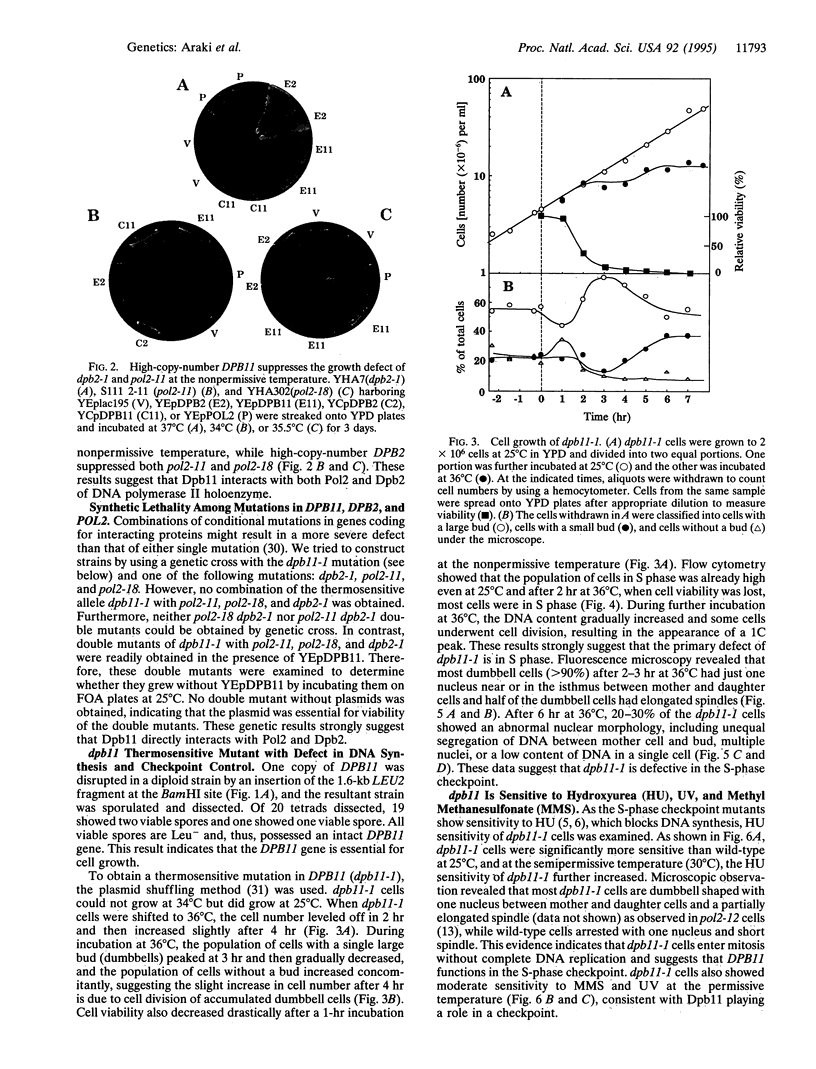
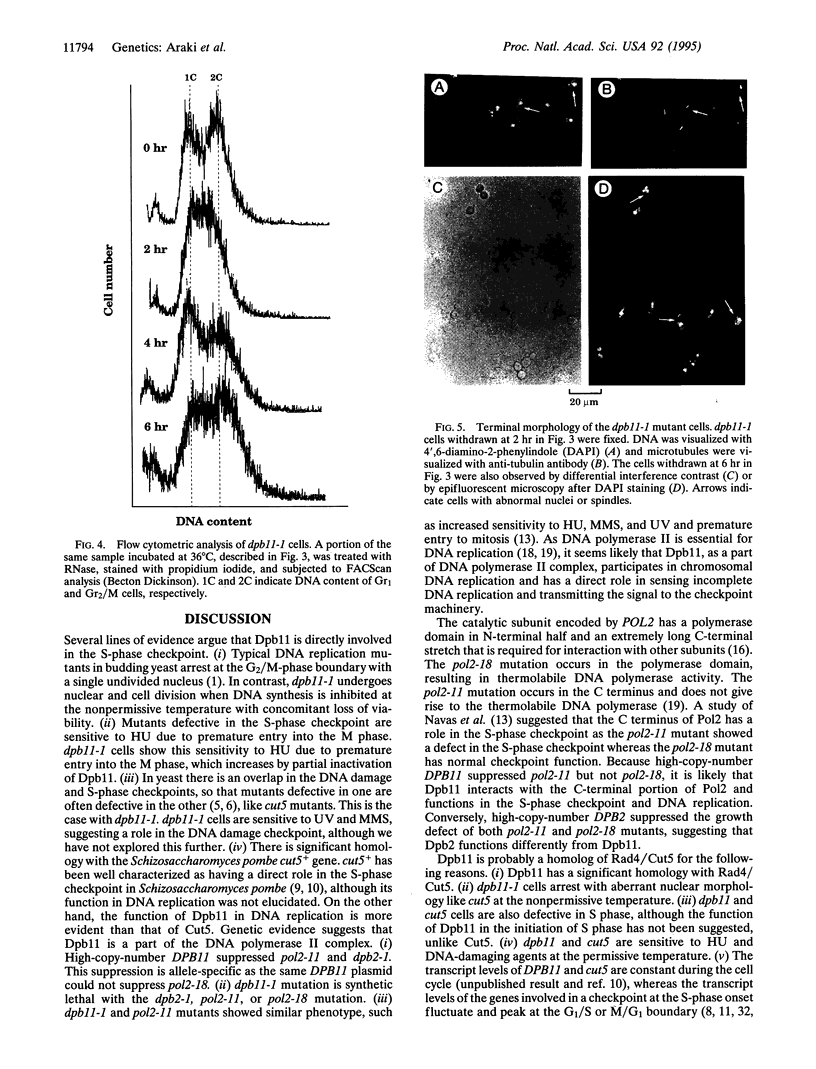
Abstract
DPB11, a gene that suppresses mutations in two essential subunits of Saccharomyces cerevisiae DNA polymerase II(epsilon) encoded by POL2 and DPB2, was isolated on a multicopy plasmid. The nucleotide sequence of the DPB11 gene revealed an open reading frame predicting an 87-kDa protein. This protein is homologous to the Schizosaccharomyces pombe rad4+/cut5+ gene product that has a cell cycle checkpoint function. Disruption of DPB11 is lethal, indicating that DPB11 is essential for cell proliferation. In thermosensitive dpb11-1 mutant cells, S-phase progression is defective at the nonpermissive temperature, followed by cell division with unequal chromosomal segregation accompanied by loss of viability.dpb11-1 is synthetic lethal with any one of the dpb2-1, pol2-11, and pol2-18 mutations at all temperatures. Moreover, dpb11 cells are sensitive to hydroxyurea, methyl methanesulfonate, and UV irradiation. These results strongly suggest that Dpb11 is a part of the DNA polymerase II complex during chromosomal DNA replication and also acts in a checkpoint pathway during the S phase of the cell cycle to sense stalled DNA replication.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki H., Hamatake R. K., Johnston L. H., Sugino A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Hamatake R. K., Morrison A., Johnson A. L., Johnston L. H., Sugino A. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991 Sep 25;19(18):4867–4872. doi: 10.1093/nar/19.18.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Ropp P. A., Johnson A. L., Johnston L. H., Morrison A., Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992 Feb;11(2):733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Kremenstova E., Luo L. Complete transposition requires four active monomers in the mu transposase tetramer. Genes Dev. 1994 Oct 15;8(20):2416–2428. doi: 10.1101/gad.8.20.2416. [DOI] [PubMed] [Google Scholar]
- Berben G., Dumont J., Gilliquet V., Bolle P. A., Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991 Jul;7(5):475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Budd M. E., Campbell J. L. DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):496–505. doi: 10.1128/mcb.13.1.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno A., Russell P. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992 Jun;11(6):2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M., Carr A. M., Murray J., Watts F. Z., Lehmann A. R. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 1991 Dec 25;19(24):6737–6741. doi: 10.1093/nar/19.24.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Guarente L. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 1993 Oct;9(10):362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- Hamatake R. K., Hasegawa H., Clark A. B., Bebenek K., Kunkel T. A., Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J Biol Chem. 1990 Mar 5;265(7):4072–4083. [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 1994 Jan 15;13(2):425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993 Jul 30;74(2):371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Leem S. H., Ropp P. A., Sugino A. The yeast Saccharomyces cerevisiae DNA polymerase IV: possible involvement in double strand break DNA repair. Nucleic Acids Res. 1994 Aug 11;22(15):3011–3017. doi: 10.1093/nar/22.15.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990 May 18;61(4):549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Morrison A., Sugino A. DNA polymerase II, the epsilon polymerase of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1993;46:93–120. doi: 10.1016/s0079-6603(08)61019-3. [DOI] [PubMed] [Google Scholar]
- Murray A. W. Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 1992 Oct 15;359(6396):599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- Navas T. A., Zhou Z., Elledge S. J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995 Jan 13;80(1):29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Fantes P., Sutani T., McInerny C., Creanor J., Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994 Nov 15;13(22):5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Fantes P., Yanagida M. Coupling of DNA replication and mitosis by fission yeast rad4/cut5. J Cell Sci Suppl. 1994;18:57–61. doi: 10.1242/jcs.1994.supplement_18.8. [DOI] [PubMed] [Google Scholar]
- Saka Y., Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell. 1993 Jul 30;74(2):383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993 May;134(1):63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994 Mar 15;8(6):652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Zhou C., Jong A. CDC6 mRNA fluctuates periodically in the yeast cell cycle. J Biol Chem. 1990 Nov 15;265(32):19904–19909. [PubMed] [Google Scholar]
- Zwerschke W., Rottjakob H. W., Küntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem. 1994 Sep 16;269(37):23351–23356. [PubMed] [Google Scholar]






