Abstract
Sarcoidosis is an immune-mediated multisystem disease characterized by the formation of non-caseating granulomas. The pathogenesis of sarcoidosis is unclear, with proposed infectious or environmental antigens triggering an aberrant immune response in susceptible hosts. Multiple pro-inflammatory signaling pathways have been implicated in mediating macrophage activation and granuloma formation in sarcoidosis, including IFN-γ/STAT-1, IL-6/STAT-3, and NF-κB. It is difficult to distinguish sarcoidosis from other granulomatous diseases or assess disease severity and treatment response with histopathology alone. Therefore, development of improved diagnostic tools is imperative. Herein, we describe an efficient and reliable technique to classify granulomatous disease through selected gene expression and identify novel genes and cytokine pathways contributing to the pathogenesis of sarcoidosis. We quantified the expression of twenty selected mRNAs extracted from formalin-fixed paraffin embedded (FFPE) tissue (n=38) of normal lung, suture granulomas, sarcoid granulomas, and fungal granulomas. Utilizing quantitative real-time RT-PCR we analyzed the expression of several genes, including IL-6, COX-2, MCP-1, IFN-γ, T-bet, IRF-1, NOX2, IL-33, Eotaxin-1 and revealed differential regulation between suture, sarcoidosis, and fungal granulomas. This is the first study demonstrating that quantification of target gene expression in FFPE tissue biopsies is a potentially effective diagnostic and research tool in sarcoidosis.
Keywords: sarcoidosis, pathology autoimmunity, cytokines, gene expression, transcription factor, reactive oxygen species, formalin-fixed paraffin embedded tissue, granuloma, RT-PCR
INTRODUCTION
Sarcoidosis is a multisystemic immune-mediated granulomatous disease of uncertain etiology with varying relationships to environmental exposures, demographic factors, and genetic predisposition (Iannuzzi et al., 2007). Sarcoidosis predominately involves the lungs and the lymphatic system is diagnosed by a combination of clinical, radiologic, and pathologic findings. Sarcoidosis remains a diagnosis of exclusion, with a delay in diagnosis due to non-specific disease criteria and need for invasive tissue biopsy. The disease is typically diagnosed in the third or fourth decade of life, and incidence of sarcoidosis is three times higher in blacks than whites (36 compared to 11 patients per 100,000) (Rybicki et al., 1997). Two-thirds of sarcoid patients go into remission before three years of disease involvement, while the remaining one-third develops chronic disease. Mortality in patients with sarcoidosis is higher than that of the general population, mainly due to pulmonary fibrosis. Treatment of sarcoidosis is limited, with corticosteroids being the mainstay of management (Iannuzzi and Fontana, 2011).
Sarcoidosis has a genetic predisposition evident by familial clustering and higher concordance among monozygotic compared to diazygotic twins. (Grunewald, 2008; Rybicki et al., 2001; Rybicki et al., 1997). Genome-wide associations studies (GWAS) have identified multiple genes involved in sarcoidosis including the butyrophillin-like 2 (BTNL2) gene, a member of the B7 costimulatory receptor family (Valentonyte et al., 2005) and annexin A11, a calcium-dependent phospholipid-binding protein involved in cell division and vesicle trafficking (Hofmann et al., 2008). Importantly, mutations within NOD2, which induces constitutive activation of the transcription factor NF-kB, were identified in early-onset sarcoidosis, (Kanazawa et al., 2005). Other candidate genes predicting susceptibility to sarcoidosis include genes involved in antigen processing and presentation by macrophages, factors affecting macrophage activation, T cell activation, and in injury repair (Kriegova et al., 2011). Candidate antigens that may trigger granulomatous inflammation in sarcoidosis include bacterial proteins from Mycobacterium tuberculosis and propionibacteria and β-glucan, a cell wall constituent of fungi (Ishige et al., 1999; Song et al., 2005; Terčelj et al., 2014).
The pathogenic hallmark of sarcoidosis is non-caseating granulomas that accumulate in multiple affected organs and are pathogenic through mass effect or tissue destruction. Sarcoid granulomas contain central macrophages that aggregate and differentiate into epitheliod histiocytes that fuse to form multinucleated giant cells (Saidha et al., 2012). Oligoclonal αβ+ CD4+ lymphocytes have been predominately located in the center and CD8+ T cells at the periphery of granulomas (Saidha et al., 2012). Granuloma formation in sarcoidosis is postulated to be a complex process involving initially antigen processing and presentation with activation of CD4+ T cells by macrophages followed by additional macrophage accumulation and continuous production of inflammatory mediators (Iannuzzi et al., 2007).
Sarcoidosis is a systemic diseaseevident by the upregulation of inflammatory molecules in serum that are secreted from peripheral leukocytes. (Zhou et al., 2012). Microarray analysis from peripheral leukocytes of sarcoidosis patients revealed a unique gene expression signature involving genes implicated in T cell differentiation and activation and cytokine signaling (Zhou et al., 2012). In addition, inflammatory mediators have been elevated in the bronchoalveolar lavage fluid (BALF) of sarcoid patients including the cytokine IFN-γ and the chemokine IP-10 (Miotto et al., 2001; Terčelj et al., 2014).
Specification of naïve CD4+ T cells and macrophages to effector subset lineages is critical to skewing immune responses in response to antigenic triggers and involves specific activation of transcription factors. The immune response in sarcoidosis is primarily mediated by the type I inflammatory cytokine pathway, which is promoted by NF-kB and STAT1 dependent signaling (Iannuzzi and Fontana, 2011; Miotto et al., 2001). The transcription factor T-bet is elevated in sarcoidosis and promotes Th1 specification through IL-12 and IFNγ signaling (Kriegova et al., 2011). Importantly, IFNγ is elevated in sarcoidois and mediates activation of signal transducer and activator of transcription-1 (STAT1) that is central in mounting a type I response by promoting expression of interferon-regulatory factor I (IRF-1) and downstream inflammatory mediators including the chemokine IP-10, and Nox2 (Miotto et al., 2001), (Rosenbaum et al., 2009). Non-caseating epithelioid granulomas of sarcoidosis include Th1 CD4+ lymphocytes that promote potent immune response via IL-2 and IFN-γ release and subsequent activation and differentiation of macrophages (Hunninghake and Crystal, 1981; Pinkston et al., 1983; Saltini et al., 1986).
The gold standard for diagnosing sarcoidosis is tissue biopsy and the presence of these non-caseating granulomas. However, non-caseating granulomas are non-specific, and can be found in other pathologic conditions including infections such as tuberculosis and fungal infections or by environmental exposures (Iannuzzi and Fontana, 2011). The cellular morphology of granuloma from bacterial, fungal, or in sarcoidosis cannot be distinguished from one another by histology alone (Zaim et al., 1990). Interestingly, the biology of granuloma formation in sarcoidosis differs from reactions to foreign materials. Suture granulomas are formed by adsorption of plasma proteins to the suture material, local complement activation and cell apoptosis leading to macrophage recruitment, phagocytosis, and formation of multinucleated giant cells (Anderson et al., 2008).
This study aims to identify gene expression patterns that differentiate sarcoid granulomas from other causes of granulomatous inflammation. We postulated that the gene expression profile is unique between different granuloma types and wanted to perform the first study to examine the utility of gene profiling as a novel diagnostic and research tool. We quantified the expression of twenty selected mRNAs from formalin-fixed paraffin embedded (FFPE) tissue of normal lung, suture granulomas, sarcoid granulomas, and fungal granulomas. Utilizing quantitative real-time RT-PCR we analyzed the expression of several genes, including IL-6, COX-2, MCP-1, IFN-γ, T-bet, IRF-1, NOX2, IL-33, Eotaxin-1 and revealed differential regulation between suture, sarcoidosis, and fungal granulomas. This study introduces a novel potential diagnostic tool in sarcoidosis and furthers our current understanding of signaling pathways involved in granulomatous inflammation
MATERIALS & METHODS
Sample Selection
The study was approved by the Upstate Medical University Institutional Review Board for the Protection of Human Subjects. Retrospectively reviewed archived FFPE biopsy and resection pathology specimens from the department of pathology at State University of New York Upstate Medical University diagnosed as sarcoid granulomas, infectious granulomas, suture granulomas, and normal (non-granulomatous) lung tissue were used for the study. The following pathologic inclusion criteria were used to select samples for each diagnostic category: 1) normal samples (NS) included tissue with no evidence of necrotizing or non-necrotizing epithelioid granulomas, birefringent or foreign material, vasculitis, abundant background inflammation. Normal lung specimens were obtained from segments of normal lung that were procured from lung cancer resection specimens. 2) suture granuloma were composed of tissue with granulomatous inflammation surrounding suture material with no evidence of necrotizing granulomas, vasculitis, abundant background inflammation, nor demonstrable microorganisms on Ziehl-Neelsen (AFB) and Grocott’s methenamine silver (GMS) stains. 3) sarcoid granulomas were composed of tissue with non-necrotizing epithelioid granulomas and no evidence of necrotizing granulomas, birefringent or foreign material, vasculitis, abundant background inflammation, nor demonstrable microorganisms on AFB and GMS stains (Rosen, 2007), 4) Infectious granuloma were categorized by tissue with necrotizing or non-necrotizing epithelioid granulomas with demonstrable microorganisms on AFB or GMS stains but no evidence of birefringent or foreign material, abundant background inflammation, nor vasculitis. Two pathologists confirmed the diagnoses and determined the suitability for gene expression profiling.
RNA isolation and cDNA conversion
RNA was extracted from FFPE tissue using the High Pure FFPE RNA Micro Kit and based on the manufacturer’s recommendations (Roche cat # 04823125001). Briefly, five shavings 10 μm thick from a paraffin block of colon biopsies were deparaffinized by washing the tissue with Xylene and ethanol (Antica et al., 2010). Tissue was then digested in sodium dodecyl sulfate and protease K and allowed to adhere to the RNA-binding column. The columns were incubated in DNase and washed with ethanol before RNA was eluted in 50 µl of water. The quantity of the purified RNA was assessed using the RNA Pico Lab Chip Kit with the Agilent Technologies Bioanalyzer and agarose gel electrophoresis. In addition, RNA was quantified spectrophotometrically and only samples with sufficient quantity (500ng total yield) and appropriate optical density (OD 260/280 ratio = 1.7–2.1) were used for subsequent analysis(Christophi et al., 2012b). Samples that did not meet the RNA quality and quantity requirements were excluded from the study. For cDNA synthesis, 0.5 µg of total RNA from each sample was used. Briefly, 25 µl RNA and random hexameric primers (Invitrogen, Carlsbad, CA) were incubated at 72°C for 10 minutes. Reverse transcription was performed using the Superscript II RT enzyme (Invitrogen, Carlsbad, CA) and followed the specifications of the manufacturer and were incubated for 30 min at 50°C followed by 70°C for 15 minutes. cDNA was diluted with sterile water into a volume of 150 µl (Christophi et al., 2009a).
Real time quantitative PCR
Primers were designed to amplify amplicons of 50–150 bp from specific NCBI reference sequences. Primers were designed to span exons and were blasted through NCBI GenBank to ensure lack of homology to other known human cDNA sequences. Quantitative real time PCR was performed with the SYBR Green kit (Abgene, Epson, UK) and using 2 µl of the cDNA mixture and 10 nM gene-specific DNA primers in a 10 µl reaction. The Roche LightCycler® 480 Real-Time PCR System with 384-well block was used for amplification and the PCR parameters were 15 minutes for 95°C, and 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Serial dilutions of cDNA containing a known copy number of each gene were used in each quantitative PCR run to generate a standard curve relating copy number with threshold amplification cycle (Christophi and Massa, 2010; Christophi et al., 2009a). Gene expression levels were calculated during the logarithmic amplification phase by determining the initial mRNA copy number using the standard curve. Amplification of each gene specific fragment was confirmed both by examination of peaks of melting curves and by agarose gel electrophoresis. To further control for equal amount of total RNA loading, parallel examination of the housekeeping genes glyceraldehyde phosphate 3-dehydrogenase (GAPDH), the ribosomal RNA 18S, and β-actin were quantified in each sample (Christophi et al., 2009a).
Statistical Analysis
Histograms contain statistical means with the standard error values. SigmaSTAT software was used to generate p values, utilizing the unpaired Student’s t-test and two-way analysis of variance (ANOVA) as tests to determine statistical significance. A p-value of less than 0.05 was chosen to indicate statistical significance between two sample means. The data are presented in histograms showing the average value with error bars representing standard error of the mean.
RESULTS
New techniques including more efficient RNA isolation and enzymes, cDNA conversion using random hexamers, and gene-specific primers generating short amplicons (50–200bp) have allowed successful quantification of gene expression using formalin-fixed paraffin embedded tissue. This approach has demonstrated clinical utility (Clark-Langone et al., 2010). Our study utilized several additional measures to validate the technique, including primers designed to span introns, primers that are non-homologous with other human cDNA sequences, and inclusion of negative and positive controls (cDNA vectors with verified DNA sequence of each gene) for each reaction (April et al., 2009; Green et al., 2009; Sanchez-Navarro et al., 2010). Furthermore, the generated PCR product was confirmed with examination of melting peaks and by agarose gel electrophoresis (Christophi et al., 2012b). Quantification of abundant internal control genes including β-actin (Figure 2), GAPDH, and S18 rRNA (data not shown) showed similar expression levels among the different samples, suggesting valid RNA quantification and equal input for RT-PCR. Additionally, we have previously documented that gene expression between fresh tissue and formalin-fixed tissue was similar using the same technique (Christophi et al., 2012b).
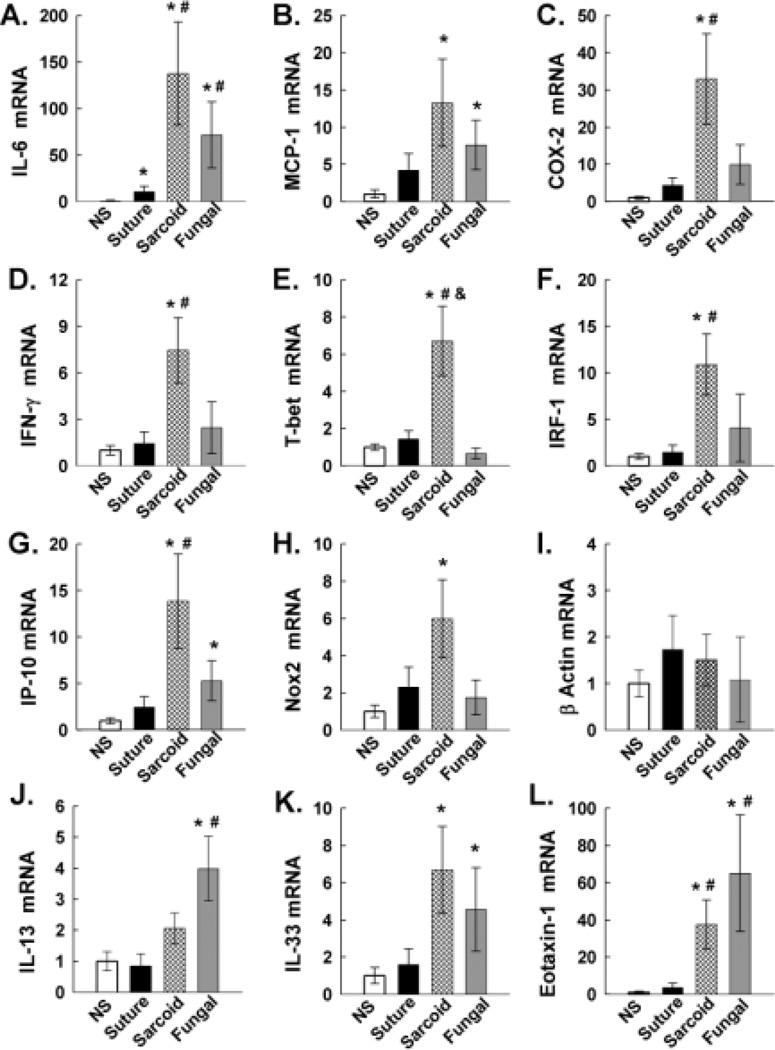
Figure 2. Gene expression analysis.
mRNA expression of gene targets, including IL-6 (A), MCP-1 (B), COX-2 (C), IFNγ (D), T-bet (E), IRF-1 (F), IP-10 (G), Nox2 (H), β-actin (I), IL-13 (J), IL-33 (K), and eotaxin 1 (L) were examined by quantitative RT-PCR from cDNA templates prepared from RNA extracted from normal samples (NS) of lung tissue, suture granuloma tissues, sarcoid granuloma tissues, and fungal granulomata. Histograms represent fluorescence data normalized to normal samples of lung tissue run in parallel on the same PCR plates. An asterisk (*), pound (#), or dollar ($) symbol represents p<0.05, representing results of two-way ANOVA measured on SigmaSTAT software. All asterisked comparisons show differences to the NS group. All pound symbols represent significant changes compared to the suture granuloma group. All dollar signs represent significant changes comparing sarcoid to fungal granulomas. Error bars represent SEM +/− 0.05.
Histology
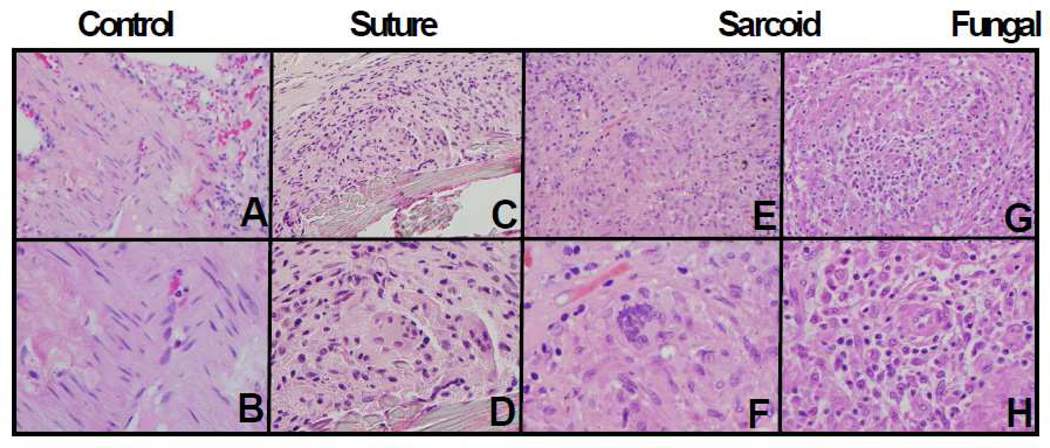
The morphology of sarcoid granulomas is very similar to infectious granulomas or foreign body reactions to suture material (Figure 1). The histopathologic features of excision, biopsy, or resection specimens were reviewed and recorded by examination of hematoxylin and eosin (H & E) stained sections of FFPE tissues. The samples were independently examined by two pathologists. To identify granulomas due to infection, staining for acid fast bacilli (AFB) and Grocott’s methenamine silver stain (GMS) were done on all samples. Granulomas of infectious etiology were diagnostic of or consistent with Histoplasmosis.
Figure 1. Histology suture, sarcoid, and infectious granulomata.
Light microscopic images of representative hematoxylin and eosin stained tissue sections are shown. Control lung parenchyma is shown in panels A and B, suture granuloma in panels C and D, sarcoid granuloma in panels E and F, and fungal granuloma in panels G and H. Multinucleated giant cells were seen in all granulomas. Images are shown at magnifications of 200× (top panels; A, C, E, and G) and 400 × (bottom panels; B, D, F, and H).
Gene expression
This study takes a selective approach to quantify cytokines, genes mediating signaling, or downstream effector molecules shown to be previously important in sarcoidosis or novel genes implicated in these signaling networks (Figure 2). Selected gene expression from FFPE biopsies of normal lung (n=12), suture granulomas (n=8), sarcoid granulomas (n=13), and fungal granulomas (n=5) was analyzed with real time RT-PCR (Figure 2). We partly selected genes that are regulated by the pro-inflammatory NF-κB pathway, mediate the type I immune response and STAT-1 signaling or type II immune response and STAT-6 signaling.
We analyzed the expression of the cytokine IL-6, the chemokine MCP-1, and the cyclooxygenase COX-2 as expression of these genes is driven by the transcription factor NF-kB (Figure 2A–C). IL-6 is a pro-inflammatory cytokine found to be elevated in BAL from sarcoidosis patients that activates the transcription factor STAT3 and is involved in cell proliferation and fibrosis (Takizawa et al., 1997). IL-6 was elevated 10-fold in suture granuloma compared to normal subject (NS) lung tissue. Sarcoid and fungal granulomas had 120-fold and 70-fold higher expression of IL-6 than NS, respectively. Also, IL-6 expression in sarcoid and fungal granulomas was significantly higher compared to suture granulomas. The monocyte chemoattractant protein MCP-1 mediates recruitment of monocytes to foci of active inflammation and stimulates bone marrow egress of monocytes (Deshmane et al., 2009). MCP-1 was significantly elevated in sarcoid and fungal granulomas compared to NS (Figure 2B). Cyclooxygenase-2 (COX-2), a prostanoid-synthesizing enzyme, is involved in macrophage activation, reactive oxygen species generation, and tissue inflammation (Hamada et al., 2008). COX-2 was significantly upregulated 30-fold in sarcoid compared to NS and suture granulomas (Figure 2C). COX-2 was 3-fold higher in sarcoid tissue compared to fungal granulomas.
In addition we examined the expression of the cytokine IFN-γ and the transcription factors T-bet and IRF-1, which are central in promoting a type I immune response (Figure 2D–F). IFN-γ is the Th1 hallmark cytokine and is induced by IL-12/STAT4, and in turn activates the transcription factor STAT-1 (Morinobu et al., 2002). IFN-γ was significantly and specifically unregulated 8-fold in sarcoid granulomas compared to NS and suture granulomas. IFN-γ was 3-fold higher in sarcoid compared to fungal granulomas, although did not reach significance. T-bet was shown to be highly expressed in sarcoid granulomas and promotes a type I immune response and IFN-γ expression (Kriegova et al., 2011). T-bet was uniquely and significantly elevated 7-fold in sarcoid granulomas compares to all other groups (Figure 2E). IRF-1 was also significantly elevated 10-fold in sarcoid compared to suture granuloma (Figure 2F).
Furthermore, we examined the expression of the chemokine IP-10 and the NADPH subunit Nox2 whose expression is primarily driven by the transcription factors STAT-1 and NF-kB (Semple and Moore, 2009). IP-10 is a chemokine that primarily chemoattracts activated Th1 T-cells to sites of inflammation and was previously shown to be elevated in in the serum and BAL of sarcoidosis patients (Dufour et al., 2002; Geyer et al., 2013; Miotto et al., 2001). IP-10 was significantly upregulated 14-fold in sarcoid compared to suture granulomas (Figure 2G). Nox2 acts to coordinate innate and adaptive immune defenses and generates reactive oxygen species to cause tissue injury (Rokutan et al., 2008; Sokolovska et al., 2013). Nox2 was 6-fold elevated in sarcoid compared to normal subjects but not significantly elevated compared to suture or fungal granulomas (Figure 4H).
In addition we examined the expression of the cytokine IL-13, IL-33, and the chemokine eotaxin I that are implicated in type II immune responses (Figure 2J–L). Interleukin-13 is a pro-inflammatory cytokine that promotes a type II immune and alternatively activated macrophages through STAT6 signaling (Christophi et al., 2009b). IL-13 was significantly and specifically elevated 4-fold in fungal granuloma compared to suture granulomas (Figure 2J). IL-33 is an IL-1-family pro-inflammatory cytokine that activates MAPK and NF-κB pathways in myeloid cells and has been implicated in allergic reactions and cell necrosis (Christophi et al., 2012a). IL-33 was significantly elevated 6-fold in sarcoid and fungal granulomas compared to normal subjects (Figure 2K). Eotaxin-1 is a potent chemoattractant for eosinophils, basophils, and macrophages and has been classically been associated with type II tissue inflammation and asthma (Miotto et al., 2001). Eotaxin-1 was significantly elevated 30-fold in sarcoid tissue and 60-fold in fungal granulomas compared to NS and suture granulomas (Figure 2L).
DISCUSSION
Through a focused approach this study quantified the expression of several cytokines, respective signaling molecules, and their downstream inflammatory gene expression. Utilizing improved RNA isolation techniques, cDNA conversion enzymes and random primers, as well as modified quantitative real time PCR, we quantified the expression of twenty genes from FFPE samples of sarcoid patients compared to patients with infectious granulomata or foreign body reactions. We demonstrate that several genes are differentially regulated in sarcoid in comparison to suture and infectious granulomas.
Sarcoidosis is predominantly mediated by a type I immune response. One important factor driving CD4+ T cell skewing is expression of T-bet, a transcription factor that promotes specification of CD4+ T cells to the Th1 lineage, in sarcoid patients. This increased expression was previously reported in mononuclear cells from BALF specimens (Kriegova et al., 2011). This study documents that T-bet mRNA is specifically elevated in FFPE tissue from sarcoid granulomas and would be an excellent diagnostic marker. Increased Th1 lineage commitment is also mediated by increased IFNγ expression mediating STAT1 phosphorylation and effector gene expression including the IRF-1 and IP-10 (Rosenbaum et al., 2009). In BAL of sarcoid patients there were previously documented higher levels of IFNγ and IP-10 (Hill et al., 2008). The chemokine IP-10 was previously shown to be elevated in serum of sarcoidosis patients compared to normal subjects and correlated with severity of stage I and II sarcoidosis. [15, 58]. Here we demonstrate a specific upregulation of IFNγ and IP-10 and for the first time upregulation of IRF -1 in sarcoidosis compared to normal subjects.
IL-6 is an important pro-inflammatory NF-kB- induced cytokine that causes fibroblast proliferation and has been previously elevated in the plasma of sarcoidosis patients compared to healthy controls (Balamugesh et al., 2006; Bihl et al., 2006). Interestingly, candidate triggers for granuloma formation in sarcoidosis Mycobacterium tuberculosis and fungal antigen P-glucan are potent stimulators of IL-6 by activating NF-kB (Ahmadzai et al., 2012; Terčelj et al., 2011). IL-6 expression correlates with disease severity and is reduced by steroid treatment (Mastorakos et al., 2013). We demonstrate that IL-6 mRNA is specifically elevated in granulomas of sarcoid patients and can also serve as a potential diagnostic factor.
In addition, we examined the expression of novel effector genes in sarcoidosis that can contribute to tissue damage through oxidative stress including COX2 and Nox-2. Nox2 an NADPH oxidase is unregulated in activated macrophages and mediates H2O2 production and bacterial clearance (Rada and Leto, 2008). COX2 is important in mediating prostaglandin production through fatty acid oxidation and radical reactions and potentially inhibiting T-regulatory cells(Akasaki et al., 2004)[49][51]. For the first time, we document that both COX2 and Nox2 are elevated in granulomas of sarcoid patients compared to suture granulomas.
In contrast, we also examined the expression of genes IL-13, IL-33, Eotaxin-1 shown to mediate type II immune responses and hypothesized that those genes would be specifically elevated in fungal granulomas. IL-13 was only significantly elevated in fungal granulomas. IL-33, an IL-1-family pro-inflammatory cytokine that activates the NF-κB pathway and effector genes and has been implicated in allergic reactions and cell necrosis, was both elevated in sarcoid and fungal granulomas. Eotaxin, an NF-kB and STAT6-induced chemokine that acts as a potent chemoattractant of macrophages, eosinophils, and basophils was also significantly elevated in sarcoid and fungal granulomas. NF-kB activation might explain elevation of IL-33 and eotaxin-1 in sarcoidosis.
This study demonstrates for the first time the potential of utilizing FFPE specimens to provide a feasible and practical approach to develop diagnostic markers in sarcoidosis. Improving the sensitivity and specificity to diagnose sarcoidosis is extremely important, as currently there is a delay in diagnosis with more than half of the patients requiring more than four physician visits to obtain a diagnosis (Newman et al., 2004). Using FFPE tissue is convenient; tissues do not require storage and results could be obtained retrospectively. Evolving molecular diagnostic tests could be simplified to examine specific genes including, IL-6, T-bet, IFN-γ, IP-10, IL-13, and β-actin in a single real time RT-PCR reaction. The specificity of diagnosing sarcoidosis can be improved by also combining histopathologic and molecular techniques. Importantly, examining excised tissue is an invasive approach to diagnose sarcoidosis and it is plausible that this inflammatory gene signature of sarcoidosis is also specific in fine-needle aspiration tissue within affected lymph nodes. This would provide an ideal approach for early and less invasive diagnosis of sarcoidoisis.
There are limitations within this study. The tissues were all compared to normal lung parenchyma from lobectomy specimens as a control, however, multiple tissue types are represented in the suture, sarcoid, and infectious granuloma groups. Additionally, it is possible that the mRNA levels do not reflect the functionality of each cytokine or protein represented. This is due to possible differences in mRNA stability, in translation, or post-translationally through various modifications. The study lacks direct clinical correlation of disease severity and preventing evaluation of gene expression in different disease stages. Furthermore, this study utilized microtome sections of excised tissue, limiting our ability to examine the cellular localization and expression of genes. Further confirmation at the protein or mRNA level could determine localization through immunohistochemistry, in situ hybridization, western blotting, or other proteomic methods.
Here, we identified that sarcoid granulomas have a unique inflammatory gene expression signature driven by STAT1 and NF-kB-dependent cytokine signaling, which aids in differentiation from infectious granulomas or foreign body reactions. Our work validates the upregulation of type I-induced genes in sarcoidosis granuloma previously identified in BALF specimens or plasma in sarcoid patients within the target tissue. Additionally we identified novel genes upregulated in sarcoidosis granulomas including Nox2, IL-13, and IL-33. Identifying the cause of non-caseating granulomas has long been a diagnostic challenge for pathologists, while quantitative RT-PCR of FFPE specimens represents a possible solution. Further controlled blinded studies examining gene expression from granulomas and correlation of gene expression with disease severity and treatment are needed to establish real time RT-PCR of FFPE as useful diagnostic tool in sarcoidosis.
Figure 3. Intracellular signaling networks upregulated in sarcoidosis.
Cytokines generated by innate and adaptive immune activation interact with transmembrane cytokine receptors, resulting in intracellular signal transduction cascades leading to alterations in target gene transcription. Foreign antigens (fungal, bacterial, amyloid) have pathogen-associated molecular patterns (PAMPs) which activate pattern recognition receptors (PRRs) to result in activation of STAT molecules (STAT6, STAT1, and STAT4) and NF-kB. STAT6 activation yields a Th2 cytokine response through upregulation of GATA3 which promotes fibroblast proliferation by IL-4 production, and a M2 macrophage phenotype through eotaxin production. NF-kB activation elicits transcription of multiple effector cytokines involved in inflammation and T cell activation, including eotaxin 1, MCP-1, IL-6, IL-33, and IL-2. Additionally, NF-kB activation increases oxidative stress within target cells through COX2 and NOX2 expression. STAT1 activation similarly contributes to oxidative stress in sarcoidosis through NOX2, and also promotes a Th1-type cytokine response through IRF-1 activation and subsequent production of IP-10. STAT4 similarly promotes a Th1 response through T-bet expression, which in turn generates IFNγ that feeds back to upregulate STAT1 and NF-kB-dependent signaling. This positive feedback loop promotes chronic inflammation in sarcoidosis.
TABLE 1. Specimen characteristics.
Specimens were divided into subgroups on the basis of the diagnosis, including normal lung tissue (n=12), suture foreign body reactions (n=8), sarcoid granulomas (n=13), and infectious granulomas (n=5). The age and gender are shown.
| Category | Sample | Age | Gender | Tissue |
|---|---|---|---|---|
| Normal | 1 | 78 | F | Lung |
| 2 | 76 | F | Lung | |
| 3 | 64 | F | Lung | |
| 4 | 49 | F | Lung | |
| 5 | 70 | M | Lung | |
| 6 | 72 | M | Lung | |
| 7 | 60 | M | Lung | |
| 8 | 76 | M | Lung | |
| 9 | 70 | F | Lung | |
| 10 | 60 | F | Lung | |
| 11 | 69 | M | Lung | |
| 12 | 61 | M | Lung | |
| Suture | 1 | 81 | M | Peritoneum |
| 2 | 40 | F | Thyroid | |
| 3 | 16 | M | Ureter | |
| 4 | 59 | M | Skin | |
| 5 | 45 | F | Soft tissue | |
| 6 | 12 | M | Soft tissue | |
| 7 | 80 | M | Skin | |
| 8 | 62 | F | Skin | |
| Sarcoid | 1 | 38 | M | Lymph node |
| 2 | 74 | M | Lymph node | |
| 3 | 41 | M | Lymph node | |
| 4 | 70 | F | Lymph node | |
| 5 | 28 | M | Lymph node | |
| 6 | 46 | M | Lymph node | |
| 7 | 35 | F | Liver | |
| 8 | 46 | F | Nasal sinus | |
| 9 | 22 | M | Skin | |
| 10 | 44 | F | Skin | |
| 11 | 36 | M | Lung | |
| 12 | 48 | F | Soft tissue | |
| 13 | 58 | F | Lymph node | |
| Fungal | 1 | 12 | F | Lung |
| 2 | 51 | F | Lung | |
| 3 | 31 | M | Liver | |
| 4 | 48 | M | Lung | |
| 5 | 58 | F | Lung | |
ACKNOWLEDGEMENTS
This work was supported in part by research grants from an NIH grant (NS041593) to Dr. Paul T. Massa and institutional grant from Washington University in St. Louis to Dr. George Christophi and institutional grant from Upstate Medical University to Dr Steve Landas.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors has any potential financial conflict of interest related to this manuscript and the results of this report have been generated, analyzed, and interpreted independently of any outside participation or influence.
REFERENCES
- Ahmadzai H, et al. Peripheral blood responses to specific antigens and CD28 in sarcoidosis. Respiratory Medicine. 2012;106:701–709. doi: 10.1016/j.rmed.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Akasaki Y, et al. Induction of a CD4+ T regulatory type 1 response by cyclooxygenase-2-overexpressing glioma. Journal of Immunology. 2004;173:4352–4359. doi: 10.4049/jimmunol.173.7.4352. [DOI] [PubMed] [Google Scholar]
- Anderson JM, et al. Foreign body reaction to biomaterials. Seminars in Immunology. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antica M, et al. Gene expression in formalin-fixed paraffin-embedded lymph nodes. J Immunol Methods. 2010;359:42–46. doi: 10.1016/j.jim.2010.05.010. [DOI] [PubMed] [Google Scholar]
- April C, et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS One. 2009;4:e8162. doi: 10.1371/journal.pone.0008162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamugesh T, et al. Inflammatory cytokine levels in induced sputum and bronchoalveolar lavage fluid in pulmonary sarcoidosis. The Indian journal of chest diseases & allied sciences. 2006;48:177–181. [PubMed] [Google Scholar]
- Bihl MP, et al. Progressive pulmonary sarcoidosis - A fibroproliferative process potentially triggered by EGR-1 and IL-6. Sarcoidosis Vasculitis and Diffuse Lung Diseases. 2006;23:38–50. [PubMed] [Google Scholar]
- Christophi GP, et al. Interleukin-33 upregulation in peripheral leukocytes and CNS of multiple sclerosis patients. Clinical Immunology. 2012a;142:308–319. doi: 10.1016/j.clim.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, Massa PT. Central neuroinvasion and demyelination by inflammatory macrophages after peripheral virus infection is controlled by SHP-1. Viral Immunol. 2010;22:371–387. doi: 10.1089/vim.2009.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, et al. Macrophages of multiple sclerosis patients display deficient SHP-1 expression and enhanced inflammatory phenotype. Lab Invest. 2009a;89:742–759. doi: 10.1038/labinvest.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, et al. Macrophages of multiple sclerosis patients display deficient SHP-1 expression and enhanced inflammatory phenotype. Laboratory Investigation. 2009b;89:742–759. doi: 10.1038/labinvest.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophi GP, et al. Immune markers and differential signaling networks in ulcerative colitis and Crohn's disease. Inflammatory Bowel Diseases. 2012b;18:2342–2356. doi: 10.1002/ibd.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Langone KM, et al. Translating tumor biology into personalized treatment planning: analytical performance characteristics of the Oncotype DX(R) Colon Cancer Assay. BMC Cancer. 2010;10:691. doi: 10.1186/1471-2407-10-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane SL, et al. Monocyte chemoattractant protein-1 (MCP-1): An overview. Journal of Interferon and Cytokine Research. 2009;29:313–325. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour JH, et al. IFN-γ-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. Journal of Immunology. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Geyer AI, et al. Plasma level of interferon γ induced protein 10 is a marker of sarcoidosis disease activity. Cytokine. 2013;64:152–157. doi: 10.1016/j.cyto.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Green TM, et al. Validation of putative reference genes for normalization of Q-RT-PCR data from paraffin-embedded lymphoid tissue. Diagn Mol Pathol. 2009;18:243–249. doi: 10.1097/PDM.0b013e3181a06f42. [DOI] [PubMed] [Google Scholar]
- Grunewald J. Genetics of sarcoidosis. Current Opinion in Pulmonary Medicine. 2008;14:434–439. doi: 10.1097/MCP.0b013e3283043de7. [DOI] [PubMed] [Google Scholar]
- Hamada T, et al. Cyclooxygenase-2 deficiency enhances Th2 immune responses and impairs neutrophil recruitment in hepatic ischemia/reperfusion injury. Journal of Immunology. 2008;180:1843–1853. doi: 10.4049/jimmunol.180.3.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, et al. Intracellular cytokine profiles and T cell activation in pulmonary sarcoidosis. Cytokine. 2008;42:289–292. doi: 10.1016/j.cyto.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Hofmann S, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nature Genetics. 2008;40:1103–1106. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- Hunninghake GW, Crystal RG. Pulmonary sarcoidosis. A disorder mediated by excess helper T-lymphocyte activity at sites of disease activity. New England Journal of Medicine. 1981;305:429–434. doi: 10.1056/NEJM198108203050804. [DOI] [PubMed] [Google Scholar]
- Iannuzzi MC, Fontana JR. Sarcoidosis: Clinical presentation, immunopathogenesis, and therapeutics. JAMA - Journal of the American Medical Association. 2011;305:391–399. doi: 10.1001/jama.2011.10. [DOI] [PubMed] [Google Scholar]
- Iannuzzi MC, et al. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- Ishige I, et al. Quantitative PCR of mycobacterial and propionibacterial DNA in lymph nodes of Japanese patients with sarcoidosis. Lancet. 1999;354:120–123. doi: 10.1016/S0140-6736(98)12310-3. [DOI] [PubMed] [Google Scholar]
- Kanazawa N, et al. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-κB activation: Common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- Kriegova E, et al. T-helper cell type-1 transcription factor T-bet is upregulated in pulmonary sarcoidosis. European Respiratory Journal. 2011;38:1136–1144. doi: 10.1183/09031936.00089910. [DOI] [PubMed] [Google Scholar]
- Mastorakos G, et al. Inappropriately normal plasma ACTH and cortisol concentrations in the face of increased circulating interleukin-6 concentration in exercise in patients with sarcoidosis. Stress. 2013;16:202–210. doi: 10.3109/10253890.2012.715221. [DOI] [PubMed] [Google Scholar]
- Miotto D, et al. Expression of IFN-γ-inducible protein; monocyte chemotactic proteins 1, 3, and 4; and eotaxin in T H1- and T H2-mediated lung diseases. Journal of Allergy and Clinical Immunology. 2001;107:664–670. doi: 10.1067/mai.2001.113524. [DOI] [PubMed] [Google Scholar]
- Morinobu A, et al. STAT4 serine phosphorylation is critical for IL-12-induced IFN-γ production but not for cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12281–12286. doi: 10.1073/pnas.182618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LS, et al. A case control etiologic study of sarcoidosis: Environmental and occupational risk factors. American Journal of Respiratory and Critical Care Medicine. 2004;170:1324–1330. doi: 10.1164/rccm.200402-249OC. [DOI] [PubMed] [Google Scholar]
- Pinkston P, et al. Spontaneous release of interleukin-2 by lung T lymphocytes in active pulmonary sarcoidosis. New England Journal of Medicine. 1983;308:793–800. doi: 10.1056/NEJM198304073081401. [DOI] [PubMed] [Google Scholar]
- Rada B, Leto T. Oxidative innate immune defenses by Nox/Duox Family NADPH oxidases. 2008;Vol. 15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokutan K, et al. Nox enzymes and oxidative stress in the immunopathology of the gastrointestinal tract. Semin Immunopathol. 2008;30:315–327. doi: 10.1007/s00281-008-0124-5. [DOI] [PubMed] [Google Scholar]
- Rosen Y. Pathology of sarcoidosis. Seminars in Respiratory and Critical Care Medicine. 2007;28:36–52. doi: 10.1055/s-2007-970332. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JT, et al. Hypothesis: Sarcoidosis is a STAT1-mediated disease. Clinical Immunology. 2009;132:174–183. doi: 10.1016/j.clim.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki BA, et al. Familial aggregation of sarcoidosis: A Case-Control Etiologic Study of Sarcoidosis (ACCESS) American Journal of Respiratory and Critical Care Medicine. 2001;164:2085–2091. doi: 10.1164/ajrccm.164.11.2106001. [DOI] [PubMed] [Google Scholar]
- Rybicki BA, et al. Racial differences in sarcoidosis incidence: A 5-year study in a health maintenance organization. American Journal of Epidemiology. 1997;145:234–241. doi: 10.1093/oxfordjournals.aje.a009096. [DOI] [PubMed] [Google Scholar]
- Saidha S, et al. Etiology of sarcoidosis: Does infection play a role? Yale Journal of Biology and Medicine. 2012;85:133–141. [PMC free article] [PubMed] [Google Scholar]
- Saltini C, et al. Spontaneous release of interleukin 2 by lung T lymphocytes in active pulmonary sarcoidosis is primarily from the Leu3+DR+ T cell subset. Journal of Clinical Investigation. 1986;77:1962–1970. doi: 10.1172/JCI112525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Navarro I, et al. Comparison of gene expression profiling by reverse transcription quantitative PCR between fresh frozen and formalin-fixed, paraffin-embedded breast cancer tissues. Biotechniques. 2010;48:389–397. doi: 10.2144/000113388. [DOI] [PubMed] [Google Scholar]
- Semple JL, Moore GWK. Ozone exposure and mortality [3] New England Journal of Medicine. 2009;360:2786–2787. doi: 10.1056/NEJMc090738. [DOI] [PubMed] [Google Scholar]
- Sokolovska A, et al. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nature Immunology. 2013;14:543–553. doi: 10.1038/ni.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, et al. Mycobacterial catalase-peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. Journal of Experimental Medicine. 2005;201:755–767. doi: 10.1084/jem.20040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H, et al. Increased IL-6 and IL-8 in bronchoalveolar lavage fluids (BALF) from patients with sarcoidosis: Correlation with the clinical parameters. Clinical and Experimental Immunology. 1997;107:175–181. doi: 10.1046/j.1365-2249.1997.d01-905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terčelj M, et al. Inflammatory markers and pulmonary granuloma infiltration in sarcoidosis. Respirology. 2014;19:225–230. doi: 10.1111/resp.12199. [DOI] [PubMed] [Google Scholar]
- Terčelj M, et al. In vitro and in vivo reactivity to fungal cell wall agents in sarcoidosis. Clinical and Experimental Immunology. 2011;166:87–93. doi: 10.1111/j.1365-2249.2011.04456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentonyte R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nature Genetics. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- Zaim MT, et al. Non-neoplastic inflammatory dermatoses. A clinical pathologic correlative approach. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 1990;3:381–414. [PubMed] [Google Scholar]
- Zhou T, et al. Peripheral Blood Gene Expression as a Novel Genomic Biomarker in Complicated Sarcoidosis. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0044818. [DOI] [PMC free article] [PubMed] [Google Scholar]