Abstract
Mitochondria grow, divide, and fuse in cells. Mitochondrial division is critical for the maintenance of the structure and function of mitochondria. Alterations in this process have been linked to many human diseases, including peripheral neuropathies and aging-related neurological disorders. In this review, we discuss recent progress in mitochondrial division by focusing on molecular and in vivo analyses of the evolutionarily conserved, central component of mitochondrial division, dynamin-related protein 1 (Drp1), in the yeast and mouse model organisms.
Keywords: Mitochondria, Membrane dynamics, Membrane fission, Dynamin-related GTPase, Neurodegeneration, Yeast, Mice
1. Introduction
Mitochondria play important roles in many cellular and physiological functions, including energy production, lipid and amino acid metabolism, body temperature control, calcium signaling, and cell death [1, 2]. In many cell types, mitochondria consist of short tubular structures with occasional branches and are distributed throughout the cytoplasm. Mitochondria increase in size by importing proteins, nucleic acids, and lipids into one of four compartments - the outer membrane, inner membrane, intermembrane space, and matrix. In addition to growth, mitochondria divide and fuse to control their size, morphology, and number. Mitochondrial division and fusion are highly regulated under different physiological conditions and are mediated by three conserved dynamin-related GTPases. Dynamin-related protein 1 (Drp1) controls mitochondrial division, whereas mitofusin and optic atrophy 1 (Opa1) drive fusion [3-6]. Recent studies have shown that aberrations in these dynamic processes are associated with many human disorders [7-10]. A mutation in Drp1 leads to postneonatal death with developmental defects in the brain and eye [11]. Mutations in mitofusin 2 and Opa1 cause Charcot-Marie-Tooth disease type 2A and dominant optic atrophy 1, respectively. In addition, changes in mitochondrial division and fusion have been suggested to play major roles in the pathogenesis of aging-related diseases, such as Alzheimer's disease, Huntington's disease, and Parkinson's disease [7, 8, 10]. In this review, we will discuss recent findings on the physiological function of mitochondrial division from studies using genetic model organisms, such as yeast and mice. Mitochondrial fusion is reviewed in another article in this issue.
2. Molecular analysis of Drp1
The central components of mitochondrial division, Drp1 in mammals and Dnm1 in yeast, are high molecular weight GTPases that belong to the dynamin protein family [4, 12, 13]. Classic dynamin functions in membrane scission during vesicle formation at the plasma membrane and Golgi complex, whereas Drp1, mitofusin, and Opa1 function in membrane remodeling in mitochondria [14-18]. The majority of Drp1 molecules forms soluble dimers and tetramers in the cytosol, which are recruited to sites of mitochondrial division through interactions with several mitochondrial outer membrane proteins, including mitochondrial fission factor (Mff), fission 1 (Fis1), and two homologous proteins, mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51). MiD49 and Mid51 are also called mitochondrial elongation factor 1 and 2 (MIEF1 and 2). Mff, Fis1 and MiD/MIEF are discussed in more detail below (2. 3. Receptors for Dnm1 and Drp1).
The diameter of mitochondrial tubules (~300 nm) is larger than the necks of coated pits (~50 nm) formed during endocytosis at the plasma membrane. Consistent with the larger diameter of mitochondria, purified Drp1 assembles into spirals with a diameter (~100 nm) larger than that of dynamin spirals (~50 nm) [19, 20]. However, Drp1 spirals are still too narrow to encircle mitochondria by themselves, suggesting involvement of mechanisms upstream of Drp1-dependent constriction. Recent studies have shown that another intracellular organelle, the ER, mediates a step prior to those of Drp1 and Dnm1.
2. 1. Structure and domains
Drp1 and its yeast homolog, Dnm1, contain four domains: an N-terminal GTPase domain, middle domain, variable domain (a.k.a. B domain), and C-terminal GTPase effector domain (GED) [12, 13]. The GTPase domain hydrolyzes GTP and regulates self-assembly of Drp1 and Dnm1 [21, 22]. GTP simulates polymerization of Drp1/Dnm1 and, in turn, the GTPase activity is enhanced in assembled Drp1. During GTP hydrolysis, the diameters of Drp1/Dnm1 spirals decrease which potentially drives constriction of mitochondria.
The middle domain is important for the self-assembly of Drp1 into dimers and tetramers, as well as higher-order oligomers. Crystal structures of dynamin and Drp1 show that three helices from the middle domain and one from the GED form a four-helix bundle that mediates Drp1-Drp1 interactions at the dimerization interface [23-25]. The surface of the bundle located opposite the dimerization interface may bring two dimers together to form tetramers and mediate further oligomerization of Drp1. Consistent with the structural data, mutations in this domain block oligomerization of Drp1 and, thereby, assembly-stimulated GTPase activity [26]. A spontaneous dominant negative mutation in this domain (A395D) leads to postneonatal death in humans, with defective brain development [11].
The variable domain, which is located next to the middle domain, is less conserved among different organisms [27]. In Dnm1, the corresponding region contains the pleckstrin homology (PH) domain and targets Dnm1 to the plasma membrane through interactions with the phosphoinositide PIP2 [14, 15]. Drp1 binds to a mitochondria-specific phospholipid, cardiolipin; however, the interaction depends on a positively-charged residue in the GTPase domain [28]. The role of the variable domain in cardiolipin association remains to be determined. The variable domain may, therefore, function in providing the specificity for the localization of Drp1 to mitochondria through proteins located in the mitochondrial outer membrane; however, its molecular function is unclear.
The C-terminal GED folds back and binds to the GTPase domain and stimulates GTPase activity [29, 30]. Unlike other GTPases, such as ras-related GTPases, no guanine nucleotide exchange factors (GEFs) or GTPase activating proteins (GAPs) have been identified for Dnm1 and Drp1, perhaps due to the GED and relatively low affinities for GDP. Self-assembly of Dnm1 carrying a mutation in this domain is increased in yeast, perhaps as a results of slowed disassembly of oligomers, due to reduced GTPase activity [30]. In contrast, when expressed in mammalian cells, GED mutants connect mitochondrial tubules, suggesting a defect in mitochondrial division [29]. Part of the GED contributes a helix to the self-association interface together with the middle domain.
2. 2. Isoforms
The Drp1 gene contains three alternative exons and expresses multiple isoforms [31-33]. One of the alternative exons is located in the GTPase domain, and the other two are in the variable domain [34]. Whereas most Drp1 isoforms are present in the cytosol and mitochondria, isoforms expressing the third, but not the second, alternative exon are associated with microtubules [34]. Percentages of the microtubule-associated Drp1 isoforms vary depending on cell type, with immune cells expressing high levels of these forms. Instead of engaging mitochondrial fission, these two isoforms stabilize the microtubule cytoskeleton, and this activity is regulated via phosphorylation of Drp1 by cyclin-dependent protein kinase [34]. Because the microtubule-associated isoforms and mitochondria-associated isoforms copolymerize, these different isoforms may function cooperatively to anchor mitochondria to microtubules or to use microtubules as templates for Drp1 spirals to encircle mitochondria for division.
2. 3. Receptors for Dnm1 and Drp1
Because yeast Dnm1 and mammalian Drp1 are present as unassembled forms in the cytosol, key mechanisms for mitochondrial division are mitochondrial recruitment of Dnm1/Drp1 and their subsequent assembly. These steps are mediated by their receptors and adapters located in the outer membrane.
2. 3. 1. Yeast
Dnm1 is recruited to mitochondria through the outer membrane protein Fis1 and two homologous, functionally-redundant adaptor proteins, Mdv1 and Caf4 [35-38]. Both proteins lack transmembrane domains, but are constitutively associated with the outer membrane through Fis1. The adapter proteins assemble Dnm1 on the mitochondrial outer membrane. Purified Mdv1 stimulates the assembly of Dnm1 in vitro [39]. Therefore, the role of Fis1 may simply be to anchor Mdv1 to mitochondria. In support of this model, addition of a transmembrane domain of a mitochondrial outer membrane protein to Mdv1 bypasses the requirement of Fis1 for Dnm1 recruitment and mitochondrial division [40].
Dnm1 is also attached to mitochondria through another mechanism for mitochondrial division and distribution. This mechanism involves a cortical protein, Num1, which binds to the cortical actin cytoskeleton, and Mdm36, which connects Dnm1 to Num1 [41, 42]. In budding yeast, short actin filaments form the actin cortex beneath the plasma membrane [43]. Yeast mitochondria are mainly associated with the actin cytoskeleton, in contrast to mammalian mitochondria, which are associated with microtubules [44]. The Dnm1-Mdm36-Num1 complex is required for both mitochondrial division and mitochondrial anchoring to the actin cortex [41, 42]. In particular, this complex functions in the retention of mitochondria in mother cells [41, 45]. Interestingly, this anchoring mechanism can be substituted by synthetic linker molecules that connect mitochondria to either the plasma membrane or the cortical ER, suggesting that mitochondrial dynamics and distribution coordinate with other cellular structures [42, 45].
2. 3. 2. Mammals
To recruit cytosolic Drp1 to the site of mitochondrial division in mammalian cells, several outer membrane proteins have been identified, including Mff, Fis1, MiD49, and MiD51. These individual transmembrane proteins are diverse in their amino acid sequences and topologies. Supporting their roles as receptor proteins, knockout and knockdown of Mff, Fis1, and MiDs decreases mitochondrial fission and elongates mitochondria. The combined loss of these proteins additively affects mitochondrial division [46]. Interestingly, when Drp1 is expressed together with Mff or MiD49 in yeast cells null for DNM1, FIS1, MDV1 and CAF4, four genes for mitochondrial division, each pair of proteins can induce mitochondrial fission [40]. These data suggest that Mff, Fis1, and MiDs function in mitochondrial fission independently. These Drp1-recruiting proteins may be regulated under different conditions in response to diverse physiological cues. Unlike yeast, no adaptor proteins have been identified for Drp1 in mammals. It appears that Drp1 receptor proteins have the ability to assemble on mitochondria after recruitment of Drp1.
Mff is a 33-kDa protein anchored to the outer membrane via its C-terminal transmembrane domain [47, 48]. Mff forms punctate structures along mitochondrial tubules, and many of these are located at the contact site between mitochondria and the ER [49] (roles of the contact sites in mitochondrial division is discussed below - 2.4. Constriction of mitochondria by the ER and actin cytoskeleton). Purified Mff slightly enhances the GTPase activity of Drp1 [40].
MiD49 and 51 (MIEF1 and 2) are homologous proteins of ~50 kDa and are inserted into the mitochondrial outer membrane via an N-terminal transmembrane domain [50, 51]. The role of these proteins has been controversial, and it has been suggested that they block mitochondrial division by sequestering Drp1 at the mitochondrial surface in nonfunctional forms [51, 52]. Purified MiD proteins co-assemble with Drp1, slightly stimulate the GTPase activity of Drp1, and modify the structure of Drp1 filaments [40].
Fis1 is a 17-kDa protein that is also anchored to the outer membrane via its C-terminal tail and is uniformly distributed along mitochondrial tubules [53-55]. The exact function of Fis1 is unknown, and its contribution to mitochondrial division appears to be smaller than those of Mff and MiD proteins under basal conditions [46]. Interaction of Fis1 with Drp1 changes upon phosphorylation of Drp1 in response to extracellular stimuli; therefore, Fis1 may function under specific physiological conditions that stimulate mitochondrial division during the cell cycle, hyperglycemia, or neuronal stimulation [56, 57].
2. 4. Constriction of mitochondria by the ER and actin cytoskeleton
Mitochondria and the ER form a structural connection called mitochondria-ER contact sites (a.k.a. mitochondria-associated membrane) [58]. It has been shown that mitochondria and the ER communicate through calcium signaling at the contact sites and through phospholipid transfer during lipid biosynthesis. In addition to these functions, recent studies have suggested the ER promotes mitochondrial division at the interorganelle contact site [49]. ER tubules surround mitochondrial tubules and appear to squeeze mitochondria. In addition, an ER-associated protein, inverted formin 2 (INF2), plays important roles in polymerization of actin filaments at the contact site and recruitment of Drp1 to mitochondria [59]. There are two isoforms of INF2; one is a soluble, cytosolic form and the other is anchored to the ER membrane by prenylation at the CAAX motif of the C-terminus. Formin is an actin-binding protein and promotes actin nucleation. It has been suggested that the ER-associated form of INF2 polymerizes actin filaments, the actin cytoskeleton mediates initial constriction of mitochondria, and then Drp1 drives completion of the constriction, physically separating two daughter mitochondria. The involvement of the actin cytoskeleton in mitochondrial division is reminiscent of actin polymerization at the coated pits to generate endocytic vesicles by dynamins.
Implying a role of ER-mediated mitochondrial division in human disease, mutations in the INF2 gene can cause dominant intermediate Charcot-Marie-Tooth neuropathy, which leads to degeneration of motor and sensory neurons. Other genes related to mitochondrial division and fusion, such as GDAP1 and mitofusin 2, are also mutated in Charcot-Marie-Tooth diseases. Similar to INF2, GDAP1 is located and functions at the mitochondria-ER contact site [60], whereas mitofusin 2 connects these two organelles [61], in addition to its function in mitochondrial fusion. Therefore, Charcot-Marie-Tooth diseases caused by mutations in INF2, mitofusin 2, and GDAP1 might result from defects in mitochondrial division at the mitochondria-ER contacts sites. Deciphering the functional relationship between these contact site proteins is important and awaits further studies.
Moreover, involvement of the actin cytoskeleton in mitochondrial division has been implied as part of the pathogenesis of Alzheimer’s disease. Hyperphosphorylation and proteolytic cleavage of a microtubule-binding protein, tau, lead to its aggregation in Alzheimer’s disease. Overexpression of truncated versions of tau fragments mitochondria [62]. Similarly, fragmentation of mitochondria has been observed in mouse models for Alzheimer's disease which accumulate amyloid-β [63]. Interestingly, overexpression of tau causes stabilization of the actin cytoskeleton and elongation of mitochondria possibly by sequestering Drp1 from mitochondria to actin filaments [64, 65]. Therefore, endogenous tau may negatively regulate interactions of Drp1 with mitochondria through the actin cytoskeleton while hyperphosphorylation and proteolytic cleavage of tau cause activation of Drp1 in Alzheimer's disease. Supporting this idea, Drp1 was associated with phosphorylated tau [66].
3. Physiological studies of Drp1
3.1. Yeast
Mitochondrial morphology
In budding yeast, mitochondria continuously fuse and divide during growth, sporulation, and mating (Figure 1). The loss of Dnm1 connects mitochondrial tubules and generates a single net-like structure [67-69]. Despite the dramatic morphological changes, there are no obvious growth defects in dnm1Δ cells. dnm1Δ mitochondria maintain their respiratory function and are inherited normally by daughter cells. During mitosis, a portion of mitochondria extends toward daughter cells and is divided between daughter and mother cells by an unknown mechanism. This division process occurs without loss of mitochondrial function. The force of cytokinesis may separate mitochondria. However, during meiosis, mitochondrial segregation into spores is more dependent on Dnm1, and dnm1Δ cells are defective in proper segregation of mitochondria into four spores [70].
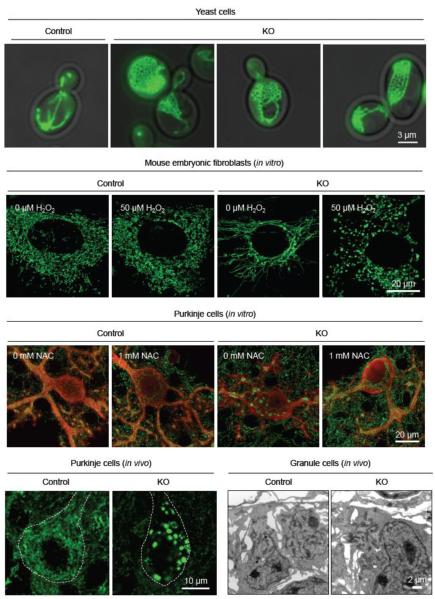
Figure 1. Mitochondrial morphology in yeast and mammalian cells.
Mitochondria were visualized in yeast cells (wild-type and dnm1Δ), cultured mouse embryonic fibroblasts (MEFs) (wild-type and Drp1KO) and cultured Purkinje cells in vitro (wild-type and Drp1KO). Mitochondria were also examined in Purkinje cells and granule cells in vivo. Mitochondria formed short tubules in wild-type cells and net-like structures in yeast dnm1Δ cells. In Drp1KO MEFs, elongated mitochondrial tubules are highly connected. Upon addition of reactive oxygen species (50 μM H2O2) to the culture medium, elongated mitochondria became large spheres in Drp1KO MEFs. In Drp1KO Purkinje cells, mitochondria formed large spheres, which looked similar to those seen in H2O2-treated Drp1KO MEFs. When Drp1KO Purkinje cells were treated with antioxidants (1 mM N-acetylcysteine, NAC), mitochondria showed elongated tubules without forming spheres, which were similar to mitochondria in Drp1KO MEFs. These observations suggest that the loss of Drp1 induces oxidative damage in Drp1KO Purkinje cells and transforms elongated tubules into large spheres. In Drp1KO granule cells, mitochondria appeared normal. © Wakabayashi et al. (2009) and Kageyama et al. (2013).
Simultaneous loss of Dnm1 and fusion components, such as Fzo1, Ugo1 and Mgm1, restores tubular mitochondria similar in morphology to wild-type mitochondria [67, 71, 72]. In addition to the overall morphology, structure of the inner membrane cristae is lost in cells defective in mitochondrial fusion, but these phenotypes are rescued by additional loss of Drp1 in double mutants lacking both Dnm1 and Mgm1 [72]. Although the exact role of Dnm1 in crista morphology remains to be determined, Dnm1 may regulate inner membrane organization through the intramitochondrial contact site between the outer membrane and inner membrane. Similarly, it has been shown that mammalian Drp1 also remodels inner membrane cristae during apoptosis [73].
Mitochondrial DNA (mtDNA) nucleoids
Mitochondrial division is also important for the morphology of mtDNA nucleoids in the budding yeast. Whereas dnm1Δ cells normally maintain mtDNA nucleoids, when inner membrane crista structure is disorganized and cristae junctions are lost in cells lacking the protein complex containing Fcj1 [74-77], a yeast homolog of mitofilin, the lack of Dnm1 leads to the aggregation of mtDNA nucleoids and loss of mtDNA [78]. Therefore, inner membrane cristae and mitochondrial division may perform overlapping functions to partition mtDNA nucleoids in the matrix. Consistent with this model, mitochondrial division is spatially linked to nucleoids, resulting in their distribution into newly generated tips in the mitochondrial network [79]. Similarly, in the fission yeast, mitochondrial division takes place between two mtDNA nucleoids and appears to ensure that each mitochondrion contains mtDNA. The role of mitochondrial division in mtDNA nucleoids is conserved because Drp1 knockout (KO) mouse embryonic fibroblasts showed clusters of mtDNA [80], and Drp1-depleted cells aggregated mtDNA nucleoids [81].
Mitophagy
Mitophagy has been suggested as a mechanism for removal of damaged mitochondria. Because mitochondria are constantly exposed to oxidative stress, efficient clearance of damaged mitochondria is critical for the maintenance of mitochondrial functional competence. Mitochondria become smaller by division and are engulfed by autophagosomes as dnm1Δ cells showed decreased delivery of mitochondria to lysosomes [82]. In one possible mechanism, the scaffold protein for autophagy Atg11 binds to the mitochondrial outer membrane protein Atp32 and the Atp32-Atg11 complex marks mitochondria for degradation [83-85]. In addition, Atg11 binds to and recruits Dnm1 to promote mitochondrial division [83]. Interestingly, the Dnm1-Atg11-Atg32 complex is located at mitochondria-ER contact sites [83]. Both steady-state mitochondrial division and mitophagy-induced mitochondrial division may occur at the organelle contact site.
Aging
Considering the role of Dnm1 in mitophagy, the lack of mitochondrial division is expected to decrease turnover of mitochondria and, therefore, lead to accumulation of damaged mitochondria. This may potentially result in shortening of life span. However, it is surprising that life span is extended in dnm1Δ cells [86]. In aged wild-type cells, mitochondrial tubules become fragmented, whereas, in dnm1Δ cells, aging-related mitochondrial fragmentation and increases in reactive oxygen species were suppressed [86]. Similarly, extension of life span was observed in drp1 mutants of C. elegans. Knockdown of drp1 leads to dramatic extension of life span in daf16 or age1 mutants, in which insulin signaling is reduced [87]. In contrast, these phenotypes are in contrast to those observed in mammalian neurons, where the loss of mitochondrial division increases oxidative damage in mitochondria and causes cell death due to impaired respiratory function [7, 88, 89].
3.1. Mice
Mitochondrial morphology
In contrast to elongation and excessive connection of mitochondria in dnm1Δ cells, the effect of loss of mammalian Drp1 on mitochondrial morphology varies depending on cell type (Figure 1). In cerebellar Purkinje cells, where Drp1 is highly expressed, the lack of Drp1 transforms organelle morphology in two phases [88]. First, an imbalance between mitochondrial fusion and division results in elongation and connection of mitochondrial tubules. These elongated mitochondria gradually accumulate oxidative damage and undergo additional morphological changes from elongated tubules into large spheres. The formation of mitochondrial spheres can be suppressed by antioxidants, such an N-acetylcysteine and coenzyme Q10 [88]. In contrast, granule cells, another type of neuron in the cerebellum, in which Drp1 levels are much lower than in Purkinje cells [88, 90], maintain normal mitochondrial morphology after the loss of Drp1 [91]. Mitochondria in granule cells may divide at a very low frequency or use Drp1-independent mechanisms for mitochondrial division. In neurons of the forebrain, mitochondria become large spheres similar to those seen in Purkinje cells [92]. Interestingly, mouse embryonic fibroblasts lacking Drp1 show interconnection of elongated tubules, similar to yeast dnm1Δ cells, but enlarged spheres are rarely observed. These morphological differences may be explained by the level of oxidative stress, because addition of reactive oxygen species, such as hydrogen peroxide, transforms elongated tubules into spheres in Drp1KO mouse embryonic fibroblasts [88].
Embryonic and brain development
Drp1KO causes embryonic lethality in mice [91, 92]. Drp1KO embryos die at approximately E 11.5 and are smaller than wild-type embryos, suggesting the requirement of Drp1 for cell growth, proliferation, and differentiation [91]. As a possible cause of the lethality, giant cells are missing in the placenta of Drp1KO embryos. In contrast, blood vessels are not grossly affected [91]. The architecture of the heart appears normal, although isolated cardiomyocytes show decreased beating rate, suggesting that functional defects in the heart may also contribute to embryonic death. Heterozygous Drp1KO mice have levels of Drp1 that are decreased by 20%–30%, but these animals are normal in birth, growth, and mating [91, 93]. Mitochondrial morphology also appears normal in heterozygous Drp1KO mice, suggesting that mitochondrial division is not severely affected [93]. Partial decreases in Drp1 levels are, therefore, tolerated in mice.
Drp1 is highly expressed in the brain. To determine the effect of Drp1KO on brain development, Drp1 was knocked out in the cerebellum and surrounding regions using brain-specific Drp1KO with En1-Cre recombinase and floxed alleles of Drp1 [91]. Development of the cerebellum was dramatically decreased, and the mice died within 24 hours of birth. In Drp1KO cerebella, proliferation of neurons was greatly decreased, and the number of Purkinje cells was dramatically decreased [91]. Similarly, when Drp1 was deleted in a broad region of the brain using Nes-Cre recombinase, brain development was inhibited and many apoptotic cells were observed in the premature, superficial-layer neurons and the deep cortical layers [92]. In neurons isolated from Nes-Cre Drp1KO mice, the size of mitochondria was increased and the number was decreased [92]. In particular, synapses lacked mitochondria, and synapse formation was defective in these neurons in culture [92]. These observations are consistent with findings in Drosophila mutants carrying mutations in Drp1 [94]. drp1 mutant flies failed to distribute mitochondria at the neuromuscular junctions, leading to defects in mobilization of the vesicle reserve pool, likely due to decreased amounts of ATP in this region [94]. As a result, when Drp1 mutants were exposed to high frequency stimulation to induce continuous neurosecretion, the mutants were unable to maintain normal levels of neurotransmission [94]. In mammals, Drp1 may have a more direct role in endocytosis of synaptic vesicles. Drp1 forms protein complexes with components of clathrin-coated vesicles and controls the size of endocytic vesicles in response to synaptic stimulation [95].
The role of Drp1 in apoptosis seems to vary depending on cell type and physiological context. In Drp1KO embryos, developmentally-regulated apoptosis of neural crest cells was decreased during neural tube closure [91]. In contrast, apoptosis was increased in the neuroepithelium of the brain [92]. When apoptosis was examined in isolated Drp1KO mouse embryonic fibroblasts and embryonic stem cells, the release of cytochrome c and apoptotic cell death in response to different death stimuli were not affected in the absence of Drp1 [91, 92]. In culture, Drp1KO neurons are sensitive to apoptotic induction.
It should be noted that a spontaneous dominant negative mutation in human Drp1 in the middle domain resulted in developmental defects in the brain and eye, and postneonatal death one month after birth [11]. The patient had elevated lactate levels in blood and brain fluid, suggesting decreased mitochondrial respiration. However, skin fibroblasts isolated from the patient showed normal respiratory capacity, even though mitochondria were elongated, as expected from failure of division. This observation is consistent with findings in fibroblasts isolated from Drp1KO mouse embryos [91, 92]. Fibroblasts may tolerate the loss of Drp1 and mitochondrial division in terms of mitochondrial functions. The structure and respiratory function of mitochondria in the skeletal muscle of the patient also appeared normal.
Survival of postmitotic neurons
In addition to its role in brain development, Drp1 ensures the survival of postmitotic neurons in mice [88, 89]. When Drp1 is knocked out in postmitotic Purkinje cells in the cerebellum using L7-Cre recombinase, mitochondria accumulated oxidative damage, became defective in respiration, and gradually degenerated over 6 months. As a consequence of the loss of Purkinje cells, L7-Drp1KO mice became defective in motor coordination behavior. Mitochondria became elongated and then became large spheres due to oxidative damage [88, 89]. On the other hand, the distribution of mitochondria was not affected. These spherical mitochondria lacked a subunit of the electron transport chain complex IV, which is encoded by mtDNA, suggesting that the respiratory defect at least partly results from abnormalities in mtDNA. Mitochondria also accumulated components related to mitophagy, such as LC3 and p62, suggesting that mitophagy may be slowed, with accumulation of its intermediates [88, 89]. Similarly, mitochondria are highly decorated with ubiquitin. The E3 ubiquitin ligase parkin has been suggested to ubiquitinate mitochondrial proteins during mitophagy to signal engulfment of mitochondria by autophagosomes [96]. However, when L7-Drp1KO mice were crossed with parkinKO mice, L7-Drp1–parkin double-KO mice maintained ubiquitination of mitochondria in the absence of parkin. Therefore, ubiquitination of mitochondria was mediated by other E3 ubiquitin ligases under these conditions. Another study has also suggested a parkin-independent mechanism for mitophagy [97]. It is of interest to identify the enzymes that ubiquitinate mitochondrial protein during mitophagy. When Drp1KO Purkinje cells were treated with the antioxidants N-acetylcysteine and coenzyme Q10, the degeneration of Drp1KO Purkinje cells was suppressed, suggesting that oxidative damage is a downstream event that leads to neurodegeneration when mitochondrial division is blocked [88, 89].
In addition to mitochondrial division, Drp1 also controls peroxisomal division [98-100]. Consistent with this role, peroxisomes were elongated in Drp1KO mouse embryonic fibroblasts [91]. However, the morphology of peroxisomes was not affected in Drp1KO Purkinje cells [88]. Peroxisomal division may be less dependent on Drp1 in Purkinje cells. The degeneration of Drp1KO Purkinje cells likely results from defects in mitochondria division.
4. Concluding remarks
Studies have shown the physiological importance of mitochondrial division and revealed many aspects of the molecular mechanism underlying this process. The knowledge has helped us better understand the pathogenesis of human diseases that are linked to mitochondrial division. In addition, these studies have raised many important questions. How do signaling mechanisms couple mitochondrial division and mitophagy? What is the exact mechanism by which the ER and actin cytoskeleton constrict mitochondria? What is the mechanism of Drp1-independent mitochondrial division? Many dynamic processes appear to be coordinated in mitochondria. It would be exciting to understand how multiple processes, such as mitochondrial fusion and division, mtDNA nucleoid biogenesis and movement, mitochondrial movement, and degradation coordinate. Addressing these questions will broaden our knowledge of mitochondrial dynamics and human diseases.
Highlights.
-
-
Many neurodegenerative diseases are linked to mitochondrial division.
-
-
Mitochondrial division controls mitochondrial morphology and function.
-
-
Drp1 is a central component of mitochondrial division.
-
-
Genetic studies of Drp1 using yeast and mice enhance understanding of physiological roles of mitochondrial division.
Acknowledgements
We thank many scientists who advanced our understanding of mitochondrial dynamics, and apologize that we were unable to cite all of the relevant research due to space restrictions.
This work was supported by NIH grants to M.I. (GM084015) and H.S. (GM089853 and NS084154).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. Bioenergetic origins of complexity and disease. Cold Spring Harb Symp Quant Biol. 2011;76:1–16. doi: 10.1101/sqb.2011.76.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamura Y, Itoh K, Sesaki H. SnapShot: Mitochondrial dynamics. Cell. 2011;145:1158–1151. doi: 10.1016/j.cell.2011.06.018. 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shutt TE, McBride HM. Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochimica et biophysica acta. 2013;1833:417–424. doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 6.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annual review of genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 7.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends in Cell Biology. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy PH, Reddy TP. Mitochondria as a therapeutic target for aging and neurodegenerative diseases. Curr Alzheimer Res. 2011;8:393–409. doi: 10.2174/156720511795745401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. The New England journal of medicine. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 12.Otera H, Ishihara N, Mihara K. New insights into the function and regulation of mitochondrial fission. Biochimica et biophysica acta. 2013;1833:1256–1268. doi: 10.1016/j.bbamcr.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochimica et biophysica acta. 2013;1833:150–161. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annual review of cell and developmental biology. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase, Nature reviews. Molecular cell biology. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageyama Y, Zhang Z, Sesaki H. Mitochondrial division: molecular machinery and physiological functions. Current opinion in cell biology. 2011;23:427–434. doi: 10.1016/j.ceb.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westermann B. Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Seminars in cell & developmental biology. 2010;21:542–549. doi: 10.1016/j.semcdb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto K, Shaw JM. Mitochondrial Morphology and Dynamics in Yeast and Multicellular Eukaryotes. Annu Rev Genet. 2005 doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 19.Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nature structural & molecular biology. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nature cell biology. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 21.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. The EMBO journal. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 26.Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285:32494–32503. doi: 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strack S, Cribbs JT. Allosteric modulation of Drp1 mechanoenzyme assembly and mitochondrial fission by the variable domain. The Journal of biological chemistry. 2012;287:10990–11001. doi: 10.1074/jbc.M112.342105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, Martinou JC. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]
- 30.Fukushima NH, Brisch E, Keegan BR, Bleazard W, Shaw JM. The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol Biol Cell. 2001;12:2756–2766. doi: 10.1091/mbc.12.9.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon Y, Pitts KR, Dahan S, McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol. 1998;140:779–793. doi: 10.1083/jcb.140.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howng SL, Sy WD, Cheng TS, Lieu AS, Wang C, Tzou WS, Cho CL, Hong YR. Genomic organization, alternative splicing, and promoter analysis of human dynamin-like protein gene. Biochemical and biophysical research communications. 2004;314:766–772. doi: 10.1016/j.bbrc.2003.12.172. [DOI] [PubMed] [Google Scholar]
- 33.Uo T, Dworzak J, Kinoshita C, Inman DM, Kinoshita Y, Horner PJ, Morrison RS. Drp1 levels constitutively regulate mitochondrial dynamics and cell survival in cortical neurons. Experimental neurology. 2009;218:274–285. doi: 10.1016/j.expneurol.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strack S, Wilson TJ, Cribbs JT. Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. The Journal of cell biology. 2013;201:1037–1051. doi: 10.1083/jcb.201210045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated Mitochondrial Fission Is a Multi-step Process Requiring the Novel Integral Membrane Component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerveny KL, Jensen RE. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell. 2003;14:4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1342–1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerveny KL, Studer SL, Jensen RE, Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Developmental cell. 2007;12:363–375. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Hammermeister M, Schodel K, Westermann B. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Molecular biology of the cell. 2010;21:2443–2452. doi: 10.1091/mbc.E10-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Current biology : CB. 2011;21:R560–569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boldogh IR, Yang HC, Pon LA. Mitochondrial inheritance in budding yeast. Traffic. 2001;2:368–374. doi: 10.1034/j.1600-0854.2001.002006368.x. [DOI] [PubMed] [Google Scholar]
- 45.Klecker T, Scholz D, Fortsch J, Westermann B. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. Journal of cell science. 2013;126:2924–2930. doi: 10.1242/jcs.126045. [DOI] [PubMed] [Google Scholar]
- 46.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Molecular biology of the cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO reports. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. The EMBO journal. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T, Yu R, Jin SB, Han L, Lendahl U, Zhao J, Nister M. The mitochondrial elongation factors MIEF1 and MIEF2 exert partially distinct functions in mitochondrial dynamics. Experimental cell research. 2013 doi: 10.1016/j.yexcr.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 55.Jofuku A, Ishihara N, Mihara K. Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem Biophys Res Commun. 2005;333:650–659. doi: 10.1016/j.bbrc.2005.05.154. [DOI] [PubMed] [Google Scholar]
- 56.Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W, Wang Y, Long J, Wang J, Haudek SB, Overbeek P, Chang BH, Schumacker PT, Danesh FR. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 2012;15:186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochimica et biophysica acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 59.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pla-Martin D, Rueda CB, Estela A, Sanchez-Piris M, Gonzalez-Sanchez P, Traba J, de la Fuente S, Scorrano L, Renau-Piqueras J, Alvarez J, Satrustegui J, Palau F. Silencing of the Charcot-Marie-Tooth disease-associated gene GDAP1 induces abnormal mitochondrial distribution and affects Ca2+ homeostasis by reducing store-operated Ca2+ entry. Neurobiology of disease. 2013;55:140–151. doi: 10.1016/j.nbd.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 61.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 62.Quintanilla RA, Dolan PJ, Jin YN, Johnson GV. Truncated tau and Abeta cooperatively impair mitochondria in primary neurons. Neurobiology of aging. 2012;33:619–635. doi: 10.1016/j.neurobiolaging.2011.02.007. e625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie H, Guan J, Borrelli LA, Xu J, Serrano-Pozo A, Bacskai BJ. Mitochondrial Alterations near Amyloid Plaques in an Alzheimer's Disease Mouse Model. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:17042–17051. doi: 10.1523/JNEUROSCI.1836-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DuBoff B, Gotz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75:618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DuBoff B, Feany M, Gotz J. Why size matters - balancing mitochondrial dynamics in Alzheimer's disease. Trends in neurosciences. 2013;36:325–335. doi: 10.1016/j.tins.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Manczak M, Reddy PH. Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer's disease neurons: implications for mitochondrial dysfunction and neuronal damage. Human molecular genetics. 2012;21:2538–2547. doi: 10.1093/hmg/dds072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gorsich SW, Shaw JM. Importance of mitochondrial dynamics during meiosis and sporulation. Mol Biol Cell. 2004;15:4369–4381. doi: 10.1091/mbc.E03-12-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sesaki H, Southard SM, Yaffe MP, Jensen RE. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol Biol Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Germain M, Mathai JP, McBride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. Embo J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. The Journal of cell biology. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. The Journal of cell biology. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, Hutu DP, Zerbes RM, Schulze-Specking A, Meyer HE, Martinou JC, Rospert S, Rehling P, Meisinger C, Veenhuis M, Warscheid B, van der Klei IJ, Pfanner N, Chacinska A, van der Laan M. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Developmental cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 77.Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. The EMBO journal. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itoh K, Tamura Y, Iijima M, Sesaki H. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Molecular biology of the cell. 2013;24:1842–1851. doi: 10.1091/mbc.E13-03-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ban-Ishihara R, Ishihara T, Sasaki N, Mihara K, Ishihara N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11863–11868. doi: 10.1073/pnas.1301951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, Klionsky DJ. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Molecular biology of the cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Developmental cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Developmental cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 85.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Developmental cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nystrom T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 87.Yang CC, Chen D, Lee SS, Walter L. The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate Caenorhabditis elegans longevity. Aging Cell. 2011;10:724–728. doi: 10.1111/j.1474-9726.2011.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. The Journal of cell biology. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z, Kageyama Y, Sesaki H. Mitochondrial division prevents neurodegeneration. Autophagy. 2012;8:1531–1533. doi: 10.4161/auto.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin HW, Takatsu H, Mukai H, Munekata E, Murakami K, Nakayama K. Intermolecular and interdomain interactions of a dynamin-related GTP-binding protein, Dnm1p/Vps1p-like protein. J Biol Chem. 1999;274:2780–2785. doi: 10.1074/jbc.274.5.2780. [DOI] [PubMed] [Google Scholar]
- 91.Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 93.Manczak M, Sesaki H, Kageyama Y, Reddy PH. Dynamin-related protein 1 heterozygote knockout mice do not have synaptic and mitochondrial deficiencies. Biochimica et biophysica acta. 2012;1822:862–874. doi: 10.1016/j.bbadis.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 95.Li H, Alavian KN, Lazrove E, Mehta N, Jones A, Zhang P, Licznerski P, Graham M, Uo T, Guo J, Rahner C, Duman RS, Morrison RS, Jonas EA. A Bcl-xL-Drp1 complex regulates synaptic vesicle membrane dynamics during endocytosis. Nature cell biology. 2013;15:773–785. doi: 10.1038/ncb2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Youle RJ, Narendra DP. Mechanisms of mitophagy, Nature reviews. Molecular cell biology. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 99.Kaur N, Hu J. Dynamics of peroxisome abundance: a tale of division and proliferation. Curr Opin Plant Biol. 2009;12:781–788. doi: 10.1016/j.pbi.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 100.Opalinski L, Veenhuis M, van der Klei IJ. Peroxisomes: membrane events accompanying peroxisome proliferation. The international journal of biochemistry & cell biology. 2011;43:847–851. doi: 10.1016/j.biocel.2011.03.006. [DOI] [PubMed] [Google Scholar]