Abstract
The term autophagy refers to the engulfment and degradation of cytoplasmic components within the lysosome. This process can benefit cells and organisms by removing damaged, superfluous, or harmful cellular components, and by generating a supply of recycled macromolecules that can support biosynthesis or energy production. Recent interest in autophagy has been driven by its potential role in several disease-related phenomena including neurodegeneration, cancer, immunity and aging. Drosophila provides a valuable animal model context for these studies, and work in this system has also begun to identify novel developmental and physiological roles of autophagy. Here, we provide an overview of methods for monitoring autophagy in Drosophila, with a special emphasis on the larval fat body. These methods can be used to investigate whether observed vesicles are of autophagic origin, to determine a relative rate of autophagic degradation, and to identify specific step(s) in the autophagic process in which a given gene functions.
Keywords: Drosophila, autophagy, autophagosome, fat body, Atg8, Ref(2)P
1. Introduction
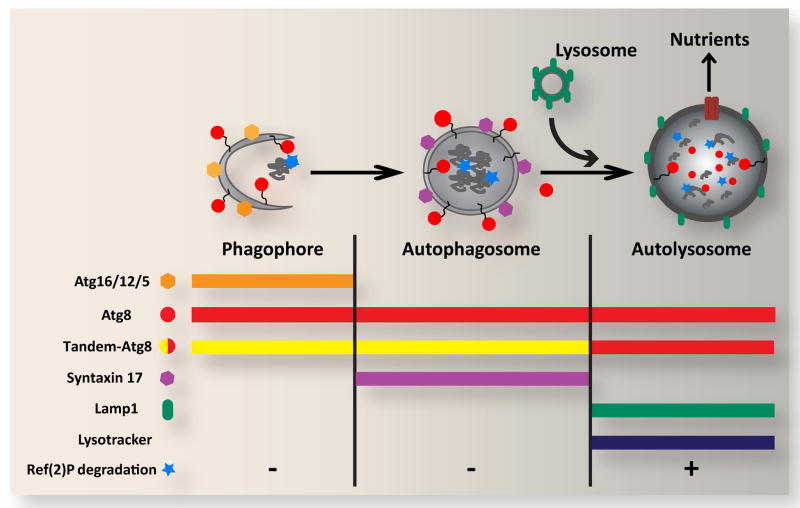
Although several routes of access into the lysosome have been described, the process of autophagy (specifically, macroautophagy) is unique in its ability to deliver nearly any cytoplasmic component to the lysosome, from whole organelles to large cytoplasmic aggregates. This versatility is made possible by the de novo production of engulfing vesicles known as autophagosomes, which are double membrane-bound organelles generated through the activity of a group of regulators collectively referred to as ATG proteins. Many of the tools and methods for studying autophagy are based on the selective activation or inactivation of these proteins, and on analysis of their intracellular localization (Fig. 1). As is often the case, most of these factors are conserved and represented in single copy in Drosophila, which serves as a useful intermediate model system between the complexity and redundancy of mammalian systems and the ease and simplicity of yeast [1,2].
Figure 1. Overview of autophagic structures, markers and assays.
In response to cellular stress and other inductive signals, cytoplasm, protein aggregates and organelles are engulfed within double-membraned autophagosomes, which form through growth of the cup-shaped phagophore/isolation membrane. Fusion of the outer autophagosomal membrane with endosomes (not shown) and lysosomes results in exposure of the sequestered material to the hydrolytic lysosomal compartment. Degraded materials are ultimately exported to the cytoplasm as nutrients for energy and biosynthesis. Each of these steps can be identified and distinguished based on their specific constellation of associated proteins, as well as by pH and degradative properties. Atg5, Atg12 and Atg16 form a complex on the outer surface of the phagophore, and are removed upon its closure to form the autophagosome. Syntaxin 17 is recruited to the mature autophagosome, and is released upon fusion with the lysosome. In contrast, Atg8 is associated with the entire autophagic pathway from phagophore to autolysosome. Autophagosome-lysosome fusion can be monitored through loss of GFP fluorescence from the tandem GFP-mCherry-Atg8 marker. General lysosomal markers such as Lamp1 and LysoTracker can also be used to identify later autophagic vesicles. The degradative capacity of autophagy can be monitored through the cytoplasmic levels or specific cleavage of the autophagy substrate Ref(2)P.
Autophagy proceeds constitutively at a low basal rate in most cells, and can be induced to high levels under conditions of nutrient limitation, organelle damage, infection and other cellular stresses [3,4]. These responses are particularly apparent and well studied in the Drosophila larval fat body, which uses autophagy to mobilize nutrients throughout the animal in response to starvation. Many of the tools and assays we describe here have been developed and most extensively tested in fat body cells, but are likely to be useful under a wide array of conditions. For a comprehensive overview of the use and interpretation of autophagy assays across multiple experimental systems, please see Klionsky et al. [5].
2. Steady-state markers and methods
The characteristic double membrane morphology and unique protein and lipid composition of autophagosomes makes these vesicles highly suitable as specific markers of autophagy, and methods based on their identification have served as autophagy assays for several decades. It is important to note that changes in the presence, number or size of autophagic vesicles provides little information as to the rate of autophagic degradation: for example, a cell may contain a large number of autophagosomes due to either a high rate of production or a defect in turnover. Nonetheless, such static measurements can play a crucial role in determining whether observed structures are of autophagic origin.
2.1 Electron microscopy
Transmission electron microscopy (TEM) has long been considered the gold standard for identification of autophagic vesicles, and still provides the highest resolution view of autophagosome structure and cargo. Standard TEM fixation and staining methods are appropriate for the study of autophagy by electron microscopy, although the high content of unsaturated fatty acids in autophagic vesicle membranes allows them to be preferentially stained through protocols using imidazole-buffered osmium tetroxide impregnation [6]. Autophagic vesicles are identified in electron micrographs as intact cytoplasmic components enclosed within a double membrane (autophagosome) or partially degraded cellular material bound by a single membrane (autolysosome). It is important to recognize and avoid potentially confusing similar structures such as swollen mitochondria, circular traces of endoplasmic reticulum, and multivesicular bodies [7]. Quantification of the cellular volume occupied by autophagosomes and autolysosomes and/or the number of such compartments can provide a snapshot of the autophagic capacity of a cell at a given time point. Methods for unbiased sampling, volume estimation and point counting have been described [8] and are generally applicable for studies of Drosophila samples. Analysis of autophagy using TEM-based methods has been used extensively in Drosophila to characterize responses to viral and bacterial infection, starvation, amyloid beta expression, ecdysone signaling and developmental cell death [9–14].
2.2 Atg8
The identification of proteins that localize specifically to the surface of autophagic vesicles has provided the means to monitor autophagy through fluorescence microscopy and biochemical approaches. The most widely used such protein is Atg8 (homologous to MAP-LC3, GABARAP and GATE16 proteins in mammals), an ubiquitin-like protein that becomes conjugated to lipids of the inner and outer membranes of the autophagosome as it forms [15]. Upon fusion of the autophagosome with the lysosome, the population of Atg8 associated with the inner autophagosomal membrane is internalized and eventually degraded within the autolysosome, whereas Atg8 bound to the outer membrane is de-lipidated and released into the cytosol. Thus, Atg8 can potentially mark all stages of the autophagy pathway (Fig. 1).
The Atg8 family is represented by two members in Drosophila, Atg8a and Atg8b. Both proteins (as well as human MAP-LC3) have been localized to autophagic vesicles in a variety of Drosophila cells and tissue types, and Atg8a has been shown to be required genetically for autophagosome formation [16]. Transgenic lines expressing Atg8 fused with GFP or RFP/mCherry at its amino terminus have been used extensively to monitor autophagy. Several such lines have been described, driven by UAS, Hsp70, fat body-specific and endogenous Atg8a promoters [16–19]. In addition, autophagic vesicles can be visualized using antibodies that recognize endogenous Atg8a [20, 21]. In each of these cases, the occurrence of autophagy in a cell can be inferred by the presence of cytoplasmic Atg8 punctae (spots) visualized by fluorescence microscopy. A number of programs such as ImageJ are available to help count punctae in an automated and objective manner. As autophagy and several of its regulators can influence cell size, it is important to control for this variable when quantifying autophagic punctae on a per cell basis.
Due in part to the distinct properties of different compartments along the autophagic pathway, the localization patterns of GFP-Atg8, mCherry-Atg8 and endogenous Atg8 can differ in important ways. For example, because the fluorescence of GFP is rapidly quenched in the acidic environment of the autolysosome, GFP-Atg8 labels the autophagosome only prior to its fusion with the lysosome to form the autolysosome (see section 3.1). In contrast, mCherry fluorescence is relatively pH independent, and thus will label autophagic vesicles both before and after this fusion event. Indeed, cells undergoing autophagy will usually display considerably more punctae marked with mCherry-Atg8 (autophagosomes and autolysosomes) than with GFP-Atg8 (autophagosomes only). mCherry also appears to be more resistant to lysosomal proteases than Atg8 itself, and as a result mCherry-Atg8 can be expected to display a broader pattern of distribution than the endogenous protein. In addition, the appearance of Atg8- labeled vesicles can differ depending on the duration and nature of the inducing stimuli, generally becoming larger, rounder and more uniform over time (Fig. 2A)
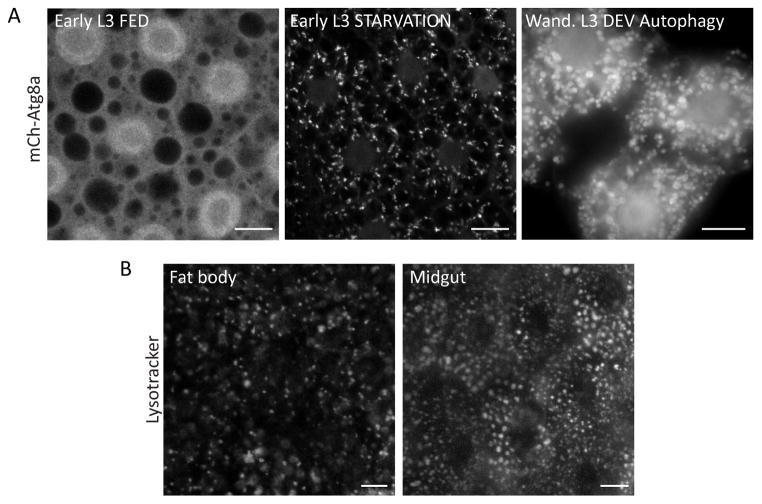
Figure 2. Static markers of autophagic vesicles.
A. Localization of Atg8a in the larval fat body. During the early L3 instar, autophagy proceeds at a low basal rate in well-fed animals, and mCherry-Atg8a is distributed evenly throughout the cell, with accumulation in the nucleus and exclusion from lipid droplets evident. In response to starvation, Atg8a is recruited to autophagic vesicles, forming discrete punctae of varying size and shape. In wandering stage L3 larvae, Atg8a is constitutively associated with autophagic vesicles, which are larger and rounder than the structures observed at earlier developmental stages. Scale bars, 10μm.
B. The acidophilic dye LysoTracker can be used to visualize the growth and activation of the lysosomal compartment in response to autophagy induction. Fat body and midgut cells display a distinct distribution pattern and appearance of autolysosomes, which in these images were induced in response to the oxidative stressors hydrogen peroxide and paraquat, respectively. Scale bars, 10μm.
It is important to be aware of the potential for artifacts when using these markers. In some instances Atg8 proteins can form aggregates that appear similar to autophagic vesicles, particularly when expressed at high levels or in the presence of other aggregate-prone proteins [22, 23]. Thus it is recommended to confirm that the formation of Atg8 punctae in response to a given stimulus is dependent on the function of other genes required for autophagy, and it is important to validate such findings using additional independent assays. A second consideration is whether fluorescent Atg8 proteins act strictly as neutral markers or whether they have an impact on the rate of autophagy, as overexpression of some ATG genes has been shown to either block or stimulate autophagy [16, 24]. Although such effects are not readily apparent in cells expressing Atg8 markers, it has been reported that neuronal overexpression of Atg8a can have a beneficial effect on lifespan of adult Drosophila, consistent with an enhanced rate of autophagy [25].
The processing (lipid conjugation) of Atg8 proteins that drives their association with the autophagosome can also be visualized by immunoblotting. The processed protein (Atg8-II) migrates at a faster rate on polyacrylamide gels than its unprocessed form (Atg8-I), and can accumulate to high levels in cells with high rates of autophagy. LC3 immunoblotting is a widely used autophagy assay in mammalian cells, and similar methods to detect processed Atg8a have recently been described in Drosophila [20, 21]. In general, the relative level of processed Atg8 measured biochemically correlates well with the number of Atg8 spots visualized by microscopy.
2.3 Other autophagy markers
A number of other proteins can be used to mark the autophagosome at different stages of its development. Several autophagy regulatory proteins have been shown to localize to sites of nascent autophagosome formation, prior to the recruitment of Atg8 [26, 27]. In Drosophila these include components of the Atg1 protein kinase complex [24, 28]. Another protein, Atg5, assembles into a complex with Atg12 and Atg16 at the outer surface of immature autophagosomes (referred to as isolation membranes or phagophores). This complex subsequently disassembles upon closure and completion of the mature autophagosome. Both Atg5 antibodies and tagged Atg5 transgenic and protein trap lines have been used to visualize these early structures in Drosophila, and these markers overlap with only a subset of Atg8a-positive vesicles, as expected [13, 20, 21, 29]. Finally, the Qa-SNARE Syx17 was recently shown to be recruited to mature (Atg5-negative) autophagosomes in larval fat body cells, where it promotes autophagosome-lysosome fusion [21]. Syx17 immunostaining showed a partial colocalization with Atg8-positive autophagosomes but not with Atg5 nor the lysosomal marker LAMP, consistent with Syx17 labeling mature autophagosomes prior to their fusion with lysosomes. Together, the suite of reagents available in the fly are sufficient to visualize and discriminate multiple compartments along the autophagy pathway, including sites of autophagosome induction, expansion, maturation, and fusion (Fig. 1). Such resolution can be essential in elucidating the nature of specific autophagy defects.
2.4 Lysosomal assays
Autophagy produces a net flow of membrane and cytoplasmic material to the lysosome, which can result in a temporary expansion of this compartment. Changes in lysosomal size, number and activity have thus been exploited as convenient assays to monitor autophagy. In some tissues such effects can be quite dramatic. In the fat body of well fed early third instar larvae, lysosomes are small and stain poorly with pH-dependent probes such as LysoTracker and Neutral Red. Upregulation of autophagy by starvation or other stimuli leads to a marked enlargement of lysosomes in these cells within a few hours [11]. In addition to this effect on lysosomal size, autophagic induction also increases lysosomal acidity and activity in an ATG gene-dependent manner, presumably through activation of lysosomal vATPases [30]. Thus, induction of autophagy in the fat body causes an abrupt switch from essentially negative to strongly positive punctate staining with LysoTracker and other activity-dependent markers (Fig. 2B; for detailed protocols please refer to references [31] and [32]. This assay has also been successfully applied to flow cytometric analysis of autophagy in a larval hemocyte cell line [33]. It should be noted that lysosomal size, abundance and activity are not specific indicators of autophagy, and these markers can also reflect rates of endocytosis and other routes to the lysosome, as well as changes in lysosomal turnover and reformation. Furthermore, in cell types with a high basal rate of lysosomal activity, effects of autophagy on activation of the lysosomal compartment are less noticeable. Nonetheless, such measurements of the lysosome can provide a useful assay that strongly complements methods based on autophagosome formation. Other lysosomal markers including GFP- and HRP-tagged lysosome-associated membrane protein (LAMP) transgenic lines, antibodies against lysosomal hydrolases, and fluorescent cathepsin substrates have been used to monitor the size and activity of autolysosomes [17, 29, 34].
3. Dynamic measurements of autophagic flux
The markers of autophagosomes and autolysosomes described above can provide a valuable snapshot of autophagic compartments within a cell, but are limited in their ability to measure rates of autophagic activity, often referred to as autophagic flux. In particular, these markers can behave similarly in response to either induction or inhibition of autophagy. One approach to this problem commonly used in cultured mammalian cells is to compare markers such GFP-LC3 or LC3-II in the presence vs. absence of lysosomal inhibitors such as bafilomycin A1, chloroquine or pepstatin A [5]. The rationale is that an experimental stimulus that increases LC3 by inducing autophagy should have an enhanced effect when degradation is disrupted by these inhibitors, whereas an accumulation of LC3 caused by a block in the pathway should not. However, the use of lysosomal inhibitors is poorly compatible with studies involving intact larvae or adult flies. Although prolonged feeding of chloroquine (24 h) has been shown to inhibit lysosomal acidity in the larval fat body [35, 36], incubations of this time scale can cause secondary effects resulting from the accumulation of undigested lysosomal cargo [5]. Therefore additional assays of autophagic flux have been developed or adapted for use in Drosophila.
3.1 Tandem tagged (GFP-mCherry) ATG8
One such assay is based on the tandem fusion of GFP and mCherry to Atg8, first described for use in transfected HeLa and MEF cells [37] and subsequently in Drosophila by Stenmark and colleagues [29]. Due to the differential pH sensitivity of GFP and mCherry noted above, the GFP-mCherry-Atg8 marker co-labels non-acidified autophagic compartments (phagophore and autophagosome) with both GFP and mCherry signals, whereas following fusion with acidic late endosomes or lysosomes the GFP fluorescence is lost and only the mCherry signal remains intact (Fig. 1 and Fig. 3A). Most cells undergoing autophagy will display a combination of both green+red autophagosomes and red-only autolysosomes, the latter of which tend to predominate at later timepoints. Juhasz and colleagues have used chloroquine feeding to show that the loss of GFP fluorescence duly reflects the low pH of the lysosome [35, 36]. This marker thus provides evidence of both biogenesis of autophagosomes and their successful fusion with a properly acidified lysosomal compartment. Alternatively, autophagosome-lysosome fusion can also be assayed through co-localization of Atg8 with endo/lysosomal markers such as LAMP1, LysoTracker or endocytic tracers (Fig. 3B; [17, 39].
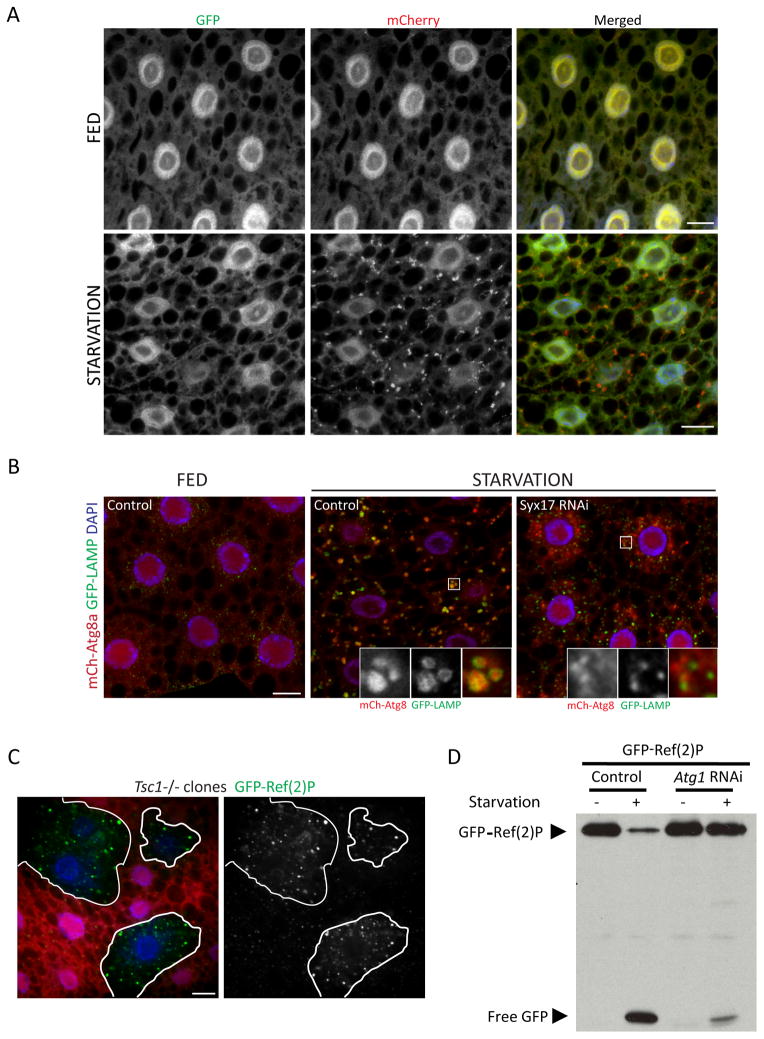
Figure 3. Assays of autophagic flux in the larval fat body.
A. Tandem-tagged GFP-mCherry-Atg8a can reveal fusion of autophagosomes with lysosomes. Under fed conditions, the green and red fluorescence of this marker display a nearly identical diffuse pattern. Upon autophagy induction by starvation, localization of this marker to punctate autophagic vesicles is evident in the red channel but not in the green channel, due to quenching of GFP fluorescence within the acidic autolysosome. Scale bars, 10μm.
B. Co-localization of Atg8a with the lysosomal marker GFP-LAMP can also be used to assess autophagosome-lysosome fusion. In these images, induction of autophagy by starvation results in accumulation of mCherry-Atg8a within vesicles marked with GFP-LAMP. Co-localization of these markers is inhibited by depletion of Syntaxin 17, a SNARE required for autophagosome-lysosome fusion. Scale bar, 10μm.
C. Accumulation of the autophagy substrate Ref(2)P can reveal a disruption of autophagy. The level of GFP-Ref(2)P within clones of Tsc1 mutant cells (marked by lack of dsRed marker; outlined in white) is increased relative to surrounding control cells, reflecting autophagy suppression in response to activated TOR signaling. Scale bar, 10μm.
D. Autophagy-dependent proteolytic cleavage of GFP-Ref(2)P, assayed by anti-GFP immunoblotting of fat body extracts. Starvation-induced autophagy results in a loss of full-length GFP-Ref(2)P and a concomitant accumulation of free GFP. Both responses are inhibited in cells depleted of Atg1.
3.2 Ref(2)P and ubiquitinated aggregates
A number of assays have been described that provide a direct indication autophagic degradation. Ref(2)P is the fly homolog of p62/sequestosome 1, an adapter protein that recruits ubiquitinated substrates to autophagosomes by virtue of its ubiquitin associated domain and LC3-interacting region. Direct interaction of p62 with Atg8 on the autophagosomal membrane results in selective uptake of p62 into the autophagosome and its eventual degradation in the autolysosome (Fig. 1). In many cell types, disruption of ATG genes or inhibition of lysosomal function leads to a significant increase in cytoplasmic levels of p62 [39, 40]. In Drosophila, autophagy-dependent clearance of Ref(2)P has been demonstrated using antibodies against the endogenous protein as well as GFP-tagged Ref(2)P transgenic lines (Fig. 3C; [24, 38]). In this case, the transgenic marker has the advantage of avoiding potential transcriptional effects on endogenous Ref(2)P, which can be upregulated in response to some autophagy-inducing stresses such as heat shock [41]. Both endogenous and GFP-Ref(2)P are prone to forming cytoplasmic aggregates, and Juhász and coworkers have shown that measurement of overall GFP-Ref(2)P protein levels provides a more reliable indication of autophagy-dependent clearance than do the number or size of GFP-Ref(2) aggregates [23].
Autophagy plays a particularly important role in promoting the health of neuronal cells, which accumulate ubiquitinated protein aggregates when autophagy is impaired. Finley and colleagues demonstrated that the level of Triton X-insoluble ubiquitinated proteins (IUP) in the fly brain increases as a function of age, and this correlates with decreased expression of a number of ATG genes [25]. Disruption or overexpression of Atg8a was shown to increase or decrease the accumulation of IUP, respectively. Thus, brain IUP levels can serve as the basis of a convenient autophagy assay, although it is important to control for potential changes in IUP formation as well as other routes of turnover. Measurement of IUP may be especially useful when studying the effects of aging or other long-term processes.
3.3 GFP liberation assays
An additional assay of autophagic degradation monitors the autophagy-dependent cleavage of GFP-Atg8 or GFP-Ref(2)P. GFP is relatively resistant to lysosomal proteolysis, presumably due to its globular, compact structure. As a result, liberation of free GFP (or mRFP/mCherry) from these fusion proteins following their autophagy-dependent delivery into the lysosome can be visualized by immunoblot analysis (Fig. 3D). This assay was developed in yeast and mammalian cell culture systems, and has recently been used to monitor autophagy in Drosophila [23]. Although formation of free GFP in these assays has been shown to be autophagy-dependent, it is important to note that the level of free GFP, both overall and relative to the full length fusion, will vary with time, and is dependent on its rate of delivery, cleavage and ultimate degradation. Therefore a timecourse analysis is recommended.
4. Perspectives and conclusions
One of the challenges in using genetic model systems such as Drosophila for studies of autophagy is the difference in timescales between the rapid dynamics of autophagy compared to the slower and more chronic effects of gene inactivation using traditional mutational tools. This difference increases the likelihood that assays such as those described above will reflect both direct and indirect effects of genetic activity. In some cases this problem is compounded by potential pleiotropic effects of a mutation, as many genes with direct and important roles in autophagy have additional functions in other processes [42, 43]. The use of mosaic approaches to activate or inactivate genes in small groups of cells can provide a partial solution to the issue of indirect effects (Fig. 3C). Nonetheless, there is a strong need for improved methods of rapid protein depletion or inactivation. An increased focus on generating temperature-sensitive or chemical-genetic reagents could be helpful in this regard. Further development of live-imaging techniques suitable for cultured Drosophila tissues will also be critical to understand the dynamics of autophagosome generation, movement and fusion in this system.
As many of the most commonly used autophagy assays are based on the processing, localization or behavior of Atg8, it is important to note that alternative mechanisms of autophagy regulation have been described that are independent of Atg8 or other ATG genes that act upstream [24, 44–46]. As a result, reliance on Atg8-centered assays is likely to underestimate the occurrence of autophagy and its contributions to a given process. It is therefore advisable to monitor autophagy using multiple independent methods whenever possible. In addition, development of additional methods that can accurately and directly measure autophagic flux through pulse-chase approaches will be of value. The improvement of such tools and experimental approaches will serve to further increase the usefulness of Drosophila to studies of autophagy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meléndez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zirin J, Perrimon N. Drosophila as a model system to study autophagy. Semin Immunopathol. 2010;32:363–372. doi: 10.1007/s00281-010-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reunanen H, Punnonen EL, Hirsimaki P. Studies on vinblastine-induced autophagocytosis in mouse liver. V. A cytochemical study on the origin of membranes. Histochemistry. 1985;83:513–517. doi: 10.1007/BF00492453. [DOI] [PubMed] [Google Scholar]
- 7.Eskelinen EL. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy. 2008;4:257–260. doi: 10.4161/auto.5179. [DOI] [PubMed] [Google Scholar]
- 8.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009;452:143–164. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 9.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Kurata S. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nature immunology. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Ling D, Song HJ, Garza D, Neufeld TP, Salvaterra PM. Abeta42-induced neurodegeneration via an age-dependent autophagic-lysosomal injury in Drosophila. PLoS One. 2009;4:e4201. doi: 10.1371/journal.pone.0004201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 15.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Stenmark H. ESCRTs and Fab1 Regulate Distinct Steps of Autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Arsham AM, Neufeld TP. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS ONE. 2009;4:e6068. doi: 10.1371/journal.pone.0006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barth JM, Szabad J, Hafen E, Kohler K. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 2011;18:915–924. doi: 10.1038/cdd.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K, Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 23.Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One. 2012;7:e44214. doi: 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 26.Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, Ktistakis NT. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci. 2013;126:5224–5238. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- 27.Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9:1491–1499. doi: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- 28.Nagy P, Kárpáti M, Varga Á, Pircs K, Venkei Z, Takáts S, Juhász G. FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014;10:453–457. doi: 10.4161/auto.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjorkoy G, Johansen T, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Shen HM. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Research. 2013;23:508–523. doi: 10.1038/cr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juhasz G, Neufeld TP. Experimental control and characterization of autophagy in Drosophila. Methods Mol Biol. 2008;445:125–133. doi: 10.1007/978-1-59745-157-4_8. [DOI] [PubMed] [Google Scholar]
- 32.Neufeld TP. Genetic manipulation and monitoring of autophagy in Drosophila. Methods Enzymol. 2008;451:653–667. doi: 10.1016/S0076-6879(08)03236-9. [DOI] [PubMed] [Google Scholar]
- 33.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juhász G, Hill JH, Yang Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Low P, Varga A, Pircs K, Nagy P, Szatmari Z, Sass M, Juhasz G. Impaired proteasomal degradation enhances autophagy via hypoxia signaling in Drosophila. BMC Cell Biol. 2013;14:29. doi: 10.1186/1471-2121-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy P, Varga A, Pircs K, Hegedus K, Juhasz G. Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet. 2013;9:e1003664. doi: 10.1371/journal.pgen.1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 38.Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE, Stenmark H. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008;4:500–506. doi: 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- 39.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leemans R, Egger B, Loop T, Kammermeier L, He H, Hartmann B, Reichert H. Quantitative transcript imaging in normal and heat-shocked Drosophila embryos by using high-density oligonucleotide arrays. Proc Natl Acad Sci U S A. 2000;97:12138–12143. doi: 10.1073/pnas.210066997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toda H, Mochizuki H, Flores R, 3rd, Josowitz R, Krasieva TB, Lamorte VJ, Tomoda T. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292–3307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140:1321–1329. doi: 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1- independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 45.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 46.Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, Harper JW, Baehrecke EH. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013;15:1067–1078. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]