Abstract
Hepatocellular carcinoma (HCC) is the third leading fatal cancer worldwide and its incidence continues to increase. Chronic viral hepatitis involving either hepatitis B virus (HBV) or hepatitis C virus (HCV) infection is the leading etiology for HCC, making HCC prevention a major goal of antiviral therapy. While recent clinical observations and translational research have enhanced our understanding of the molecular mechanisms driving the initiation and progression of HCC, much remains unknown. Current data indicates that HCC tumors are highly complex and heterogeneous resulting from the aberrant function of multiple molecular pathways. This complex biology is responsible, at least in part, for the absence of highly efficient target-directed therapies for this deadly cancer. Additionally, the direct or indirect effect of HBV and HCV infection on the development of HCC is still a contentious issue. Thus, the question remains whether viral hepatitis-associated HCC stems from virus-specific factors, and/or from a general mechanism involving inflammation and tissue regeneration. In this review we summarize general mechanisms implicated in HCC, emphasizing data generated by new technologies available today. We also highlight specific pathways by which HBV and HCV could be involved in HCC pathogenesis. However, improvements to current in vitro and in vivo systems for both viruses will be needed to rigorously define the temporal sequence and specific pathway dysregulations that drive the strong clinical link between chronic hepatitis virus infection and HCC.
Keywords: Hepatocellular carcinoma (HCC), viral hepatitis, hepatitis B virus (HBV), hepatitis C virus (HCV), oncogenesis
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common human cancers, being the fifth most prevalent tumor type and the third leading cause of cancer-related deaths worldwide [1, 2]. Chronic infection with hepatitis B virus (HBV) and hepatitis C virus (HCV), especially in the setting of established cirrhosis or advanced fibrosis, are the leading causes of HCC worldwide, and the incidence of HCC correlates with the geographic distribution of these infections [3]. Conversely, suppression of HBV replication [4, 5] and a sustained viral response (SVR) in the treatment of HCV [6], are associated with a reduction in HCC among treated populations. Men are more susceptible to HCC, most probably because the major risk factors for HCC, including chronic viral hepatitis as well as alcohol consumption and non-alcoholic fatty liver disease (NAFLD) [7, 8] are more common in males. However, there is substantial evidence for gender-related differences in cellular pathways associated with HCC that might make men more susceptible to HCC than women [9, 10]. Early diagnosis is crucial for potentially successful curative treatment. Therefore, continuous HCC surveillance is recommended for high-risk patients, including those with cirrhosis of any etiology, chronic HCV patients with advanced liver disease and chronic HBV patients even without cirrhosis [11]. Unfortunately, despite efforts aimed at early detection of HCC, a substantial number of patients are diagnosed only when their disease is at an advanced stage. These patients are usually not amenable for potentially curative treatment modalities, such as partial liver resection, liver transplantation or radiofrequency ablation (RFA) [11], but are rather offered non-curative alternatives such as transarterial chemoembolization (TACE) and/or drug therapy with Sorafenib [12]. Although associated with only a modest increase in patient survival [12], the multi-kinase inhibitor Sorafenib is the first target-directed drug therapy for liver cancer, and was developed based on understanding major intracellular and extracellular signaling pathways involved in HCC pathogenesis [13]. However, despite the progress in elucidating the general mechanism(s) of HCC as well as advances in the study of HBV and HCV virology, there is still a substantial gap in our ability to mechanistically link chronic viral hepatitis with liver carcinogenesis.
2. Pathways in HCC implicated by genomic studies
Recent advances in genetic technologies, particularly decreasing costs of sequencing have led to many studies interrogating genetic alterations in HCCs. The main pathways involved in HCC initiation or progression can be divided into several categories including cell proliferation and differentiation, inflammation, and angiogenesis, the latter being a central theme associated with this highly vascularized tumor (reviewed in [14]).
Perturbations in the epidermal growth factor (EGF) signaling pathway exemplify how cell proliferation pathways can contribute to HCC. Several studies have shown that certain polymorphisms in the EGF gene can either promote or protect from HCC [15–17], and blocking the EGF receptor has been shown to inhibit the growth of HCC cells both in vitro and in vivo [18, 19]. The ubiquitous activation of Ras and Jak/STAT signaling in HCC provide additional examples of pathways involved in cell proliferation and differentiation, that when altered, affect HCC pathogenesis. Activation of these pathways is associated with DNA hyper methylation and hence silencing of their cellular inhibitors, especially in the setting of cirrhosis [20]. The PI3K-Akt pathway, intimately involved in both cell proliferation and cellular metabolism, is also activated in a significant portion of HCC samples [21]. This pathway might link metabolic alterations in the liver, such as fat accumulation to the development of liver cancer [22]. Recently, genomic analyses of HCC samples from patients with aggressive tumors and poor prognosis revealed over-expression of the fetal oncoprotein SALL4. SALL4 normally co-represses the tumor suppressor PTEN, resulting in activation of the PI3K-Akt pathway. Importantly, blocking SALL4 function with a synthetic peptide has been shown to release PTEN from co-repression, resulting in de-phosphorylation and reduced activation of Akt, accompanied by a significant shrinkage of tumors in vivo [23]. The wnt/β-catenin signaling pathway, important in cell differentiation and proliferation, is also frequently mutated in the tumorous tissue of HCC patients [21]. Unlike in colon cancer where mutations are frequently found in the tumor suppressor gene adenomatous polyposis coli (APC) resulting in β-catenin activation, they are rarely found in HCC tumor tissue. In contrast, other mechanisms for β-catenin activation, such as promoter over-activation [24] or mutations in AXIN1 [25] are found.
Inflammation is the hallmark of chronic hepatitis of various etiologies and is thought to be a major trigger for liver carcinogenesis. NF-κB, a major player in the cellular inflammatory cascade, promotes liver cancer in the Mdr2 knockout mouse model, suggesting a link between inflammation and cancer [26].
Angiogenesis also plays a key role in HCC development and invasive potential. A major pro-angiogenesis factor, VEGF, is elevated in the sera of HCC patients, and its serum level, as well as certain VEGF polymorphisms, appear to correlate with prognosis [27, 28]. In addition, metastatic tumor antigen 1 (MTA1), a stabilizer of the angiogenesis mediator hypoxia-inducible factor-1 (HIF-1), has been found to closely correlate with post-operative recurrence of HCC and poor survival rates, especially among HBV positive HCC patients [29]. Therefore, the current evidence strongly implicates angiogenesis in HCC pathogenesis, providing clear a rationale for targeting VEGF pathways in anti-HCC therapy.
3. Hepatitis B virus is a risk factor for HCC
3.1 Epidemiology and molecular biology of HBV infection
HBV is a small DNA virus, a member of the hepadnaviridae family (reviewed in [30–32]). Transmission can occur by exposure to contaminated blood products, or alternatively by sexual or other modes of intimate contact [32]. In adults, acute infection usually resolves spontaneously, however, in newborns or small children, chronic infection is common and often leads to chronic hepatitis, cirrhosis and HCC [31].
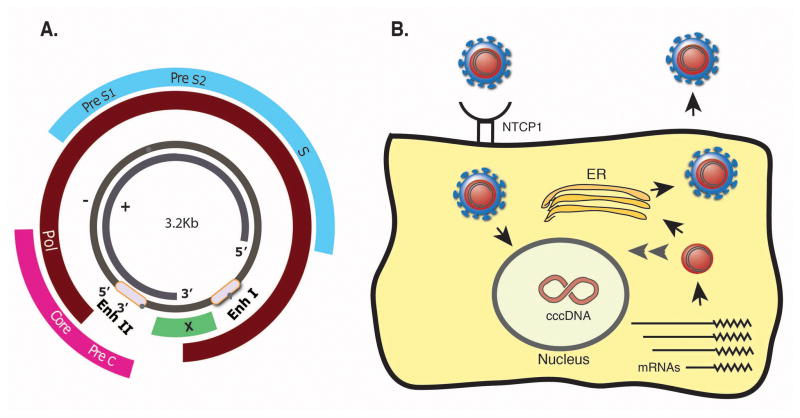
The virus 3.2kb genome contains four major open reading frames organized in a compact over-lapping gene structure (Fig 1A). HBV gene products include the polymerase (pol), which has a reverse transcriptase activity and that drives viral replication, core that forms the viral nucleocapsid and that is also cleaved to form the secreted e antigen (HBeAg), Surface (small, middle and large) proteins that are embedded in the virus envelope and are also secreted in the form of “empty” sub-viral particles and X, encoding a protein essential for viral replication. Recently, the bile acid pump sodium taurocholate co transporting polypeptide 1 (NTCP1) has been shown to be a receptor for HBV infection [33]. Following binding to its receptor and entry into the cell, the viral nucleocapsid releases the viral DNA, that is transferred to the nucleus where it is converted into a covalently closed circular DNA (cccDNA) minichromosome (reviewed in [34] and Fig 1B). The cccDNA serves as a template for viral transcription. This step in the virus life cycle is highly regulated and dependent on the presence of transcription factors, some of them are liver-enriched. Transcription is driven by distinct promoters controlled by two enhancers (Enhancer I and Enhancer II), resulting in a repertoire of viral transcripts that are capped with a common poly adenylation site. The 3.5kb pre-genomic RNA (pgRNA) serves as a template for reverse-transcription, mediated by the viral polymerase, followed by the formation of a complementary (plus) strand to create a relaxed circular form of viral DNA (rcDNA). The rcDNA, bound to the viral polymerase, is packaged to form a new progeny virus. The current view is that a portion of the newly synthesized DNA is not exported from the cell as an infectious virus but rather is imported back to the nucleus to amplify the cccDNA pool [35] (Fig 1B). This unique mechanism ensures cccDNA amplification and maintenance without integration into the host genome. Nevertheless, portions of the viral genome tend to integrate into the host’s genome, especially upon prolonged infection.
Fig 1. Gene structure and life cycle of HBV.
A. An illustration of the small HBV genome with its unique overlapping gene structure. B. Life cycle of HBV. See text for details (Enh-Enhancer, Pol-polymerase, S-Surface).
HBV infection is not cytopathic for hepatocytes, a property that may contribute to the delayed response of the immune system following HBV infection. Controversy exists regarding the role of the innate immune response in sensing and reacting to the virus (reviewed in [36]). However, cytotoxic T cells that recognize viral antigens presented on the infected cells play a major role in clearing HBV-infected cells [37]. This mechanism is responsible for complete viral clearance and clinical cure in about 95% of acutely infected adults. However, in 5% of infected adults and in a much higher proportion of newborns or small children, acute infection becomes chronic, possibly due to failure of the adaptive immune system to efficiently clear HBV infected cells. This may result from T-cell exhaustion by high antigen concentrations, promoting T-cell dysfunction [38]. Nevertheless, about 5% of chronically infected patients clear the virus spontaneously each year entering an inactive state with undetectable viremia. Those patients are usually not treated with anti-viral drugs and have a good prognosis. On the other hand, those who present with consistently high viremia and active replication are the major target population for anti-viral treatment since they are at increased risk of developing chronic liver disease and its complications.
Currently, HBV is treated with either pegylated-interferon α-2a or with reverse transcriptase (RT) inhibitors (reviewed in [39]). While these drugs effectively inhibit viral replication, they rarely lead to cure of the patient. Therefore, lifelong treatment with the RT inhibitors is usually required, especially for HBeAg negative patients [40], and preventive treatment must be initiated in silent carriers who undergo immune suppressive therapy, in order to avoid a potentially lethal HBV reactivation [41, 42]. In recent years a major advance has been made with the introduction of new generation of RT inhibitors, exemplified by Entecavir and Tenofovir, which combine excellent potency with a high resistance barrier [43, 44], making them ideal for long-term anti HBV therapy.
3.2 A strong association between chronic HBV infection and HCC
Older studies suggested chronic HBV infection might increase the risk of HCC by up to 200 fold over non-HBV carriers [45], although more recent cohort and case controlled studies indicate 10–100 fold elevated relative risk [46]. In Africa and East Asia, about two-thirds of HCC cases are attributed to HBV; in Western countries only one-fifth of HCC cases are HBV related. This difference is explained, at least in part, by the different modes of transmission (vertical versus horizontal, respectively) and more importantly, by the existence of an effective vaccine for HBV [47] and the implementation of HBV vaccination programs in most western countries. Indeed, adopting a universal pediatric vaccination program in Taiwan has led to a sharp decrease in the incidence of HCC [48].
In the classic REVEAL study, serum HBV DNA level was found to be a strong predictor for HCC, irrespective of HBeAg status, liver cirrhosis, or ALT levels [4]. Other risk factors for HCC among HBV patients include family history of HCC [49], the presence of advanced liver disease [50] as well as older age and male sex [4]. Interestingly, in contrast to chronic HCV infection, where HCC is usually associated with advanced fibrosis or cirrhosis, HBV patients can develop HCC with minimal liver damage. Thus, chronic HBV carriers who are Asian males older than 40 years or females older than 50, or those with a family history of HCC must undergo biannual ultrasound surveillance even in the absence of established cirrhosis, according to official AASLD guidelines. This recommendation is based on their calculated annual risk of developing HCC, which is greater than 0.2%, the threshold for cost effectiveness [11].
3.3 HBV and HCC- general mechanisms or virus specific factors?
HBV infection results in liver damage including inflammation, fibrosis and ultimately cirrhosis, which are all potential triggers of oncogenic transformation even without direct viral-specific mechanisms. Especially compelling is a study from Chisari and colleagues showing that HBV transgenic mice, after adoptive transfer of HBV-specific CTLs, developed HCC. Since these mice do not express the HBV X protein (HBx) and lack aberrant viral genome integration, it is likely that tumors in those animals were caused by immune-mediated liver inflammation per-se, and did not result from viral transactivation of oncogenic genes or from insertional mutagenesis, two mechanisms believed to underlie some HBV-associated HCC [51].
However, as stated previously, HBV carrier state itself is a risk factor for HCC, even in the absence of advanced liver disease and cirrhosis. This observation, as well as differences in HCC susceptibility across different HBV genotypes (genotype C conferring the greatest risk for HCC) [4, 52] and specific viral mutations associated with increased risk for HCC [53, 54], strongly imply a direct viral effect on HCC development.
A prominent obstacle in our understanding of HBV biology and the link between the virus and HCC is the lack of robust in vitro and in vivo systems that reliably mimic human HBV infection. Most in vitro studies have utilized either transiently or stably transfected hepatoma cell lines [55] that overexpress viral proteins, and are themselves transformed cells and a priori defective in many genes and growth promoting pathways, independent of virus influence. While avian and mammalian HBV-related viruses have been identified, making it possible to study their biology in natural hosts, the relevance and predictive value of these models to HBV infected humans is not always obvious (reviewed in [56]). The development of mouse models for HBV replication and infection, including HBV transgenic mice [57] and human liver chimeric mice [58, 59], respectively, have been significant, but these models are still suboptimal for unraveling HBV associated pathologies, particularly HCC (reviewed in [60]).
3.4 The role of HBV integration in the pathogenesis of HCC
The realization that in chronic infection, HBV DNA is maintained in the infected hepatocyte not only as episomal nuclear cccDNA but also integrated in the host’s genome [61] led to attempts to identify specific integration “hot spots” that might correlate with oncogenic transformation.
The most striking association between genomic integration and HCC was initially found in woodchuck hepatitis virus (WHV)-associated tumors, in which a high proportion of the tumors have viral insertions near N-myc, the proto-oncogene, resulting in its activation by the viral enhancer [62, 63]. However, although there was initially evidence for the existence of an active HBV enhancer in many HCC-associated viral integrations [64], few consistent genomic integration hot spots were identified [65, 66], and their significance in HCC pathogenesis is still debated.
More recently, next generation sequencing (NGS) has enabled large scale, highly sensitive analyses of large numbers of HCC samples. In one study, analyzing 81 tissue samples from HBV-positive HCC in parallel with HBV negative HCC and normal liver tissues, HBV integration was found in more than 80% of HBV positive cases [67]. Integration in tumor tissue was found to be more extensive and distinct from surrounding non-tumor tissue. Analysis integration breakpoints revealed that 40% of them were restricted to an 1800 bp region of the HBV genome that includes the viral enhancer, the X gene, and a portion of the core region. These specific and strategic viral break points may promote the creation of chimeric virus/human fusion gene variants upon integration, activate in-cis cellular oncogenes (specifically by the HBx) or simply disrupt tumor suppressor genes. Most importantly, three cancer-associated genes were found as frequent integration sites in HBV positive tumors: telomerase reverse transcriptase (TERT), MLL4 and CCNE1, accounting for about 40% of tumor samples with HBV integration. These integrations were accompanied by increased expression of the corresponding genes, irrespective of whether integration occurred in the promoter, exonic or intronic regions. Another important finding was that copy number variations (CNVs) were significantly higher in HBV breakpoint locations, most probably a reflection of increased genomic instability following HBV integration(s). This suggests yet another mechanistic link between HBV integration and liver carcinogenesis. In another study [68] that sequenced HBV related tumor and adjacent normal tissues using massive anchored parallel sequencing (MAPS), recurrent HBV integrations were found in 8 host genes, including TERT and FN1. For TERT, HBV integration increased expression of the gene product. Interestingly, integrations were more frequent in transcript coding units. Less diverse integration sites were found in cancer as compared to cancer-adjacent tissues, possibly as a result of post-integration clonal expansion of HCC tissue. Another analysis of HCC samples from HBV and HCV patients [69] found frequent mutations in chromatin regulator genes such as ARID1A, ARID1B, ARID2, MLL and MLL3. As seen in other studies, more frequent HBV integration was found upstream of the TERT gene, while no clonal integrations were found in the non-cancerous tissue.
These and other earlier studies [66, 70] suggest that the TERT gene locus is a common “hot spot” for HBV integration associated with HCC. Given that HBV integration is an early event in the development of HBV-associated HCC, integration at the TERT locus and the resulting overexpression of this gene may confer a clonal advantage to transformed cells and pave the way for further tumor development.
Most of these NGS-based studies confirm prior observations regarding the high frequency of breakpoints within the HBV genome downstream of the HBx region, resulting in a C-terminally truncated HBx protein [71]. These observations support the hypothesis that mutant forms of HBx may possess gain of function activities that promote carcinogenesis.
Thus, HBV integration may promote tumorgenesis via three distinct mechanisms: Genomic destabilization; cis-activation of oncogenes or altered tumor suppressor activity; and production of mutant HBV proteins with tumor-promoting potential.
3.5 The role of HBx in the pathogenesis of HCC
HBx is a 154 amino acid viral protein [72, 73] with crucial roles in HBV infection, replication [74, 75] and links to liver carcinogenesis (reviewed in [76]). Initial studies showed that HBx can transform normal murine cells in vitro [77, 78], and promote the development of liver tumors in HBx transgenic mice [79, 80].
Four mechanisms have been invoked to explain HBx oncogenic potential. These include transactivation/repression of genes implicated in cell survival and proliferation, interaction with proteins implicated in cellular response to oncogenic stress, activation of cell survival signaling pathways, and epigenetic changes, such as DNA methylation, histone modification and microRNA expression.
HBx can trans-activate cellular, as well as viral genes, by enhancing the activity of the basal transcription machinery and by binding, stabilizing and enhancing the activity of various transcription factors [72]. HBx trans-activates promoters of CREB response element (CRE)-containing genes, including the oncogene Yes-associated protein (YAP) [81], which is frequently overexpressed in HCC. In addition, HBx can alter the DNA specificity of CREB and ATF-2, resulting in binding and activation of the HBV enhancer, which contains a CRE-like rather than a perfect CRE consensus sequence [82]. Recently, Chan et al have shown HBx also modulates the DNA binding specificity of the p53 tumor suppressor, resulting in altered expression of its target genes [83]. Interestingly, p53 has been shown to repress HBV transcription [84] and HBx acts as a co-repressor of the p53 promoter [85]. Obviously, this altered activity of the “guardian of the genome” mediated by HBx may provide a mechanistic explanation for HBx induced liver carcinogenesis. However, the extent to which HBx is able to modulate the expression of cellular genes is unclear and needs to be interpreted with caution given the recently published study showing that HBx trans-activates genes exclusively from the (viral) episomal DNA, but not from chromosomal genomic loci [86].
HBx may also promote liver cell transformation via interactions with proteins implicated in the DNA damage response. One example is the UV-damage DNA-binding factor UV-DDB1. HBx interaction with UV-DDB1 stimulates viral genome replication [87, 88] but results in impaired cell viability [89]. UV-DDB1, an essential component of the first step in DNA damage repair, binds to damaged DNA. This led Becker et al to hypothesize that HBx-UV-DDB1 interaction might compromise the normal cellular response to DNA damage leading to cell transformation [90]. Alternatively, Martin-Lluesma and co-workers suggested that HBx binding to UV-DDB1 interferes with S-phase progression, leading to genomic instability and to cell transformation [91].
HBx has been reported to interact with the tumor suppressor p53, resulting in inhibition of p53 target gene expression though the mechanism for this repression is not clear. While one report suggests a direct sequestration of p53 in the cytoplasm by HBx to prevent p53 nuclear entry and target gene activation [92], another report shows that HBx-p53 interaction does not prevent p53 entry into the nucleus nor does it prevent p53 binding to the promoters of its target genes. Rather, its interaction with p53 enables HBx “hijacking” of p53 to its DNA binding sites where HBx directly co-represses genes by interaction with the basal transcription machinery, thereby overcoming p53 activity [93].
Another potential mechanism for HBx-mediated carcinogenesis is its ability to disrupt/activate cellular signaling pathways implicated in cell survival. This is exemplified by multiple reports of activation of the inflammation-associated NF-kB and STAT-3 pathways [94–96], the Ras-Raf-MAPK pathway (through activation of the Ras-GTP complex) [97], the PI3K-Akt pathway [98] and the Src-dependent pathway [99]. This latter pathway is potentially most important for HBV replication [100] and involves calcium release from mitochondria [74]. In addition, HBx can activate the wnt/β-catenin pathway [101], which is frequently mutated in HCC. One possible mechanism for this activation is interaction with the APC protein, resulting in displacement of β-catenin from its degradation complex [102]. Alternatively, ERK activation by HBx can result in phosphorylation and inactivation of GSK-3β, and up-regulation β-catenin [103].
At the current time, we are left with conflicting views as to whether HBx expression enhances cell survival or promotes apoptosis. These discrepancies may be due to differences in cell culture conditions, the use of cancerous cell lines or overexpression of HBx. Development of more reliable infection systems in primary cells will hopefully help decipher these unsolved questions.
A different mechanism for HBx-induced HCC concerns epigenetic changes induced by this protein (reviewed in [104]). These changes include hypermethylation or hypomethylation of tumor suppressor and tumor promoting genes, respectively [105–107]. In addition, HBx has been shown to promote histone acetylation and de-acetylation in tumor related genes, and to alter the expression of certain microRNAs implicated in HCC. For example, HBx upregulates the expression of miR-602, miR-143, miR-29a and miR-148a, all known regulators of genes implicated in tumorogenesis and metastasis [108–111]. Conversely, HBx downregulates the expression of other miRNAs important for cell-cell adhesion, cell migration and metastasis [104]. A recent report found that HBx suppresses transcription of the abundant liver-specific miR-122 through PPAR-γ [112]. This, together with emerging evidence of an anti-tumorigenic role for miR-122 [113], suggests a possible role of the HBx-miR-122 axis in the pathogenesis of HBV-associated HCC.
Interestingly, HBx has been recently suggested to down regulate the tumor suppressor miR 15a/miR16 cluster by sequestering those miRNAs to a binding site on its mRNA [114, 115]. This “sponge” mechanism, whereby a virus down regulates specific cellular miRNAs by its sequestering, may be common to other viruses, as well (see discussion below regarding HCV).
4. Hepatitis C virus infection is a risk factor for HCC
HCV is a blood-borne virus that was identified as the causative agent for non-A non-B hepatitis in 1989 [116]. It is a member of the Flaviviridae family of positive stranded RNA viruses and can establish a chronic infection in 50–80% of exposed individuals. HCV infection largely follows a subclinical course and can, typically after decades of infection, result in liver fibrosis and cirrhosis in a subset of those infected. Depending on the era and geographical area, it has spread largely through iatrogenic practices and intravenous drug use, resulting in approximately 170 million chronically infected people worldwide. After HBV, HCV infection is the most significant risk factor for HCC development, accounting for 10–20% of HCC cases globally [117]. Epidemiological support for an etiological link was the striking correlation between HCV spread and HCC incidence in Japan. HCV initially spread through injection treatments for schistosomiasis in the 1920s [118]. After World War II HCV incidence peaked with the burst of the parenteral amphetamine use and the paid blood donation, and significantly decreased between the 1950s to the 1960s upon establishment of a voluntary blood donation system and adopting a penalties policy against amphetamine use. HCC incidence followed this pattern but lagged for about 40 years, with its peak incidence detected between the years 1986 to 2000, followed by a sharp decrease beginning at the year 2000. Notably, these changes in HCC incidence were attributed solely to HCV-associated HCC and not to HCC from other etiologies, the rate of which remained steady [119]. Because HCV spread more recently in the United States, the number of HCC cases is still rising in this country [120].
In contrast to HBV, HCV-associated HCCs develop primarily on a background of advanced fibrosis or cirrhosis. However, in the prospective HALT-C trial of chronically infected patients in the United States, 17% of HCCs were found in the absence of advanced fibrosis [121]. Contrary to a previous report from Japan [122], this did not correlate with occult, anti-HBc positive, HBsAg negative, HBV infection [123]. These findings suggest that advanced fibrosis may not be an absolute prerequisite for HCC carcinogenesis in HCV monoinfection.
Several observations implicate HCV infection and associated inflammation as contributing to the risk of developing HCC. First, the risk of developing HCC is greater in the setting of HCV infection than other, non-viral risk factors [124, 125]. Second, there are a number of reports showing that elevated levels of transaminases, a marker for liver inflammation, correlate with HCC risk among HCV patients independently of fibrosis stage [126–128]. This is further supported by the findings from several groups that even in patients with advanced fibrosis, HCV eradication decreases the risk of HCC to 21% of that observed in patients failing therapy [129–131]. However, none of these observations distinguishes between a direct role attributed to HCV replication/gene expression in HCC development versus an indirect role for HCV through general mechanisms such as HCV-induced inflammation (for reviews on inflammation and HCC see [132, 133]). Nonetheless, there is general agreement that ongoing inflammation resulting from HCV infection appears to contribute significantly to the risk of developing HCC independently of hepatic fibrosis. This has been an important impetus for HCV eradication, as it would lower peak incidence of the still rising number of HCC cases in the United States. Recent and future approval of well-tolerated highly effective antiviral drug regimens for HCV holds great promise for achieving this goal [134], especially if they can be implemented on a global scale.
4.1 Hepatitis C virus models for the study of HCC pathogenesis
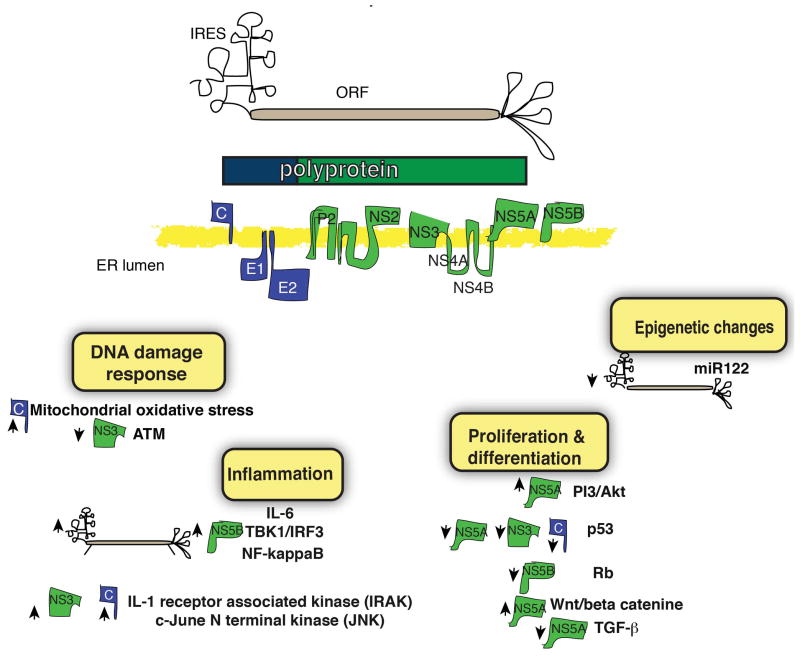
Hepatitis C virus positive stranded RNA genome of 9.6kb is translated into a long polyprotein, which is subsequently processed by host and viral proteases into 10 proteins: three structural proteins, core, envelope (E) 1 and E2, and seven non-structural (NS) proteins p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B (Fig 3). Similar to HBV, only limited tools are available to study HCV infection in large part because the authentic HCV life cycle is believed to be restricted to human hepatocytes. Most early in vitro work was limited to RNA replicons, i.e. selectable (sub)genomic viral RNAs that replicate but do not produce virus [135] or, more recently, cell-culture infectious virus (HCVcc) [136, 137] based on a limited number of genomes, the most widely used being Japanese fulminant hepatitis-1 (JFH-1) [138–141]. In addition to obvious constraints of working in vitro, these systems have the additional limitation that they almost exclusively use Huh7 hepatoma derived cell lines for HCV replication. Even with these limitations, the replicon and HCVcc systems have proven enormously useful in studying many aspects of the HCV life cycle [142], including pathways that may be important in HCC carcinogenesis.
Fig 3. The role of HCV gene products in liver carcinogenesis.
(Upper) HCV is an RNA virus encoding for a polyprotein that is cleaved to individual structural (blue) and non-structural (green) proteins, essential for viral propagation and pathogenesis. (Lower) Putative mechanisms for carcinogenesis promoted by viral products (upwards arrows=up-regulation downwards arrows=down-regulation).
Animal models of HCC development have the potential to overcome some of the limitations of in vitro systems. The only natural host beyond humans able to support HCV infection is the chimpanzee Pan troglodytes, and HCV infection in chimpanzees recapitulates most features of human infection. Despite their widespread use in the early days HCV research, infection associated HCC is rare. Recently, one animal was reported that developed multifocal HCC in the setting of chronic HCV, but notably with only minimal portal fibrosis [143]. Even though chimpanzees are undoubtedly the most relevant animal model for HCV infection, high costs, ethical concerns and low incidence of HCC have limited their use. For this reason, over twenty different HCV transgenic mice have been created that express one or several viral proteins, or the entire polyprotein under the control of a variety of liver-specific and other promoters. These have been extensively studied for HCC development. However, transgenic models have several inherent limitations. These include central immune tolerance to the transgene and subsequent lack of virus-specific adaptive immunity, random insertion of the transgene cassette, which can be particularly problematic when studying carcinogenesis, and protein expression levels that do not recapitulate those of a natural infection. In addition, depending upon the HCV proteins expressed, the type of promoter, and the genetic background of the mice, different phenotypes have been observed [144].
Nevertheless, this vast body of transgenic animal literature has found several altered pathways that suggest a possible direct role for HCV proteins in HCC carcinogenesis (for review see [145]). For example, HCV core overexpression resulted in bile duct damage, steatosis and lymphoid aggregates in the liver [146], and the frequency of HCC was increased by concomitant inflammation [147] or iron overload [148]. Such findings mimic observations in patients, and argue for the relevance of HCV transgenic mice as models for the study of HCC carcinogenesis.
A different approach to study HCV in small animal models is through the use of human hepatocyte xenografted mice. These models share some form of mouse hepatocyte injury to allow the human hepatocytes to proliferate, and lack mouse T and B lymphocytes to prevent graft rejection. The best-studied models are immunodeficient urokinase plasminogen activator transgenic mice expressed under an albumin promoter (alb-uPA) [149] and immunodeficient fumaryl actetoacetate hydrolase mice [58]. These models support infection of several HCV genotypes, and have been used for a variety of virologic and therapeutic investigations. One recent report showed that HCV infection could cause oxidative stress and induce pro-apoptotic signals in uPA mice [150], pathways that have previously been implicated in HCC carcinogenesis in vitro as well as in transgenic mice. A different liver injury model that was transplanted with both human hepatocytes and human lymphocytes showed some fibrosis [151], but to date HCC has not been reported in any xenograft-based model.
In summary, HCV studies have largely been limited to in vitro systems in HCC cell lines and animals transgenic for viral proteins. Several pathways have been implicated from these model systems and will be discussed below.
4.2 Evidence for a direct role of HCV in the pathogenesis of HCC
The epidemiological link between HCV and HCC cannot distinguish direct oncogenic effects by viral proteins from the inflammation that is elicited by its replication. For example, although HCV RNA can be detected both in HCC and in surrounding non-tumor tissue [152, 153], it remains controversial whether viral quasispecies enriched in the tumor have a direct role in HCC carcinogenesis. Moreover, and similar to the case of HBV, data implicating specific viral proteins is often conflicting and depends on the experimental system in which it was generated. Given these caveats, both HCV structural and non-structural proteins have been implicated in pro- and anti-apoptotic effects, in eliciting inflammation as well as in a variety of tumor pathways. The best studied pathways will be highlighted here (see Fig 3); for a more in-depth review of these pathways see [154].
Most people infected with HCV have hepatitis, which typically comprises a mixed lymphocyte infiltrate. In addition to a role for infiltrating T lymphocytes and NK cells [155], hepatocytes sense HCV RNA motifs through RIG-I and TLR-3, leading to activation of the NF-κB pathway and generation of interferon and other pro-inflammatory cytokines. Furthermore the viral polymerase NS5B has been shown to directly activate the inflammatory cascade through NF-κB in a MAVS and TBK-1 dependent manner, resulting in secretion of IL-6 and type I interferon [156]. Given the likely importance of inflammation in the pathogenesis of hepatic carcinogenesis (see previous sections), events possibly mediated by the viral genetic material and by at least one of its functional proteins might be a substantial trigger for hepatic transformation.
Oxidative stress is another potential trigger for oncogenic transformation via induction of DNA mutagenesis. The HCV core protein has been shown to promote formation of intracellular reactive oxygen species (ROS), both in vitro [157] and in vivo [158], possibly through its localization to the mitochondria and inhibition of electron transport [158].
Central tumor suppressor genes as well as a number of proto-oncogenes have been suggested as direct targets for alteration by HCV proteins. For example, the retinoblastoma tumor suppressor protein (Rb) has been shown to interact with the HCV NS5B protein, resulting in its poly-ubiquitination and degradation, which promotes entry into S phase [159]. Notably, this pro-oncogenic mechanism is quite common for DNA viruses, but HCV is currently the only RNA virus that has been shown to manipulate cells in this fashion.
p53 is also a target for HCV and at least three viral proteins have been shown to interact with this key tumor suppressor. Conflicting data on whether core interaction with p53 results in activation or inhibition of p53 target genes may reflect differences in the level of core expression [160]. In addition, NS3-p53 interaction can block apoptosis in vitro [161] and NS5A interaction with p53 results in p53 redistribution to the peri-nuclear membrane [162].
The wnt/β-catenin pathway may also be altered directly by HCV. Initially, NS5A was shown to promote β-catenin stabilization indirectly through inactivation of GSK3-β, which normally promotes β–catenin degradation [163, 164]. As mentioned previously, GSK3-β has also been implicated as an HBx target to promote β-catenin stabilization. However, there is some evidence that NS5A directly interacts with and stabilizes β-catenin [165].
Evidence has also emerged for HCV proteins interacting with cellular pathways implicated in liver response to damage. For example, the ataxia-telangiectasia mutated kinase (ATM) is not only required for HCV replication [166] but also interacts with the HCV NS3-4A protein complex, resulting in impaired DNA damage responses and enhanced sensitivity to ionizing irradiation [167]. The resulting increased rate of double-stranded DNA breaks is a possible direct viral role in tumorigenesis. Another example is the interaction of HCV with the TGF-β pathway, a central component of the liver proliferation and damage response. HCV NS5A has been shown to block TGF-β signaling through direct interaction with its receptor, TGF-β receptor I (TβR-I) [168], thereby promoting liver damage, fibrosis and cancer. Also, core quasispecies isolated from HCC but not from surrounding liver tissue were able to block TGF-β signaling through interactions with Smad3 [169].
Finally, HCV requires liver-specific miR-122 for replication [170], as demonstrated by a recent phase 2a clinical trial showing that sequestering miR-122 in patients leads to a dose-dependent decrease in HCV viremia [171]. An association between miR-122 and HCC was reported in two independent studies, showing high tumor incidence in mice lacking miR-122 [113, 172]. Little is known about how HCV replication affects miR-122 levels in the liver, although some initial reports support the notion that miR-122 abundance is significantly reduced in human liver with advanced fibrosis [173] and in patients who do not respond to HCV therapy [174]. Recruitment of miR-122 to the HCV genome could result in a functional depletion of this important liver-specific miR-122. As mentioned earlier with regard to the anti-tumorigenic effect of miR-122, it is possible that miR-122 sequestration by HCV might create a liver environment that ultimately contributes to HCC development.
Conclusions
Chronic HBV and HCV infections together plague more than half a billion people and are the strongest risk factors for developing HCC. The solid clinical association between these infections and HCC incidence has thus far eluded reliable mechanistic studies, in large part because of the limited tools with which we can study these viruses. Recently, functional genomics approaches have emerged that may reveal the importance of known tumor promoting pathways and facilitate the discovery of new genes implicated in HCC (reviewed in [175]). With these new technologies it will become even more important to distinguish driver mutations from the multitude of passenger mutations that are found. Further improvements in animal models to study these infections, e.g. human hepatocyte xenograft mice that display inflammation and fibrosis, would potentially allow for the manipulation of such driver mutations, and test to what extent they contribute to HCC pathogenesis. Until the functional identification of such pathways becomes feasible, the search for much-needed drugs for HCC will undoubtedly remain challenging.
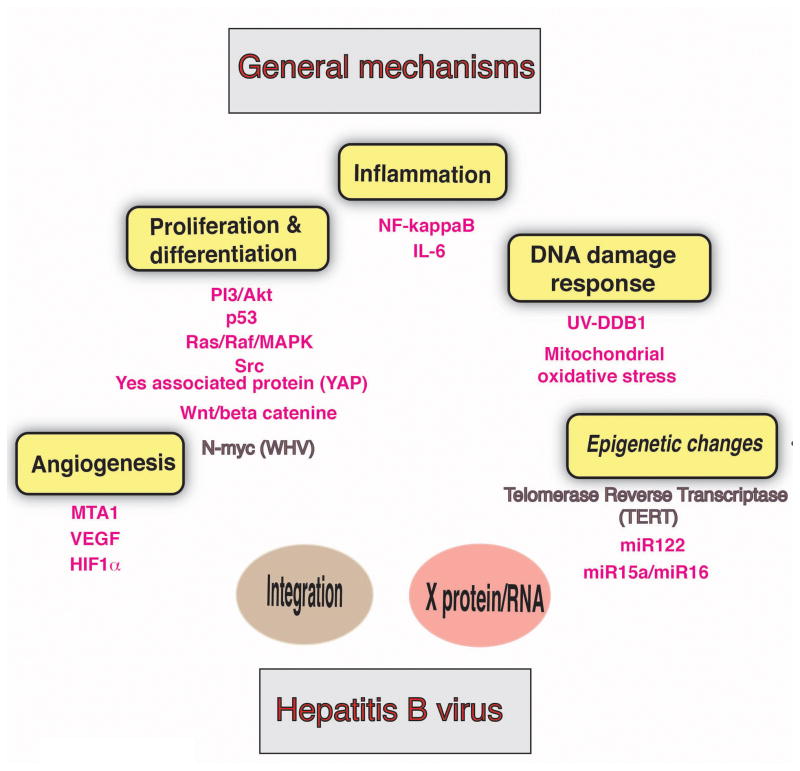
Fig 2. General mechanisms for liver carcinogenesis and the role of viral factors in this process.
Note that each mechanism implicated in liver carcinogenesis corresponds to the color of the putative viral factor triggering this mechanism.
Acknowledgments
We would like to thank Drs. Mayla Hsu, William Schneider and Timothy Sheahan for critical reading of the manuscript and helpful comments. The authors have been supported in part by grants CA057973, AI090055, AI072613, AI075099, AI099284, DK085713 and AIK08DK09057 from the U. S. National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding was also provided by the Rockefeller University Center for Clinical and Translational Science (UL1RR024143), the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust, the Greenberg Medical Research Institute, the Starr Foundation, and other generous donors and funders.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nature reviews Cancer. 2013;13:123–35. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. The New England journal of medicine. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Mittal S, El-Serag HB. Epidemiology of Hepatocellular Carcinoma: Consider the Population. Journal of clinical gastroenterology. 2013 doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CYHISJ, et al. RIsk of hepatocellular carcinoma across a biological gradient of serum hepatitis b virus dna level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 5.Singal AK, Salameh H, Kuo YF, Fontana RJ. Meta-analysis: the impact of oral anti-viral agents on the incidence of hepatocellular carcinoma in chronic hepatitis B. Alimentary pharmacology & therapeutics. 2013 doi: 10.1111/apt.12344. [DOI] [PubMed] [Google Scholar]
- 6.Morgan RL, Baack B, Smith BD, Yartel A, Pitasi M, Falck-Ytter Y. Eradication of Hepatitis C Virus Infection and the Development of Hepatocellular CarcinomaA Meta-analysis of Observational Studies. Annals of Internal Medicine. 2013;158:329–37. doi: 10.7326/0003-4819-158-5-201303050-00005. [DOI] [PubMed] [Google Scholar]
- 7.Lazo M, Hernaez R, Eberhardt MS, Bonekamp S, Kamel I, Guallar E, et al. Prevalence of Nonalcoholic Fatty Liver Disease in the United States: The Third National Health and Nutrition Examination Survey, 1988–1994. American journal of epidemiology. 2013 doi: 10.1093/aje/kws448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2012;10:1342–59. e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, et al. Gender Disparity in Liver Cancer Due to Sex Differences in MyD88-Dependent IL-6 Production. Science. 2007;317:121–4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 10.Yang W, Lu Y, Xu Y, Xu L, Zheng W, Wu Y, et al. Estrogen Represses Hepatocellular Carcinoma (HCC) Growth via Inhibiting Alternative Activation of Tumor-associated Macrophages (TAMs) Journal of Biological Chemistry. 2012;287:40140–9. doi: 10.1074/jbc.M112.348763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruix J, Sherman M. Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J-F, et al. Sorafenib in Advanced Hepatocellular Carcinoma. New England Journal of Medicine. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 13.Chan SL, Yeo W. Targeted therapy of hepatocellular carcinoma: Present and future. Journal of gastroenterology and hepatology. 2012;27:862–72. doi: 10.1111/j.1440-1746.2012.07096.x. [DOI] [PubMed] [Google Scholar]
- 14.Zender L, Villanueva A, Tovar V, Sia D, Chiang DY, Llovet JM. Cancer gene discovery in hepatocellular carcinoma. Journal of hepatology. 2010;52:921–9. doi: 10.1016/j.jhep.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abu Dayyeh BK, Yang M, Fuchs BC, Karl DL, Yamada S, Sninsky JJ, et al. A functional polymorphism in the epidermal growth factor gene is associated with risk for hepatocellular carcinoma. Gastroenterology. 2011;141:141–9. doi: 10.1053/j.gastro.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanabe KK, Lemoine A, Finkelstein DM, Kawasaki H, Fujii T, Chung RT, et al. Epidermal growth factor gene functional polymorphism and the risk of hepatocellular carcinoma in patients with cirrhosis. JAMA. 2008;299:53–60. doi: 10.1001/jama.2007.65. [DOI] [PubMed] [Google Scholar]
- 17.Zhong JH, You XM, Gong WF, Ma L, Zhang Y, Mo QG, et al. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PloS one. 2012;7:e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Poon RT, Shao W, Sun X, Chen H, Kok TW, et al. Blockage of epidermal growth factor receptor by quinazoline tyrosine kinase inhibitors suppresses growth of human hepatocellular carcinoma. Cancer letters. 2007;248:32–40. doi: 10.1016/j.canlet.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Schiffer E, Housset C, Cacheux W, Wendum D, Desbois-Mouthon C, Rey C, et al. Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology. 2005;41:307–14. doi: 10.1002/hep.20538. [DOI] [PubMed] [Google Scholar]
- 20.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–28. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 22.Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071–83. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. The New England journal of medicine. 2013;368:2266–76. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G, Jiang S, Wang C, Jiang W, Liu Z, Liu C, et al. Zinc finger transcription factor 191, directly binding to β-catenin promoter, promotes cell proliferation of hepatocellular carcinoma. Hepatology. 2012;55:1830–9. doi: 10.1002/hep.25564. [DOI] [PubMed] [Google Scholar]
- 25.Feng GJ, Cotta W, Wei XQ, Poetz O, Evans R, Jarde T, et al. Conditional disruption of Axin1 leads to development of liver tumors in mice. Gastroenterology. 2012;143:1650–9. doi: 10.1053/j.gastro.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 26.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 27.Kong S-Y, Park J-W, Lee JA, Park JE, Park KW, Hong EK, et al. Association between vascular endothelial growth factor gene polymorphisms and survival in hepatocellular carcinoma patients. Hepatology. 2007;46:446–55. doi: 10.1002/hep.21720. [DOI] [PubMed] [Google Scholar]
- 28.Poon RT, Ng IO, Lau C, Zhu LX, Yu WC, Lo CM, et al. Serum vascular endothelial growth factor predicts venous invasion in hepatocellular carcinoma: a prospective study. Annals of surgery. 2001;233:227–35. doi: 10.1097/00000658-200102000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu SH, Chung YH, Lee H, Kim JA, Shin HD, Min HJ, et al. Metastatic tumor antigen 1 is closely associated with frequent postoperative recurrence and poor survival in patients with hepatocellular carcinoma. Hepatology. 2008;47:929–36. doi: 10.1002/hep.22124. [DOI] [PubMed] [Google Scholar]
- 30.Dandri M, Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut. 2012;61:i6–i17. doi: 10.1136/gutjnl-2012-302056. [DOI] [PubMed] [Google Scholar]
- 31.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. The New England journal of medicine. 2004;350:1118–29. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 32.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–93. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 33.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012:1. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. Journal of hepatology. 2009;51:581–92. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–60. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 36.Bertoletti A, Ferrari C. Republished: Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Postgraduate Medical Journal. 2013;89:294–304. doi: 10.1136/postgradmedj-2011-301073rep. [DOI] [PubMed] [Google Scholar]
- 37.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, et al. CD8+ T Cells Mediate Viral Clearance and Disease Pathogenesis during Acute Hepatitis B Virus Infection. Journal of virology. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boni C, Fisicaro P, Valdatta C, Amadei B, Di Vincenzo P, Giuberti T, et al. Characterization of Hepatitis B Virus (HBV)-Specific T-Cell Dysfunction in Chronic HBV Infection. Journal of virology. 2007;81:4215–25. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tujios SR, Lee WM. Update in the management of chronic hepatitis B. Current opinion in gastroenterology. 2013;29:250–6. doi: 10.1097/MOG.0b013e32835ff1e9. [DOI] [PubMed] [Google Scholar]
- 40.Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–2. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 41.Hoofnagle JH. Reactivation of hepatitis B. Hepatology. 2009;49:S156–S65. doi: 10.1002/hep.22945. [DOI] [PubMed] [Google Scholar]
- 42.Lalazar G, Rund D, Shouval D. Screening, prevention and treatment of viral hepatitis B reactivation in patients with haematological malignancies. Br J Haematol. 2007;136:699–712. doi: 10.1111/j.1365-2141.2006.06465.x. [DOI] [PubMed] [Google Scholar]
- 43.Chang TT, Lai CL, Kew Yoon S, Lee SS, Coelho HS, Carrilho FJ, et al. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:422–30. doi: 10.1002/hep.23327. [DOI] [PubMed] [Google Scholar]
- 44.Gordon SC, Krastev Z, Horban A, Petersen J, Sperl J, Dinh P, et al. Efficacy of tenofovir disoproxil fumarate at 240 weeks in patients with chronic hepatitis B with high baseline viral load. Hepatology. 2013 doi: 10.1002/hep.26277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–33. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. Journal of viral hepatitis. 2009;16:453–63. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- 47.Poorolajal J, Mahmoodi M, Majdzadeh R, Nasseri-Moghaddam S, Haghdoost A, Fotouhi A. Long-term protection provided by hepatitis B vaccine and need for booster dose: a meta-analysis. Vaccine. 2010;28:623–31. doi: 10.1016/j.vaccine.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 48.Chang M-H, Chen C-J, Lai M-S, Hsu H-M, Wu T-C, Kong M-S, et al. Universal Hepatitis B Vaccination in Taiwan and the Incidence of Hepatocellular Carcinoma in Children. New England Journal of Medicine. 1997;336:1855–9. doi: 10.1056/NEJM199706263362602. [DOI] [PubMed] [Google Scholar]
- 49.Loomba R, Liu J, Yang HI, Lee MH, Lu SN, Wang LY, et al. Synergistic Effects of Family History of Hepatocellular Carcinoma and Hepatitis B Virus Infection on Risk for Incident Hepatocellular Carcinoma. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2013 doi: 10.1016/j.cgh.2013.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu MW, Hsu FC, Sheen IS, Chu CM, Lin DY, Chen CJ, et al. Prospective study of hepatocellular carcinoma and liver cirrhosis in asymptomatic chronic hepatitis B virus carriers. American journal of epidemiology. 1997;145:1039–47. doi: 10.1093/oxfordjournals.aje.a009060. [DOI] [PubMed] [Google Scholar]
- 51.Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune Pathogenesis of Hepatocellular Carcinoma. The Journal of Experimental Medicine. 1998;188:341–50. doi: 10.1084/jem.188.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toan NL, Song le H, Kremsner PG, Duy DN, Binh VQ, Koeberlein B, et al. Impact of the hepatitis B virus genotype and genotype mixtures on the course of liver disease in Vietnam. Hepatology. 2006;43:1375–84. doi: 10.1002/hep.21188. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka Y, Mukaide M, Orito E, Yuen MF, Ito K, Kurbanov F, et al. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. Journal of hepatology. 2006;45:646–53. doi: 10.1016/j.jhep.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 54.Yuan JM, Ambinder A, Fan Y, Gao YT, Yu MC, Groopman JD. Prospective evaluation of hepatitis B 1762(T)/1764(A) mutations on hepatocellular carcinoma development in Shanghai, China. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:590–4. doi: 10.1158/1055-9965.EPI-08-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sells MA, Chen M-L, Acs G. Production of Hepatitis B Virus Particles in Hep G2 Cells Transfected with Cloned Hepatitis B Virus DNA. PNAS. 1987;84:1005–9. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dandri M, Volz TK, Lutgehetmann M, Petersen J. Animal models for the study of HBV replication and its variants. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2005;34 (Suppl 1):S54–62. doi: 10.1016/s1386-6532(05)80011-3. [DOI] [PubMed] [Google Scholar]
- 57.Chisari FV. Hepatitis B virus transgenic mice: insights into the virus and the disease. Hepatology. 1995;22:1316–25. doi: 10.1016/0270-9139(95)90645-2. [DOI] [PubMed] [Google Scholar]
- 58.Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. The Journal of clinical investigation. 2010;120:924–30. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lutgehetmann M, Mancke LV, Volz T, Helbig M, Allweiss L, Bornscheuer T, et al. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology. 2012;55:685–94. doi: 10.1002/hep.24758. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Tang ZY, Hou JX. Hepatocellular carcinoma: insight from animal models. Nature reviews Gastroenterology & hepatology. 2012;9:32–43. doi: 10.1038/nrgastro.2011.196. [DOI] [PubMed] [Google Scholar]
- 61.Shaul Y, Ziemer M, Garcia PD, Crawford R, Hsu H, Valenzuela P, et al. Cloning and analysis of integrated hepatitis virus sequences from a human hepatoma cell line. Journal of virology. 1984;51:776–87. doi: 10.1128/jvi.51.3.776-787.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fourel G, Couturier J, Wei Y, Apiou F, Tiollais P, Buendia MA. Evidence for long-range oncogene activation by hepadnavirus insertion. The EMBO journal. 1994;13:2526–34. doi: 10.1002/j.1460-2075.1994.tb06542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, et al. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294–8. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 64.Shamay M, Agami R, Shaul Y. HBV integrants of hepatocellular carcinoma cell lines contain an active enhancer. Oncogene. 2001;20:6811–9. doi: 10.1038/sj.onc.1204879. [DOI] [PubMed] [Google Scholar]
- 65.Chami M, Gozuacik D, Saigo K, Capiod T, Falson P, Lecoeur H, et al. Hepatitis B virus-related insertional mutagenesis implicates SERCA1 gene in the control of apoptosis. Oncogene. 2000;19:2877–86. doi: 10.1038/sj.onc.1203605. [DOI] [PubMed] [Google Scholar]
- 66.Paterlini-Brechot P, Saigo K, Murakami Y, Chami M, Gozuacik D, Mugnier C, et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22:3911–6. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 67.Sung W-K, Zheng H, Li S, Chen R, Liu X, Li Y, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–9. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 68.Ding D, Lou X, Hua D, Yu W, Li L, Wang J, et al. Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach. PLoS Genet. 2012;8:e1003065. doi: 10.1371/journal.pgen.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–4. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 70.Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Brechot C, et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162–8. doi: 10.1136/gut.2004.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, et al. Biological Impact of Natural COOH-Terminal Deletions of Hepatitis B Virus X Protein in Hepatocellular Carcinoma Tissues. Cancer Research. 2001;61:7803–10. [PubMed] [Google Scholar]
- 72.Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. Journal of gastroenterology. 2001;36:651–60. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- 73.Wei Y, Neuveut C, Tiollais P, Buendia MA. Molecular biology of the hepatitis B virus and role of the X gene. Pathologie-biologie. 2010;58:267–72. doi: 10.1016/j.patbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 74.Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–8. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 75.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. Journal of virology. 1994;68:2026–30. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koike K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer letters. 2009;286:60–8. doi: 10.1016/j.canlet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Hohne M, Schaefer S, Seifer M, Feitelson MA, Paul D, Gerlich WH. Malignant transformation of immortalized transgenic hepatocytes after transfection with hepatitis B virus DNA. The EMBO journal. 1990;9:1137–45. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seifer M, Hohne M, Schaefer S, Gerlich WH. In vitro tumorigenicity of hepatitis B virus DNA and HBx protein. Journal of hepatology. 1991;13 (Suppl 4):S61–5. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- 79.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–20. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 80.Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. Journal of hepatology. 1999;31:123–32. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 81.Zhang T, Zhang J, You X, Liu Q, Du Y, Gao Y, et al. Hepatitis B virus X protein modulates oncogene Yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology. 2012;56:2051–9. doi: 10.1002/hep.25899. [DOI] [PubMed] [Google Scholar]
- 82.Maguire H, Hoeffler J, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–4. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 83.Chan C, Wang Y, Chow PKH, Chung AYF, Ooi LLPJ, Lee CG. Altered Binding Site Selection of p53 Transcription Cassettes by Hepatitis B Virus X Protein. Molecular and Cellular Biology. 2013;33:485–97. doi: 10.1128/MCB.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee H, Kim HT, Yun Y. Liver-specific enhancer II is the target for the p53-mediated inhibition of hepatitis B viral gene expression. The Journal of biological chemistry. 1998;273:19786–91. doi: 10.1074/jbc.273.31.19786. [DOI] [PubMed] [Google Scholar]
- 85.Lee SG, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene. 2000;19:468–71. doi: 10.1038/sj.onc.1203312. [DOI] [PubMed] [Google Scholar]
- 86.van Breugel PC, Robert EI, Mueller H, Decorsiere A, Zoulim F, Hantz O, et al. Hepatitis B virus X protein stimulates gene expression selectively from extrachromosomal DNA templates. Hepatology. 2012;56:2116–24. doi: 10.1002/hep.25928. [DOI] [PubMed] [Google Scholar]
- 87.Hodgson AAJ, Hyser JM, Keasler VV, Cang Y, Slagle BL. Hepatitis B virus regulatory HBx protein binding to DDB1 is required but is not sufficient for maximal HBV replication. Virology. 2012;426:73–82. doi: 10.1016/j.virol.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B Virus X Protein Stimulates Viral Genome Replication via a DDB1-Dependent Pathway Distinct from That Leading to Cell Death. Journal of virology. 2005;79:4238–45. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bontron S, Lin-Marq N, Strubin M. Hepatitis B Virus X Protein Associated with UV-DDB1 Induces Cell Death in the Nucleus and Is Functionally Antagonized by UV-DDB2. Journal of Biological Chemistry. 2002;277:38847–54. doi: 10.1074/jbc.M205722200. [DOI] [PubMed] [Google Scholar]
- 90.Becker SA, Lee T-H, Butel JS, Slagle BL. Hepatitis B Virus X Protein Interferes with Cellular DNA Repair. Journal of virology. 1998;72:266–72. doi: 10.1128/jvi.72.1.266-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin-Lluesma S, Schaeffer C, Robert EI, van Breugel PC, Leupin O, Hantz O, et al. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology. 2008;48:1467–76. doi: 10.1002/hep.22542. [DOI] [PubMed] [Google Scholar]
- 92.Elmore LW, Hancock AR, Chang S-F, Wang XW, Chang S, Callahan CP, et al. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proceedings of the National Academy of Sciences. 1997;94:14707–12. doi: 10.1073/pnas.94.26.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish JA. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. Journal of virology. 1995;69:1851–9. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Su F, Schneider RJ. Hepatitis B virus HBx protein activates transcription factor NF-kappaB by acting on multiple cytoplasmic inhibitors of rel-related proteins. Journal of virology. 1996;70:4558–66. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waris G, Huh KW, Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol Cell Biol. 2001;21:7721–30. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yun C, Um HR, Jin YH, Wang JH, Lee MO, Park S, et al. NF-kappaB activation by hepatitis B virus X (HBx) protein shifts the cellular fate toward survival. Cancer letters. 2002;184:97–104. doi: 10.1016/s0304-3835(02)00187-8. [DOI] [PubMed] [Google Scholar]
- 97.Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10350–4. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee YI, Kang-Park S, Do S-I, Lee YI. The Hepatitis B Virus-X Protein Activates a Phosphatidylinositol 3-Kinase-dependent Survival Signaling Cascade. Journal of Biological Chemistry. 2001;276:16969–77. doi: 10.1074/jbc.M011263200. [DOI] [PubMed] [Google Scholar]
- 99.Klein NP, Schneider RJ. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol Cell Biol. 1997;17:6427–36. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klein NP, Bouchard MJ, Wang LH, Kobarg C, Schneider RJ. Src kinases involved in hepatitis B virus replication. The EMBO journal. 1999;18:5019–27. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cha MY, Kim CM, Park YM, Ryu WS. Hepatitis B virus X protein is essential for the activation of Wnt/beta-catenin signaling in hepatoma cells. Hepatology. 2004;39:1683–93. doi: 10.1002/hep.20245. [DOI] [PubMed] [Google Scholar]
- 102.Hsieh A, Kim HS, Lim SO, Yu DY, Jung G. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/beta-catenin signaling. Cancer letters. 2011;300:162–72. doi: 10.1016/j.canlet.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 103.Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Molecular cell. 2005;19:159–70. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 104.Tian Y, Yang W, Song J, Wu Y, Ni B. HBV X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Molecular and Cellular Biology. 2013 doi: 10.1128/MCB.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771–8. doi: 10.1158/0008-5472.CAN-07-0529. [DOI] [PubMed] [Google Scholar]
- 106.Tong A, Gou L, Lau QC, Chen B, Zhao X, Li J, et al. Proteomic profiling identifies aberrant epigenetic modifications induced by hepatitis B virus X protein. Journal of proteome research. 2009;8:1037–46. doi: 10.1021/pr8008622. [DOI] [PubMed] [Google Scholar]
- 107.Zhu YZ, Zhu R, Fan J, Pan Q, Li H, Chen Q, et al. Hepatitis B virus X protein induces hypermethylation of p16(INK4A) promoter via DNA methyltransferases in the early stage of HBV-associated hepatocarcinogenesis. Journal of viral hepatitis. 2010;17:98–107. doi: 10.1111/j.1365-2893.2009.01156.x. [DOI] [PubMed] [Google Scholar]
- 108.Kong G, Zhang J, Zhang S, Shan C, Ye L, Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PloS one. 2011;6:e19518. doi: 10.1371/journal.pone.0019518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer biology & therapy. 2010;9:803–8. doi: 10.4161/cbt.9.10.11440. [DOI] [PubMed] [Google Scholar]
- 110.Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PloS one. 2012;7:e35331. doi: 10.1371/journal.pone.0035331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–9. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 112.Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M, et al. Epigenetic regulation of miR-122 by PPARgamma and hepatitis B virus X protein in hepatocellular carcinoma cells. Hepatology. 2013 doi: 10.1002/hep.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. The Journal of clinical investigation. 2012;122:2871–83. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, et al. Hepatitis B Virus Inhibits Apoptosis of Hepatoma Cells by Sponging the MicroRNA 15a/16 Cluster. Journal of virology. 2013;87:13370–8. doi: 10.1128/JVI.02130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu X-D. Hepatitis B Viral RNA Directly Mediates Down-regulation of the Tumor Suppressor MicroRNA miR-15a/miR-16-1 in Hepatocytes. Journal of Biological Chemistry. 2013;288:18484–93. doi: 10.1074/jbc.M113.458158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Houghton M. The long and winding road leading to the identification of the hepatitis C virus. Journal of hepatology. 2009;51:939–48. doi: 10.1016/j.jhep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 117.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. Journal of hepatology. 2006;45:529–38. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 118.Tanaka Y, Hanada K, Orito E, Akahane Y, Chayama K, Yoshizawa H, et al. Molecular evolutionary analyses implicate injection treatment for schistosomiasis in the initial hepatitis C epidemics in Japan. Journal of hepatology. 2005;42:47–53. doi: 10.1016/j.jhep.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 119.Tanaka H, Imai Y, Hiramatsu N, Ito Y, Imanaka K, Oshita M, et al. Declining incidence of hepatocellular carcinoma in Osaka, Japan, from 1990 to 2003. Ann Intern Med. 2008;148:820–6. doi: 10.7326/0003-4819-148-11-200806030-00004. [DOI] [PubMed] [Google Scholar]
- 120.Salomon JA, Weinstein MC, Hammitt JK, Goldie SJ. Empirically calibrated model of hepatitis C virus infection in the United States. American journal of epidemiology. 2002;156:761–73. doi: 10.1093/aje/kwf100. [DOI] [PubMed] [Google Scholar]
- 121.Lok AS, Seeff LB, Morgan TR, di Bisceglie AM, Sterling RK, Curto TM, et al. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136:138–48. doi: 10.1053/j.gastro.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ikeda K, Marusawa H, Osaki Y, Nakamura T, Kitajima N, Yamashita Y, et al. Antibody to hepatitis B core antigen and risk for hepatitis C-related hepatocellular carcinoma: a prospective study. Ann Intern Med. 2007;146:649–56. doi: 10.7326/0003-4819-146-9-200705010-00008. [DOI] [PubMed] [Google Scholar]
- 123.Lok AS, Everhart JE, Di Bisceglie AM, Kim HY, Hussain M, Morgan TR. Occult and previous hepatitis B virus infection are not associated with hepatocellular carcinoma in United States patients with chronic hepatitis C. Hepatology. 2011;54:434–42. doi: 10.1002/hep.24257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baffy G, Brunt EM, Caldwell SH. Hepatocellular carcinoma in non-alcoholic fatty liver disease: an emerging menace. Journal of hepatology. 2012;56:1384–91. doi: 10.1016/j.jhep.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 125.Yang JD, Harmsen WS, Slettedahl SW, Chaiteerakij R, Enders FT, Therneau TM, et al. Factors that affect risk for hepatocellular carcinoma and effects of surveillance. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2011;9:617–23. e1. doi: 10.1016/j.cgh.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 126.Ishiguro S, Inoue M, Tanaka Y, Mizokami M, Iwasaki M, Tsugane S. Serum aminotransferase level and the risk of hepatocellular carcinoma: a population-based cohort study in Japan. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2009;18:26–32. doi: 10.1097/CEJ.0b013e3282fa9edd. [DOI] [PubMed] [Google Scholar]
- 127.Kumada T, Toyoda H, Kiriyama S, Sone Y, Tanikawa M, Hisanaga Y, et al. Incidence of hepatocellular carcinoma in hepatitis C carriers with normal alanine aminotransferase levels. Journal of hepatology. 2009;50:729–35. doi: 10.1016/j.jhep.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 128.Lee MH, Yang HI, Lu SN, Jen CL, Yeh SH, Liu CJ, et al. Hepatitis C virus seromarkers and subsequent risk of hepatocellular carcinoma: long-term predictors from a community-based cohort study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:4587–93. doi: 10.1200/JCO.2010.29.1500. [DOI] [PubMed] [Google Scholar]
- 129.Cardoso AC, Moucari R, Figueiredo-Mendes C, Ripault MP, Giuily N, Castelnau C, et al. Impact of peginterferon and ribavirin therapy on hepatocellular carcinoma: incidence and survival in hepatitis C patients with advanced fibrosis. Journal of hepatology. 2010;52:652–7. doi: 10.1016/j.jhep.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 130.Morgan TR, Ghany MG, Kim HY, Snow KK, Shiffman ML, De Santo JL, et al. Outcome of sustained virological responders with histologically advanced chronic hepatitis C. Hepatology. 2010;52:833–44. doi: 10.1002/hep.23744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu ML, Lin SM, Chuang WL, Dai CY, Wang JH, Lu SN, et al. A sustained virological response to interferon or interferon/ribavirin reduces hepatocellular carcinoma and improves survival in chronic hepatitis C: a nationwide, multicentre study in Taiwan. Antiviral therapy. 2006;11:985–94. [PubMed] [Google Scholar]
- 132.Luedde T, Schwabe RF. NF-kappaB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nature reviews Gastroenterology & hepatology. 2011;8:108–18. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weber A, Boege Y, Reisinger F, Heikenwalder M. Chronic liver inflammation and hepatocellular carcinoma: persistence matters. Swiss medical weekly. 2011;141:w13197. doi: 10.4414/smw.2011.13197. [DOI] [PubMed] [Google Scholar]
- 134.Jesudian AB, de Jong YP, Jacobson IM. Emerging therapeutic targets for hepatitis C virus infection. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2013;11:612–9. e1. doi: 10.1016/j.cgh.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 135.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–3. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 136.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 137.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature medicine. 2005;11:791–6. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, et al. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. Journal of medical virology. 2001;64:334–9. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 139.Li YP, Ramirez S, Gottwein JM, Scheel TK, Mikkelsen L, Purcell RH, et al. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1101–10. doi: 10.1073/pnas.1203829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li YP, Ramirez S, Jensen SB, Purcell RH, Gottwein JM, Bukh J. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19757–62. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yi M, Lemon SM. Genotype 1a HCV (H77S) infection system. Methods in molecular biology. 2009;510:337–46. doi: 10.1007/978-1-59745-394-3_25. [DOI] [PubMed] [Google Scholar]
- 142.Rice CM. New insights into HCV replication: potential antiviral targets. Topics in antiviral medicine. 2011;19:117–20. [PMC free article] [PubMed] [Google Scholar]
- 143.Lanford RE, Lemon SM, Walker C. The chimpanzee model of hepatitis C infections and small animal surrogates. Hepatitis C Antiviral Drug Discovery & Development. 2011:99–132. [Google Scholar]
- 144.Rusyn I, Lemon SM. Mechanisms of HCV-induced liver cancer: What did we learn from in vitro and animal studies? Cancer letters. 2013 doi: 10.1016/j.canlet.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lerat H, Higgs M, Pawlotsky JM. Animal models in the study of hepatitis C virus-associated liver pathologies. Expert review of gastroenterology & hepatology. 2011;5:341–52. doi: 10.1586/egh.11.14. [DOI] [PubMed] [Google Scholar]
- 146.Tsutsumi T, Suzuki T, Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, et al. Alteration of intrahepatic cytokine expression and AP-1 activation in transgenic mice expressing hepatitis C virus core protein. Virology. 2002;304:415–24. doi: 10.1006/viro.2002.1702. [DOI] [PubMed] [Google Scholar]
- 147.Klopstock N, Katzenellenbogen M, Pappo O, Sklair-Levy M, Olam D, Mizrahi L, et al. HCV tumor promoting effect is dependent on host genetic background. PloS one. 2009;4:e5025. doi: 10.1371/journal.pone.0005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Furutani T, Hino K, Okuda M, Gondo T, Nishina S, Kitase A, et al. Hepatic iron overload induces hepatocellular carcinoma in transgenic mice expressing the hepatitis C virus polyprotein. Gastroenterology. 2006;130:2087–98. doi: 10.1053/j.gastro.2006.02.060. [DOI] [PubMed] [Google Scholar]
- 149.Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antiviral research. 2008;80:231–8. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 150.Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, Kneteman N, et al. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS pathogens. 2009;5:e1000291. doi: 10.1371/journal.ppat.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–44. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alam SS, Nakamura T, Naganuma A, Nozaki A, Nouso K, Shimomura H, et al. Hepatitis C virus quasispecies in cancerous and noncancerous hepatic lesions: the core protein-encoding region. Acta medica Okayama. 2002;56:141–7. doi: 10.18926/AMO/31716. [DOI] [PubMed] [Google Scholar]
- 153.Sobesky R, Feray C, Rimlinger F, Derian N, Dos Santos A, Roque-Afonso AM, et al. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology. 2007;46:1704–12. doi: 10.1002/hep.21898. [DOI] [PubMed] [Google Scholar]
- 154.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969–83. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nature medicine. 2013;19:859–68. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yu GY, He G, Li CY, Tang M, Grivennikov S, Tsai WT, et al. Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Molecular cell. 2012;48:313–21. doi: 10.1016/j.molcel.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, et al. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–75. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 158.Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. The Journal of biological chemistry. 2005;280:37481–8. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 159.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18159–64. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kao CF, Chen SY, Chen JY, Wu Lee YH. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene. 2004;23:2472–83. doi: 10.1038/sj.onc.1207368. [DOI] [PubMed] [Google Scholar]
- 161.Deng L, Nagano-Fujii M, Tanaka M, Nomura-Takigawa Y, Ikeda M, Kato N, et al. NS3 protein of Hepatitis C virus associates with the tumour suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. The Journal of general virology. 2006;87:1703–13. doi: 10.1099/vir.0.81735-0. [DOI] [PubMed] [Google Scholar]
- 162.Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. Journal of virology. 2001;75:1401–7. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, et al. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. Journal of hepatology. 2009;51:853–64. doi: 10.1016/j.jhep.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 164.Street A, Macdonald A, McCormick C, Harris M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. Journal of virology. 2005;79:5006–16. doi: 10.1128/JVI.79.8.5006-5016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Milward A, Mankouri J, Harris M. Hepatitis C virus NS5A protein interacts with beta-catenin and stimulates its transcriptional activity in a phosphoinositide-3 kinase-dependent fashion. The Journal of general virology. 2010;91:373–81. doi: 10.1099/vir.0.015305-0. [DOI] [PubMed] [Google Scholar]
- 166.Ariumi Y, Kuroki M, Dansako H, Abe K-I, Ikeda M, Wakita T, et al. The DNA Damage Sensors Ataxia-Telangiectasia Mutated Kinase and Checkpoint Kinase 2 Are Required for Hepatitis C Virus RNA Replication. Journal of virology. 2008;82:9639–46. doi: 10.1128/JVI.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lai C-K, Jeng K-S, Machida K, Cheng Y-S, Lai MMC. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology. 2008;370:295–309. doi: 10.1016/j.virol.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 168.Choi SH, Hwang SB. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. The Journal of biological chemistry. 2006;281:7468–78. doi: 10.1074/jbc.M512438200. [DOI] [PubMed] [Google Scholar]
- 169.Pavio N, Battaglia S, Boucreux D, Arnulf B, Sobesky R, Hermine O, et al. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-[beta] pathway. Oncogene. 2005;24:6119–32. doi: 10.1038/sj.onc.1208749. [DOI] [PubMed] [Google Scholar]
- 170.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 171.Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV Infection by Targeting MicroRNA. New England Journal of Medicine. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 172.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. The Journal of clinical investigation. 2012;122:2884–97. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. Journal of hepatology. 2013;58:234–9. doi: 10.1016/j.jhep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 174.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nature medicine. 2009;15:31–3. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- 175.Han ZG. Functional genomic studies: insights into the pathogenesis of liver cancer. Annual review of genomics and human genetics. 2012;13:171–205. doi: 10.1146/annurev-genom-090711-163752. [DOI] [PubMed] [Google Scholar]