat fixation protocol first developed for Drosophila embryos [96] can be employed and has been used as a fixative for adherens junction components in ovaries [97].
4.3. Microtubules
Preservation and efficient immunolabeling of the microtubule cytoskeleton can be challenging. One important consideration is the fixation method; efficient labeling of microtubules in the germ cells requires care not to over fix the tissue, which prevents efficient antibody penetration. A protocol that gives excellent results involves briefly fixing ovaries soon after dissection in a high (36%) concentration of reagent grade formaldehyde, in an effort to fix the dynamic microtubules as quickly as possible without over-fixing, followed by a detergent permeabilization step [94]. Alternative protocols for microtubule preservation rely on methanol fixation, including a recently developed protocol that includes a detergent extraction step prior to methanol fixation in an effort to allow for better penetration of antibodies [98]. In addition, better germline labeling of microtubules can be achieved by using an anti-tubulin antibody that is directly conjugated to a fluor. Many labs use DM1A, an anti-α-tubulin monoclonal antibody that can be purchased as a fluorescent conjugate (e.g., FITC-DM1A, Sigma F2168). Alternatively, microtubules can be visualized using a transgene that expresses a fluorescent protein fused to either tubulin or a microtubule binding protein (Table 4).
4.4. F-actin
Visualizing the F-actin distribution in fixed ovaries is best accomplished using fluorescently labeled phalloidin following fixation with formaldehyde [87]. Phalloidin is a small peptide that binds specifically to F-actin, and is a remarkably specific probe [99]. Inclusion of detergent and phalloidin during the fixation step has been reported to give improved results [100]. While much of the F-actin cytoskeleton appears to be faithfully preserved by these protocols, a recent report demonstrated that formaldehyde fixation introduces a striking artifact in the long (~50 μ) F-actin bundles that restrain the nurse cell nuclei during dumping [29]. In formaldehyde fixed tissue, these bundles have a striated appearance, but the striations are not observed in live tissue when the bundles are labeled with fluorescent protein fusions [29]. It therefore appears that formaldehyde fixation can result in the alteration of some F-actin structures, though based on our experience this artifact appears to be limited to the long F-actin cables that form at stage 10B. If fixation artifacts are suspected, the organization of F-actin in fixed cells can be compared to the distribution of fluorescent protein fusions that label the F-actin cytoskeleton (Table 4) in live cells, as done in Huelsmann et al [29].
4.5. Imaging live oogenesis
The ability to conduct live-cell imaging experiments has had a significant impact on cell biological studies of Drosophila oogenesis. Short term (~30 min) imaging of dissected egg chambers and ovarioles is easily accomplished by dissecting ovaries into an inert, gas-permeable halocarbon oil, and it is possible to introduce fluorescent tracers and/or experimental treatments into oocytes, nurse cells, or follicle cells using microinjection. Several excellent methods papers describe these procedures in detail, and two have excellent accompanying video tutorials [101–103].
Remarkable progress has been made during the past few years in long-term (up to ~24 hours) imaging of live ovarian tissue. Recent advances in our understanding of cell division patterns [104], morphogenetic movements [13], and cell migration [35] have all benefited from long-term live-cell imaging of oogenesis. Much of this work was made possible because of improved ex vivo culture procedures that allow Drosophila egg chambers to remain viable for up to 24 hours in culture [88]. Prasad et al (2007) present a thorough protocol for live imaging of egg chambers, and Morris et al [104] discuss special considerations for imaging germaria. Important considerations for long-term imaging involve culturing egg chambers in carefully prepared Schneider’s media that is supplemented with bovine insulin.
4.6.1 Visualization of RNA: fixed tissue
Localization of RNA during oogenesis is essential for patterning the egg chamber and for storing patterning cues for embryogenesis. In fixed ovaries, RNA localization and abundance can be assessed using colorimetric RNA in situ hybridization [105]. More recently, fluorescent in situ hybridization (FISH) techniques have been optimized for use in Drosophila that allow greater sensitivity and precision in localization [106]. In addition, a detailed procedure for simultaneous FISH and immunofluorescence in ovaries was published [89]. Finally, methods have also been described to localize both RNA and protein by electron microscopy (ISH-IEM; [107].
4.6.2 Visualization of RNA: live tissue
Many mechanistic insights into RNA localization required the ability to observe the process in vivo. Two basic strategies have been developed to observe RNA localization in live ovaries: microinjection of fluorescently labeled RNA, and the use of genetically encoded fluorescent protein fusions that bind to RNA sequences in engineered constructs. These approaches are discussed in detail in reviews from the Davis and Gavis labs [25,26], and procedures for microinjecting dissected egg chambers are included in Parton et al [108]. The use of a fluorescent protein fusion relies on the fusing the bacteriophage MS2 protein to a fluorescent protein and incorporating RNA sequence that specifically binds MS2 into the 3′ UTR of the mRNA under study. This has been done for five developmentally important RNAs (table FP reagents), and the St Johnston lab has published detailed guidelines for designing and using the MS2 system to localize RNA in oogenesis [109].
4.7. Microscopy
Conventional light and fluorescence microscopy techniques are invaluable tools in the study of Drosophila oogenesis. However, because of the thickness of egg chambers, widefield fluorescence microscopy images of ovarioles and egg chambers often exhibit substantial blurring caused by autofocus light originating from above and beneath the focal plane. Therefore, some method of optical section microscopy is highly recommended. Essentially all major forms of optical sectioning microscopy, including laser scanning confocal microscopy (conventional and multi-photon), spinning disc confocal microscopy, structured illumination, and deconvolution of widefield images are useful in reducing blur caused by out of focus light in optical sections. The Davis lab has published a brief but useful guide on choosing imaging systems that are well suited for Drosophila oogenesis [110].
In addition to these well-established techniques, newer imaging techniques are emerging that offer new opportunities for imaging oogenesis. Among them are a host of super-resolution techniques that rely on photophysics and computation to localize individual fluorescent molecules with greater precision that is possible with conventional light microscopy [111]. Applying these techniques to the relatively thick ovarian tissue presents significant challenges, but imaging is possible [112] and should improve. In addition, lightsheet, or selective plane illumination microscopy (SPIM) promises to offer an additional useful method for imaging oogenesis. Lightsheet microscopy achieves optical sectioning by separate objective lenses for illumination and detection, and these microscopes are able to achieve extremely rapid optical sectioning rates [113]. Sample preparation for lightsheet microscopy often involves embedding the tissue in a gel – e.g., low-melt agarose – and this appears to be compatible with live ovarian tissue.
4.8. Flow cytometry
The analysis specific cell populations isolated from ovaries is greatly aided by the ability to dissociate ovarian cells and perform fluorescence-activated cell sorting (FACS). FACS has been used to understand DNA replication and endoreplication through nuclear flow sorting [14] and to select specific populations of GFP expressing cells for gene expression analysis [114,115]. Dissected ovaries can be dissociated with the aid of collagenase and/or trypsin, and thorough protocols for nuclear flow sorting for DNA content analysis [116] and intact cell FACs analysis [117] have been published.
5. Conclusions
During the past several years, some of the most significant advances in our understanding of Drosophila ovarian biology have resulted from improved methods of live-cell imaging [13,20,104]. We anticipate that further improvements in live cell imaging methods combined with new genetic analysis tools will lead to many additional insights into the molecular mechanisms of ovarian development.
Acknowledgments
We thank Peter McLean for contributions to Figure 1, and members of the Cooley lab for comments on the manuscript. Research in the Cooley lab is supported by NIH grant GM043301.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. Development of Drosophila melanogaster. Plainview, New York: Cold Spring Harbor Press; 1993. pp. 1–70. [Google Scholar]
- 2.Bastock R, St Johnston D. Drosophila oogenesis. Curr Biol. 2008;18:R1082–7. doi: 10.1016/j.cub.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez-Valentín R, López-González I, Jorquera R, Labarca P, Zurita M, Reynaud E. Oviduct contraction in Drosophila is modulated by a neural network that is both, octopaminergic and glutamatergic. J Cell Physiol. 2006;209:183–98. doi: 10.1002/jcp.20722. [DOI] [PubMed] [Google Scholar]
- 4.Middleton CA, Nongthomba U, Parry K, Sweeney ST, Sparrow JC, Elliott CJH. Neuromuscular organization and aminergic modulation of contractions in the Drosophila ovary. BMC Biol. 2006;4:17. doi: 10.1186/1741-7007-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudson AM, Petrella LN, Tanaka AJ, Cooley L. Mononuclear muscle cells in Drosophila ovaries revealed by GFP protein traps. Dev Biol. 2008;314:329–40. doi: 10.1016/j.ydbio.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H, Spradling AC. Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev Biol. 1993;159:140–52. doi: 10.1006/dbio.1993.1228. [DOI] [PubMed] [Google Scholar]
- 7.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–4. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 8.Kirilly D, Xie T. The Drosophila ovary: an active stem cell community. Cell Res. 2007;17:15–25. doi: 10.1038/sj.cr.7310123. [DOI] [PubMed] [Google Scholar]
- 9.Huynh J-R, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol. 2004;14:R438–49. doi: 10.1016/j.cub.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 10.Hudson AM, Cooley L. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol. 2002;156:677–87. doi: 10.1083/jcb.200109065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Reyes A, Elliott H, St Johnston D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature. 1995;375:654–8. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- 12.Kugler J-M, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- 13.Haigo SL, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 2011;331:1071–4. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilly MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–26. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- 15.López-Schier H, St Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001;15:1393–405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng WM, Althauser C, Ruohola-Baker H. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development. 2001;128:4737–46. doi: 10.1242/dev.128.23.4737. [DOI] [PubMed] [Google Scholar]
- 17.Woodruff RI, Tilney LG. Intercellular bridges between epithelial cells in the Drosophila ovarian follicle: a possible aid to localized signaling. Dev Biol. 1998;200:82–91. doi: 10.1006/dbio.1998.8948. [DOI] [PubMed] [Google Scholar]
- 18.Haglund K, Nezis IP, Lemus D, Grabbe C, Wesche J, Liestøl K, et al. Cindr interacts with anillin to control cytokinesis in Drosophila melanogaster. Curr Biol. 2010;20:944–50. doi: 10.1016/j.cub.2010.03.068. [DOI] [PubMed] [Google Scholar]
- 19.Airoldi SJ, McLean PF, Shimada Y, Cooley L. Intercellular protein movement in syncytial Drosophila follicle cells. J Cell Sci. 2011;124:4077–86. doi: 10.1242/jcs.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLean PF, Cooley L. Protein equilibration through somatic ring canals in Drosophila. Science. 2013;340:1445–7. doi: 10.1126/science.1234887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean PF, Cooley L. Bridging the divide: Illuminating the path of intercellular exchange through ring canals. Vol. 8. Fly Landes Bioscience Inc; 2014. pp. 0–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzalupo S, Cooley L. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 2006;13:1950–9. doi: 10.1038/sj.cdd.4401892. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins VK, Timmons AK, McCall K. Diversity of cell death pathways: insight from the fly ovary. Trends Cell Biol. 2013;23:567–74. doi: 10.1016/j.tcb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasko P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;136:2493–503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weil TT, Parton RM, Davis I. Making the message clear: visualizing mRNA localization. Trends Cell Biol. 2010;20:380–90. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–82. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- 28.Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69:173–84. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- 29.Huelsmann S, Ylänne J, Brown NH. Filopodia-like Actin Cables Position Nuclei in Association with Perinuclear Actin in Drosophila Nurse Cells. Dev Cell. 2013;26:604–15. doi: 10.1016/j.devcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tootle TL, Spradling AC. Drosophila Pxt: a cyclooxygenase-like facilitator of follicle maturation. Development. 2008;135:839–47. doi: 10.1242/dev.017590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr-Weaver TL, Spradling AC. Drosophila chorion gene amplification requires an upstream region regulating s18 transcription. Mol Cell Biol. 1986;6:4624–33. doi: 10.1128/mcb.6.12.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12:734–44. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Smith L, Armento A, Deng W-M. Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J Cell Biol. 2008;182:885–96. doi: 10.1083/jcb.200802084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 35.Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Peters NC, Thayer NH, Kerr SA, Tompa M, Berg CA. Following the “tracks”: Tramtrack69 regulates epithelial tube expansion in the Drosophila ovary through Paxillin, Dynamin, and the homeobox protein Mirror. Dev Biol. 2013;378:154–69. doi: 10.1016/j.ydbio.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rørth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–8. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 38.Neelakanta G, Hudson AM, Sultana H, Cooley L, Fikrig E. Expression of Ixodes scapularis antifreeze glycoprotein enhances cold tolerance in Drosophila melanogaster. PLoS ONE. 2012;7:e33447. doi: 10.1371/journal.pone.0033447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109:6626–31. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, et al. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn. 1997;209:310–22. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 41.Ward EJ, Thaipisuttikul I, Terayama M, French RL, Jackson SM, Cosand KA, et al. GAL4 enhancer trap patterns during Drosophila development. Genesis. 2002;34:46–50. doi: 10.1002/gene.10138. [DOI] [PubMed] [Google Scholar]
- 42.Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–12. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- 43.Quinlan ME. Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development. 2013;140:4417–25. doi: 10.1242/dev.097337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skora AD, Spradling AC. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:7389–94. doi: 10.1073/pnas.1003180107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 46.Hsu H-J, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci USA. 2009;106:1117–21. doi: 10.1073/pnas.0809144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gans M, Audit C, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohler JD. Developmental genetics of the Drosophila egg. I. Identification of 59 sex-linked cistrons with maternal effects on embryonic development. Genetics. 1977;85:259–72. doi: 10.1093/genetics/85.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komitopoulou K, Gans M, Margaritis LH, Kafatos FC, Masson M. Isolation and Characterization of Sex-Linked Female-Sterile Mutants in DROSOPHILA MELANOGASTER with Special Attention to Eggshell Mutants. Genetics. 1983;105:897–920. doi: 10.1093/genetics/105.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrimon N, Mohler D, Engstrom L, Mahowald AP. X-linked female-sterile loci in Drosophila melanogaster. Genetics. 1986;113:695–712. doi: 10.1093/genetics/113.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–17. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–36. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tearle R, Nüsslein-Volhard C. Tübingen mutants and stocklist. Drosophila Information Service. 1987;66:209–69. [Google Scholar]
- 54.Underwood EM, Caulton JH, Allis CD, Mahowald AP. Developmental fate of pole cells in Drosophila melanogaster. Dev Biol. 1980;77:303–14. doi: 10.1016/0012-1606(80)90476-5. [DOI] [PubMed] [Google Scholar]
- 55.Technau G, Campos-Ortega J. Lineage analysis of transplanted individual cells in embryos of Drosophila melanogaster III. Commitment and proliferative capabilities of pole cells and midgut progenitors. Roux’s Arch. Dev. Biol. 1986;195:489–98. doi: 10.1007/BF00375889. [DOI] [PubMed] [Google Scholar]
- 56.Silver-Morse L, Li WX. The role of receptor tyrosine kinases in primordial germ cell migration. Methods Mol Biol. 2011;750:291–306. doi: 10.1007/978-1-61779-145-1_20. [DOI] [PubMed] [Google Scholar]
- 57.Illmensee K. The potentialities of transplanted early gastrula nuclei of Drosophila melanogaster. Production of their imago descendants by germ-line transplantation. Wilhelm Roux’s Arch. Entwickslungmech. Org. 1973;171:331–43. doi: 10.1007/BF00577730. [DOI] [PubMed] [Google Scholar]
- 58.Van Deusen EB. Sex determination in germ line chimeras of Drosophila melanogaster. J Embryol Exp Morphol. 1977;37:173–85. [PubMed] [Google Scholar]
- 59.Xu T, Rubin GM. The effort to make mosaic analysis a household tool. Development. 2012;139:4501–3. doi: 10.1242/dev.085183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 61.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 62.Theodosiou NA, Xu T. Use of FLP/FRT system to study Drosophila development. Methods. 1998;14:355–65. doi: 10.1006/meth.1998.0591. [DOI] [PubMed] [Google Scholar]
- 63.Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–53. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–69. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 65.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–9. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–92. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morris JZ, Navarro C, Lehmann R. Identification and analysis of mutations in bob, Doa and eight new genes required for oocyte specification and development in Drosophila melanogaster. Genetics. 2003;164:1435–46. doi: 10.1093/genetics/164.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yohn CB, Pusateri L, Barbosa V, Lehmann R. l(3)malignant brain tumor and three novel genes are required for Drosophila germ-cell formation. Genetics. 2003;165:1889–900. doi: 10.1093/genetics/165.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 70.Yan Y, Denef N, Schüpbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wieschaus E, Szabad J. The development and function of the female germ line in Drosophila melanogaster: a cell lineage study. Dev Biol. 1979;68:29–46. doi: 10.1016/0012-1606(79)90241-0. [DOI] [PubMed] [Google Scholar]
- 72.Margolis J, Spradling A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121:3797–807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- 73.Harrison DA, Perrimon N. Simple and efficient generation of marked clones in Drosophila. Curr Biol. 1993;3:424–33. doi: 10.1016/0960-9822(93)90349-s. [DOI] [PubMed] [Google Scholar]
- 74.Griffin R, Sustar A, Bonvin M, Binari R, del Valle Rodriguez A, Hohl AM, et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods Nature Publishing Group. 2009;6:600–2. doi: 10.1038/nmeth.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H-H, Chen C-H, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12:947–53. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 77.Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, et al. A Drosophila Resource of Transgenic RNAi Lines for Neurogenetics. Genetics. 2009;182:1089–100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–9. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 79.Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, Yang-Zhou D, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–7. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perrimon N, Ni J-Q, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2:a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yan D, Neumüller RA, Buckner M, Ayers K, Li H, Hu Y, et al. A regulatory network of Drosophila germline stem cell self-renewal. 2014 doi: 10.1016/j.devcel.2014.01.020. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci USA. 2001;98:15050–5. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quiñones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, et al. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–31. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCall K, Peterson JS, Pritchett TL. Detection of cell death in Drosophila. Methods Mol Biol. 2009;559:343–56. doi: 10.1007/978-1-60327-017-5_24. [DOI] [PubMed] [Google Scholar]
- 86.Timmons AK, Meehan TL, Gartmond TD, McCall K. Use of necrotic markers in the Drosophila ovary. Methods Mol Biol. 2013;1004:215–28. doi: 10.1007/978-1-62703-383-1_16. [DOI] [PubMed] [Google Scholar]
- 87.Verheyen E, Cooley L. Looking at oogenesis. Methods Cell Biol. 1994;44:545–61. [PubMed] [Google Scholar]
- 88.Prasad M, Jang AC-C, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nature protocols. 2007;2:2467–73. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- 89.Zimmerman SG, Peters NC, Altaras AE, Berg CA. Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nature protocols. 2013;8:2158–79. doi: 10.1038/nprot.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singleton K, Woodruff RI. The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev Biol. 1994;161:154–67. doi: 10.1006/dbio.1994.1017. [DOI] [PubMed] [Google Scholar]
- 91.Ashburner M. Drosophila: A laboratory manual. Cold Spring Harbor Laboratory Pr; 1989. [Google Scholar]
- 92.Haack T, Bergstralh DT, Johnston DS. Damage to the Drosophila follicle cell epithelium produces “‘false clones’” with apparent polarity phenotypes. Biology Open. doi: 10.1242/bio.20134671. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Máthé E. Immunocytological Analysis of Oogenesis. In: Henderson D, editor. Methods in Molecular Biology. Totowa, NJ: Humana Press Inc; 2004. pp. 89–127. [DOI] [PubMed] [Google Scholar]
- 94.Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- 95.Sokol NS, Cooley L. Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr Biol. 1999;9:1221–30. doi: 10.1016/s0960-9822(99)80502-8. [DOI] [PubMed] [Google Scholar]
- 96.Miller KG, Field CM, Alberts BM. Actin-binding proteins from Drosophila embryos: a complex network of interacting proteins detected by F-actin affinity chromatography. J Cell Biol. 1989;109:2963–75. doi: 10.1083/jcb.109.6.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peifer M, Orsulic S, Sweeton D, Wieschaus E. A role for the Drosophila segment polarity gene armadillo in cell adhesion and cytoskeletal integrity during oogenesis. Development. 1993;118:1191–207. doi: 10.1242/dev.118.4.1191. [DOI] [PubMed] [Google Scholar]
- 98.Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, Guichet A. The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development. 2006;133:129–39. doi: 10.1242/dev.02179. [DOI] [PubMed] [Google Scholar]
- 99.Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–8. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frydman HM, Spradling AC. The receptor-like tyrosine phosphatase lar is required for epithelial planar polarity and for axis determination within drosophila ovarian follicles. Development. 2001;128:3209–20. doi: 10.1242/dev.128.16.3209. [DOI] [PubMed] [Google Scholar]
- 101.Parton RM, Vallés AM, Dobbie IM, Davis I. Isolation of Drosophila egg chambers for imaging. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5402. pdb.prot5402. [DOI] [PubMed] [Google Scholar]
- 102.Weil TT, Parton RM, Davis I. Preparing individual Drosophila egg chambers for live imaging. J Vis Exp. 2012 doi: 10.3791/3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pokrywka NJ. Live imaging of GFP-labeled proteins in Drosophila oocytes. J Vis Exp. 2013 doi: 10.3791/50044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morris LX, Spradling AC. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development. 2011;138:2207–15. doi: 10.1242/dev.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol. 1994;44:575–98. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- 106.Lécuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol. 2008;420:289–302. doi: 10.1007/978-1-59745-583-1_18. [DOI] [PubMed] [Google Scholar]
- 107.Herpers B, Xanthakis D, Rabouille C. ISH-IEM: a sensitive method to detect endogenous mRNAs at the ultrastructural level. Nature protocols. 2010;5:678–87. doi: 10.1038/nprot.2010.12. [DOI] [PubMed] [Google Scholar]
- 108.Parton RM, Vallés AM, Dobbie IM, Davis I. Live cell imaging in Drosophila melanogaster. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.top75. pdb.top75. [DOI] [PubMed] [Google Scholar]
- 109.Belaya K, St Johnston D. Using the mRNA-MS2/MS2CP-FP system to study mRNA transport during Drosophila oogenesis. Methods Mol Biol. 2011;714:265–83. doi: 10.1007/978-1-61779-005-8_17. [DOI] [PubMed] [Google Scholar]
- 110.Davis I, Parton RM. Selection of appropriate imaging equipment and methodology for live cell imaging in Drosophila. CSH Protoc. 2006 doi: 10.1101/pdb.ip21. [DOI] [PubMed] [Google Scholar]
- 111.Toomre D, Bewersdorf J. A new wave of cellular imaging. Annu Rev Cell Dev Biol. 2010;26:285–314. doi: 10.1146/annurev-cellbio-100109-104048. [DOI] [PubMed] [Google Scholar]
- 112.Zobel T, Bogdan S. A high resolution view of the fly actin cytoskeleton lacking a functional WAVE complex. J Microsc. 2013;251:224–31. doi: 10.1111/jmi.12020. [DOI] [PubMed] [Google Scholar]
- 113.Weber M, Huisken J. Light sheet microscopy for real-time developmental biology. Curr Opin Genet Dev. 2011;21:566–72. doi: 10.1016/j.gde.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 114.Bryant Z, Subrahmanyan L, Tworoger M, LaTray L, Liu CR, Li MJ, et al. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc Natl Acad Sci USA. 1999;96:5559–64. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kai T, Williams D, Spradling AC. The expression profile of purified Drosophila germline stem cells. Dev Biol. 2005;283:486–502. doi: 10.1016/j.ydbio.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 116.Calvi BR, Lilly MA. Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary. Methods Mol Biol. 2004;247:203–13. doi: 10.1385/1-59259-665-7:203. [DOI] [PubMed] [Google Scholar]
- 117.Lim RSM, Osato M, Kai T. Isolation of undifferentiated female germline cells from adult Drosophila ovaries. Curr Protoc Stem Cell Biol. 2012;Chapter 2(Unit2E.3) doi: 10.1002/9780470151808.sc02e03s22. [DOI] [PubMed] [Google Scholar]
- 118.Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–64. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 119.Cabrera GR, Godt D, Fang P-Y, Couderc J-L, Laski FA. Expression pattern of Gal4 enhancer trap insertions into the bric à brac locus generated by P element replacement. Genesis. 2002;34:62–5. doi: 10.1002/gene.10115. [DOI] [PubMed] [Google Scholar]
- 120.Hartman TR, Zinshteyn D, Schofield HK, Nicolas E, Okada A, O’Reilly AM. Drosophila Boi limits Hedgehog levels to suppress follicle stem cell proliferation. J Cell Biol. 2010;191:943–52. doi: 10.1083/jcb.201007142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pfeiffer BD, Jenett A, Hammonds AS, Ngo TTB, Misra S, Murphy C, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–20. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sahai-Hernandez P, Nystul TG. A dynamic population of stromal cells contributes to the follicle stem cell niche in the Drosophila ovary. Development. 2013;140:4490–8. doi: 10.1242/dev.098558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–70. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- 124.Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- 125.Tracey WD, Ning X, Klingler M, Kramer SG, Gergen JP. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–84. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–6. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 127.Queenan AM, Ghabrial A, Schüpbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–80. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 128.Zhu C-H, Xie T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development. 2003;130:2579–88. doi: 10.1242/dev.00499. [DOI] [PubMed] [Google Scholar]
- 129.Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–71. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- 130.Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- 131.Wilder EL, Perrimon N. Dual functions of wingless in the Drosophila leg imaginal disc. Development. 1995;121:477–88. doi: 10.1242/dev.121.2.477. [DOI] [PubMed] [Google Scholar]
- 132.Goentoro LA, Reeves GT, Kowal CP, Martinelli L, Schüpbach T, Shvartsman SY. Quantifying the Gurken morphogen gradient in Drosophila oogenesis. Dev Cell. 2006;11:263–72. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–71. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 134.Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–57. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 135.Niepielko MG, Hernáiz-Hernández Y, Yakoby N. BMP signaling dynamics in the follicle cells of multiple Drosophila species. Dev Biol. 2011;354:151–9. doi: 10.1016/j.ydbio.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 136.Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, et al. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–78. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- 137.Dodson GS, Guarnieri DJ, Simon MA. Src64 is required for ovarian ring canal morphogenesis during Drosophila oogenesis. Development. 1998;125:2883–92. doi: 10.1242/dev.125.15.2883. [DOI] [PubMed] [Google Scholar]
- 138.Robinson DN, Cooley L. Examination of the function of two kelch proteins generated by stop codon suppression. Development. 1997;124:1405–17. doi: 10.1242/dev.124.7.1405. [DOI] [PubMed] [Google Scholar]
- 139.Forrest KM, Clark IE, Jain RA, Gavis ER. Temporal complexity within a translational control element in the nanos mRNA. Development. 2004;131:5849–57. doi: 10.1242/dev.01460. [DOI] [PubMed] [Google Scholar]
- 140.Sano H, Nakamura A, Kobayashi S. Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech Dev. 2002;112:129–39. doi: 10.1016/s0925-4773(01)00654-2. [DOI] [PubMed] [Google Scholar]
- 141.Serano TL, Cheung HK, Frank LH, Cohen RS. P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene. 1994;138:181–6. doi: 10.1016/0378-1119(94)90804-4. [DOI] [PubMed] [Google Scholar]
- 142.Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–79. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fehon RG, Oren T, LaJeunesse DR, Melby TE, McCartney BM. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics. 1997;146:245–52. doi: 10.1093/genetics/146.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brummel TJ, Twombly V, Marqués G, Wrana JL, Newfeld SJ, Attisano L, et al. Characterization and relationship of Dpp receptors encoded by the saxophone and thick veins genes in Drosophila. Cell. 1994;78:251–61. doi: 10.1016/0092-8674(94)90295-x. [DOI] [PubMed] [Google Scholar]
- 145.Royou A, Field C, Sisson JC, Sullivan W, Karess R. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol Biol Cell. 2004;15:838–50. doi: 10.1091/mbc.E03-06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–90. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–49. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kelso RJ, Hudson AM, Cooley L. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J Cell Biol. 2002;156:703–13. doi: 10.1083/jcb.200110063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lighthouse DV, Buszczak M, Spradling AC. New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev Biol. 2008;317:59–71. doi: 10.1016/j.ydbio.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Röper K, Mao Y, Brown NH. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J Cell Sci. 2005;118:3937–48. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- 151.Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–17. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- 152.Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjørkøy G, Johansen T, et al. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5:298–302. doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- 153.Lerit DA, Gavis ER. Transport of Germ Plasm on Astral Microtubules Directs Germ Cell Development in Drosophila. Curr Biol. 2011 doi: 10.1016/j.cub.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.LaJeunesse DR, Buckner SM, Lake J, Na C, Pirt A, Fromson K. Three new Drosophila markers of intracellular membranes. BioTechniques. 2004;36:784-8–790. doi: 10.2144/04365ST01. [DOI] [PubMed] [Google Scholar]
- 155.Snapp EL, Iida T, Frescas D, Lippincott-Schwartz J, Lilly MA. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol Biol Cell. 2004;15:4512–21. doi: 10.1091/mbc.E04-06-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–74. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 157.Shimada Y, Burn KM, Niwa R, Cooley L. Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev Biol. 2011;355:250–62. doi: 10.1016/j.ydbio.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev Cell. 2006;10:209–22. doi: 10.1016/j.devcel.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 159.Viktorinová I, Dahmann C. Microtubule polarity predicts direction of egg chamber rotation in Drosophila. Curr Biol. 2013;23:1472–7. doi: 10.1016/j.cub.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 160.Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–64. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- 161.Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell. 2006;11:251–62. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 162.Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schüpbach T. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci. 2008;121:887–94. doi: 10.1242/jcs.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–53. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–68. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 165.Lu N, Guarnieri DJ, Simon MA. Localization of Tec29 to ring canals is mediated by Src64 and PtdIns(3,4,5)P3-dependent mechanisms. EMBO J. 2004;23:1089–100. doi: 10.1038/sj.emboj.7600127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, et al. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–22. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]