Abstract
Synthesis of a novel class of compounds and their biophysical studies with TAR-RNA are presented. The synthesis of these compounds was achieved by conjugating neomycin, an aminoglycoside, with benzimidazoles modeled from a B-DNA minor groove binder, Hoechst 33258. The neomycin-benzimidazole conjugates have varying linkers that connect the benzimidazole and neomycin units. The linkers of varying length (5-23 atoms) in these conjugates contain one to three triazole units. The UV thermal denaturation experiments showed that the conjugates resulted in greater stabilization of the TAR-RNA than either neomycin or benzimidazole used in the synthesis of conjugates. These results were corroborated by the FID displacement and tat-TAR inhibition assays. The binding of ligands to the TARRNA is affected by the length and composition of the linker. Our results show that increasing the number of triazole groups and the linker length in these compounds have diminishing effect on the binding to TAR-RNA. Compounds that have shorter linker length and fewer triazole units in the linker displayed increased affinity towards the TAR RNA.
Keywords: TAR, Neomycin, Aminoglycosides, tat-TAR inhibition, Bnzimidazole
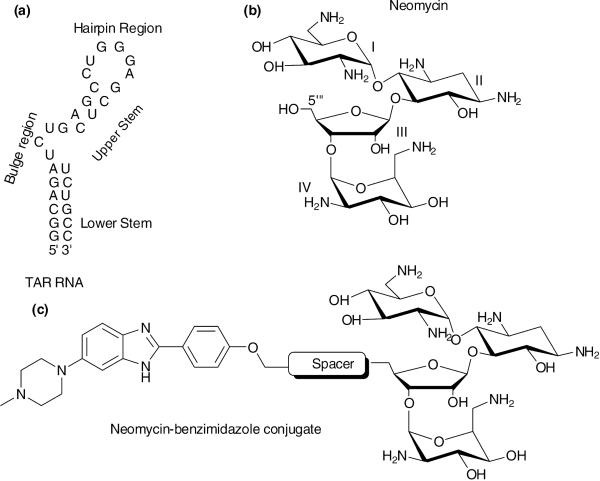
Since the first reports of HIV-AIDS in the United States in the early 1980s, research towards its cure have identified an RNA directed strategy which targets the interactions of tat protein with the Trans Activating Region (TAR) of the viral RNA.1 A 29-mer oligonucleotide, which is a model of a full 59 mer TAR region of the viral RNA, contains two of the most commonly found structural features in the RNA-namely the hairpin loop and the short trinucleotide bulge (Figure 1a). The trinucleotide bulge region has a wide major groove that is accessed by the tat protein for viral replication and thus inhibition of this interaction has developed into a viable approach to stop viral growth.2,3
Figure 1.
(a) Illustration of 29 mer short oligonucleotide mimic of the TAR- RNA. (b,c) Chemical structures of neomycin and neomycin-benzimidazole conjugates.
Several DNA and RNA binders have been investigated to inhibit tat-TAR interactions. These binders include polyamines (argininamide)3, polyamides,4 peptides,5,6 peptidomimetics,7 intercalators,8 quinoline derivatives,9,10 quinolones,11 DNA minor groove binders,2 aminoglycosides12-14 and their derivatives.15-17 Neomycin is an aminosugar (Figure 1b) that has been known for decades for its RNA binding.18,19 Neomycin has been shown to inhibit the tat-TAR interaction by binding to the trinucleotide pyrimidine bulge of the TAR-RNA14 and has the highest affinity among the selected aminoglycosides studied.12 Neomycin has been observed to bind with the TAR-RNA making contacts in the minor groove present in the lower stem using its ring III and IV while rings I and II have been found to interact with the trinucleotide bulge (Figure 1a and 1b). The bis-benzimidazole Hoechst 33258, a known B-DNA minor groove binding molecule, has also been shown to interact with the TAR-RNA at a site opposite to the bulge region where it recognizes the helical region below the hairpin loop (Figure 1a). RNAase A footprinting analysis has suggested that Hoechst 33258 binds to GCUCU bases of the TAR RNA in the upper stem.2
Aminoglycosides have emerged as versatile nucleic acid binders over the past decade.20-26 Several aminoglycoside conjugates have been synthesized27-30 and shown to have enhanced binding to variety of DNA,31-34 RNA,35 and DNA:RNA hybrid36 structures. We have recently reported recognition of TAR-RNA using a series of dimeric neomycin conjugates.37,38 The neomycin dimers have shown significant enhancement in the protection of MT-2 cells from the cytopathic effects of HIV infection in comparison to neomycin alone.37 These studies have opened new avenues for multi-valent approaches for the recognition of TAR RNA by aminoglycoside based small molecules.
In our continuing effort to develop novel small molecules for TAR RNA recognition, we herein report a series of dye Hoechst 33258. Hoechst 33258 has been shown to interact with the TAR RNA through intercalative binding.39 However, the planar surface of Hoechst 33258 is wider than that of RNA base pairs. Therefore, a smaller benzimidazole was hypothesized to be a more complimentary surface that favors facile entry between the helical bases in addition to reducing the molecular weight and polarity of the conjugated ligand.
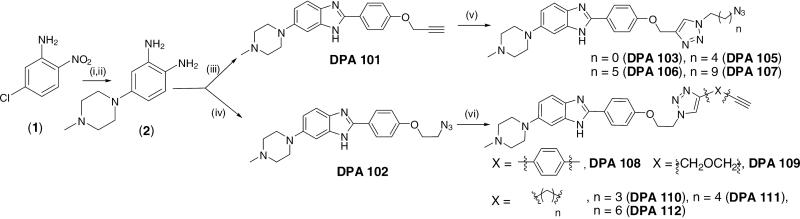
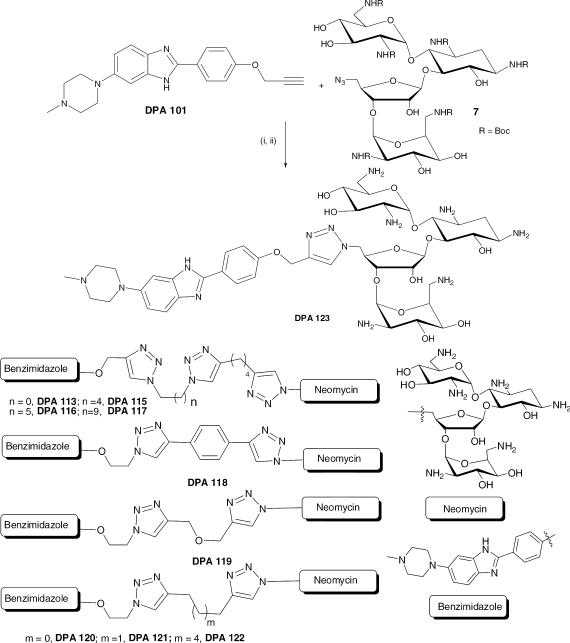
The monobenzimidazole derivatives modeled from Hoechst 33258 (referred to as benzimidazoles henceforth), like its parent structure, penetrates easily in the cell and can be neomycin–benzimidazole conjugates modeled from the bisbenzimidazole fluorescently detected (unpublished results). The benzimidazoles (DPA 101 and DPA 102, Scheme 1) lack one benzimidazole unit in comparison to Hoechst 33258 which is a bisbenzimidazole dye. These smaller benzimidazoles are much easier to synthesize and purify than the larger and more polar bisbenzimidazoles. The synthesis of neomycin-benzimidazole conjugates was achieved using a divergent strategy. The neomycin units (azide40,41/alkyne units) and the benzimidazole units (containing the complementary alkyne/azide) were prepared separately (Scheme 1, supporting information Scheme S1). Scheme 1 shows the synthesis of alkyne/azide terminated benzimidazoles. The synthesis of clickable benzimidazoles rested on selective incorporation of the terminal alkyne or azide units. DPA 101 was synthesized in three steps from commercially available 5-chloro-2-nitroaniline (1) as shown in Scheme 1. This transformation was achieved by substitution of the chloro substituent in 1 with N-methyl piperazine in the presence of K2CO3. The synthesized compound 5-(4-methylpiperazin-1-yl)-2-nitroaniline was then hydrogenated to its corresponding diamine422 followed by condensation with 4-(prop-2-ynyloxy) benzaldehyde (3)43 in the presence of Na2S2O5 to afford the desired ligand DPA 101. Similarly, DPA 102 was synthesized by condensation of the above mentioned diamine (2) with 4-(2-azidoethoxy) benzaldehyde (4)44 in the presence of the oxidant sodium metabisulfite45. DPA 101 and DPA 102 were then reacted with various bisazides (5a-d) or bisalkynes (6a-e) to afford the azide or alkyne terminated benzimidazole derivatives (DPA 103-DPA 112) containing various linker lengths. The ligands DPA 103-DPA 112 were then reacted with Boc protected neomycin azide (7) or Boc protected neomycin alkyne (8) (see supporting information S2a,b for the reaction scheme) under click chemistry conditions leading to the formation of Boc protected neomycin benzimidazole conjugates (DPA 113-123). Deprotection of the Boc groups was achieved with 4M HCl in dioxane (Scheme 2, S2a,b). The purity of new compounds was checked with TLC and 1HNMR; the identity of the synthesized compounds was further corroborated by ( 1H/13C) NMR and mass (MALDI-TOF) spectroscopic analysis.. A reaction scheme for the synthesis of DPA 123 and the structures of all ligands synthesized are displayed in Scheme 2.
Scheme 1.
Reagents and conditions (i) DMF, N-methylpiperazine, 100 °C, 5h, 60% (ii) Pd-C (10%), ethanol, H2, 6h, qaunt. (iii) 4-(prop-2-ynyloxy) benzaldehyde (3), Na2S2O5, H2O, reflux, 12h, 65% (iv) 4-(2-azidoethoxy) benzaldehyde (4), Na2S2O5, H2O, reflux, 12h, 82% (v) sodium ascorbate, copper (II) sulfate, 50 fold excess bisazides [N3-CH2-(CH2)n-N3; n = 0(5a); n = 4(5b), n = 5(5c), n = 9(5d)], ethanol, room temperature, overnight,65-80 % (vi) sodium ascorbate, copper (II) sulfate, 50 fold excess bisalkynes [1, 4 diethynyl benzene (6a), propargyl ether (6b), HC-(CH2)n-CH; n = 3(6c), n = 4(6d), n = 6(6e)] ethanol, temperature, overnight, 75-90 %.
Scheme 2.
(i) Sodium ascorbate, copper (II) sulfate, ethanol, room temperature, overnight (ii) Dichloromethane, 4M HCl in 1,4 dioxane, 49 % cumulative yield for two steps.
These neomycin–benzimidazole conjugates (DPA 113-123) were then evaluated for their affinity towards TAR RNA.
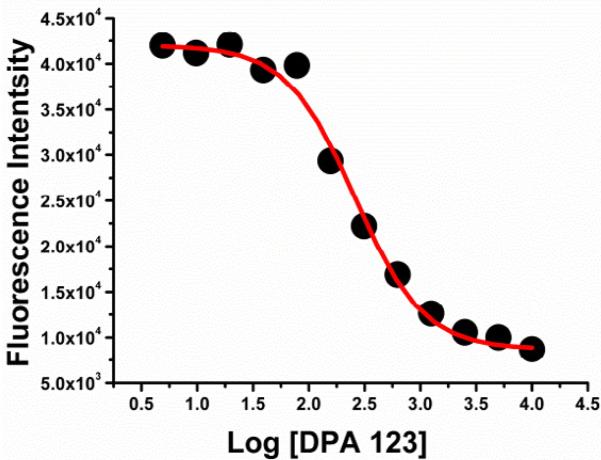
The relative affinities of these ligands were evaluated by two methods. In the FRET analysis,46 a tat peptide containing fluorescein and rhodamine as FRET pair was complexed with TAR RNA. The tat-TAR complex was incubated with ligand of interest to displace bound tat protein which is reflected by the quenching of fluorescein emission. The tat-TAR inhibitory affinity can be expressed as IC50 which reflects a ligand's ability to displace 50% bound tat peptide from TAR. In another assay based on fluorescent intercalator displacement assay (FID),47 a TAR bound intercalator (EtBr) was successively displaced by the addition of ligand of interest. The resulting binding isotherm can then be analyzed to obtain DC50 values (DC50 corresponds to the concentration of the ligand required to displace 50% of the bound intercalator). The results of these assays are summarized in Table 1 and a representative plot is shown in Figure 2. The data shown in Table 1 reveals that affinity of these ligands towards TAR is dependent on both linker length and their composition. In general, ligands with shorter linker and fewer triazoles (eg DPA 123, DPA 120) displayed a stronger affinity to TAR RNA in comparison to conjugates with longer linker length and more triazole units (DPA 113-DPA117) as displayed by their DC50 and IC50 values. DPA 123, with a short linker and a single triazole, emerges as the best ligand in binding to the TAR with DC50 and IC50 values of 33 nM and 257 nM, respectively. To study the selectivity of DPA123 for TAR over other HIV RNA targets, we investigated its ability to affect the Rev-RRE (rev response element) complexation. We observed that DPA 123 displays much better ability to affect tat-TAR binding, when compared to its ability to displace Rev from RRE, another RNA target of HIV virus. The IC50 value for inhibition of RRE RNA-HIV-1 Rev peptide complex was (768 ± 309) nM (see supporting information), significantly higher than IC50 value for inhibition of tat-TAR complexation.
Table 1.
A table showing the DC50, IC50 and thermal stabilization afforded by ligands used in the study.
| Ligand | Linker Lengtha | No. of triazoles | DC50(nM) | IC50(nM) | ΔTm (°C) |
|---|---|---|---|---|---|
| Neomycin | NA | NA | 1474 ± 79 | 0.2 | |
| DPA 101 | NA | NA | ND | ND | 1.0 |
| DPA 123 | 5 | 1 | 33 | 272 ± 8 | 5.6 |
| DPA 119 | 11 | 2 | 78 | 1285 ± 155 | 3.0 |
| DPA 120 | 12 | 2 | 83 | 337 ± 43 | 3.4 |
| DPA 121 | 13 | 2 | 140 | 507 ± 9 | 4.1 |
| DPA 118 | 13 | 2 | 180 | 379 ± 20 | 3.0 |
| DPA 122 | 15 | 2 | 81 | 949 ± 92 | 5.4 |
| DPA 113 | 17 | 3 | 184 | 411 ± 51 | 2.5 |
| DPA 115 | 20 | 3 | 150 | 851 ± 80 | 2.1 |
| DPA 116 | 21 | 3 | 200 | 1368 ± 50 | 3.8 |
| DPA 117 | 23 | 3 | 180 | 1380 ± 150 | 4.0 |
The linker length was assigned by counting the number of main chain atoms between the 5’”-carbon on ribose ring (ring III, see Figure 1) of neomycin and oxygen atom on the phenolic end of the benzimidazole.
Figure 2.
A representative plot showing the decrease in the fluorescence of fluorescein labeled tat-TAR complex upon ligand (DPA 123) binding of tat-TAR RNA complex. A 1:1 mixture of tat peptide (100 nM) and TAR RNA (100 nm) were mixed in Tris buffer containing 50 mM Tris, 20 mM KCl at pH 7.4. The resulting complex was titrated with DPA 123 until no change in fluorescence was observed. The resulting binding isotherm was fitted with a dose response curve using Origin 5.0 (see supporting information for fitting details).
The nucleic acid binding of these ligands was also evaluated using thermal denaturation experiments. The thermal denaturation temperatures are listed in Table 1. In the presence of benzimidazole DPA 101 alone, the thermal stabilization afforded was 1.0 °C while the same with neomycin was 0.2 °C. All conjugates afforded higher thermal stabilization of TAR RNA than any of the building blocks used to build the conjugates, with DPA 123 providing the highest thermal stabilization (5.6 °C). The highest thermal stabilization afforded by DPA 123 also corroborated the results obtained from FID experiments and the tat-TAR inhibition assay. In some cases, however, we did not observe correlation between the thermal binding data and the DC50/IC50 values. As these DC50/IC50 values are obtained by evaluating two entirely different assays, the lack of correlation could be attributed to the differences in the binding sites of the two probes bound TAR in the two assays. EtBr intercalates nonspecifically whereas the fluorescein tagged tat protein binds more specifically in a 1:1 fashion. The thermal denaturation profiles of the RNA, on the other hand, reflect a more complex temperature dependence of ligand:RNA equilibrium constants and are not expected to always correlate with isothermal room temperature Kd or IC50 measurements.
We also performed circular dichroism studies with DPA 123 to investigate the ligand induced changes in the conformation of the TAR RNA. As shown in Figure 3, the titration of DPA 123 led to increasingly diminished signal at 210 nm and 262 nm which also displayed the presence of isobestic points at 231 nm and 272 nm. The changes in the CD intensity upon ligand binding are indicative of the complexation of ligand with the host RNA duplex that is accompanied by slight structural alterations of the nucleic acid structure.48 However, the CD spectrum displays the characteristic A-form features (a negative band at 210 nm and a positive band at 263 nm)49 during the entire course of titration suggesting that overall structural features of the RNA are retained upon binding of DPA 123.
Figure 3.
CD titration profile of TAR RNA with DPA 123. The ligand was added serially to the RNA solution in small increments as displayed on the graph. After each successive addition, the nucleic acid ligand-complex was stirred and equilibrated for five minutes before the spectrum was recorded. Each spectrum shown in the figure is an average of three scans. The experiments were performed in cacodylate buffer containing 10 mM sodium cacodylate, 0.5 mM EDTA, 100 mM KCl at pH 6.8. (T = 20 °C). The boxed regions have been expanded for clarity.
Hoechst 33258 binding to TAR has previously been reported to occur in an intercalative way39 and thus a similar mode of binding is likely for the benzimidazoles. We, however, did not observe any induced CD in the benzimidazole chromophore absorption at low drug:RNA ratios and the use of higher than 1.6 equivalents of DPA 123 led to the precipitation which prevented us from studying the induced CD at higher drug concentrations.
In conclusion, our studies show that conjugation of neomycin to benzimidazoles modeled from Hoechst 33258 results in improved RNA binding and improved inhibition of RNA- protein interactions. Conjugates with short linkers (DPA 123) displayed a better and selective binding (TAR vs. RRE) than compounds with longer linkers, and the binding was affected by the number and placement of triazole units. Effect of these conjugates on HIV inhibition is currently being explored and will be reported in due course.
Supplementary Material
Acknowledgments
Financial support to this work was provided by National Institute of Health grants (R15CA125724 and 1R41GM100607).
Abbreviations
- UV
Ultra Violet
- FID
Fluorescent Intercalators Displacement
- CD
Circular Dichroism
- TAR
Trans Activating Region
- NMR
Nuclear Magnetic Resonance
- MALDI-TOF
Matrix Assisted Laser Desorption and Ionization- Time of Flight
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Details of experimental procedures and synthesis of DPA 123 is provided. Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/XXXX
References and notes
- 1.Zapp ML, Stern S, Green MR. Cell. 1993;74:969. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
- 2.Dassonneville L, Hamy F, Colson P, Houssier C, Bailly C. Nucleic Acids Res. 1997;25:4487. doi: 10.1093/nar/25.22.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peytou V, Condom R, Patino N, Guedj R, Aubertin AM, Gelus N, Bailly C, Terreux R, Cabrol-Bass D. J. Med. Chem. 1999;42:4042. doi: 10.1021/jm980728e. [DOI] [PubMed] [Google Scholar]
- 4.Mischiati C, Jeang KT, Feriotto G, Breda L, Borgatti M, Bianchi N, Gambari R. Antisense Nucleic Acid Drug Dev. 2001;11:209. doi: 10.1089/108729001317022214. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig V, Krebs A, Stoll M, Dietrich U, Ferner J, Schwalbe H, Scheffer U, Durner G, Gobel MW. Chembiochem. 2007;8:1850. doi: 10.1002/cbic.200700232. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Hyun S, Kim HJ, Yu J. Angew. Chem. Int. Ed Engl. 2008;47:134. doi: 10.1002/anie.200703090. [DOI] [PubMed] [Google Scholar]
- 7.Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Biochemistry (N. Y. ) 2007;46:741. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 8.Gelus N, Hamy F, Bailly C. Bioorg. Med. Chem. 1999;7:1075. doi: 10.1016/s0968-0896(99)00030-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Chen R, He M, Pang R, Tan Z, Yang M. Bioorg. Med. Chem. 2009;17:1948. doi: 10.1016/j.bmc.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 10.He M, Yuan D, Lin W, Pang R, Yu X, Yang M. Bioorg. Med. Chem. Lett. 2005;15:3978. doi: 10.1016/j.bmcl.2005.01.068. [DOI] [PubMed] [Google Scholar]
- 11.Gatto B, Tabarrini O, Massari S, Giaretta G, Sabatini S, Del Vecchio C, Parolin C, Fravolini A, Palumbo M, Cecchetti V. ChemMedChem. 2009;4:935. doi: 10.1002/cmdc.200800437. [DOI] [PubMed] [Google Scholar]
- 12.Mei H, Galan AA, Halim NS, Mack DP, Moreland DW, Sanders KB, Truong HN, Czarnik AW. Bioorg. Med. Chem. Lett. 1995;5:2755. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Huber PW, Cui M, Czarnik AW, Mei HY. Biochemistry. 1998;37:5549. doi: 10.1021/bi972808a. [DOI] [PubMed] [Google Scholar]
- 14.Faber C, Sticht H, Schweimer K, Rösch P. J. Biol. Chem. 2000;275:20660. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 15.Riguet E, Desire J, Boden O, Ludwig V, Gobel M, Bailly C, Decout JL. Bioorg. Med. Chem. Lett. 2005;15:4651. doi: 10.1016/j.bmcl.2005.07.082. [DOI] [PubMed] [Google Scholar]
- 16.Riguet E, Tripathi S, Chaubey B, Desire J, Pandey VN, Decout JL. J. Med. Chem. 2004;47:4806. doi: 10.1021/jm049642d. [DOI] [PubMed] [Google Scholar]
- 17.Elson-Schwab L, Tor Y. In: Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery. Arya DP, editor. Wiley; New York: 2007. pp. 267–287. [Google Scholar]
- 18.Arya DP. Top. Curr. Chem. 2005;253:149. [Google Scholar]
- 19.Willis B, Arya DP. Adv Carbohyd Chem Biochem. 2006;60:251. doi: 10.1016/S0065-2318(06)60006-1. [DOI] [PubMed] [Google Scholar]
- 20.Arya DP. Acc. Chem. Res. 2011;44:134. doi: 10.1021/ar100113q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charles I, Davis E, Arya DP. Biochemistry. 2012;51:5496. doi: 10.1021/bi3004507. [DOI] [PubMed] [Google Scholar]
- 22.Xi H, Davis E, Ranjan N, Xue L, Hyde-Volpe D, Arya DP. Biochemistry. 2011;50:9088. doi: 10.1021/bi201077h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjan N, Andreasen KF, Kumar S, Hyde-Volpe D, Arya DP. Biochemistry. 2010;49:9891. doi: 10.1021/bi101517e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xi H, Gray D, Kumar S, Arya DP. FEBS Lett. 2009;583:2269. doi: 10.1016/j.febslet.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Willis B, Arya DP. Curr. Org. Chem. 2006;10:663. [Google Scholar]
- 26.Kirk SR, Luedtke NW, Tor Y. J. Am. Chem. Soc. 2000;122:980. [Google Scholar]
- 27.Charles I, Xue L, Arya DP. Bioorg. Med. Chem. Lett. 2002;12:1259. doi: 10.1016/s0960-894x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 28.Arya DP, Xue L, Tennant P. J Am Chem Soc. 2003;125:8070. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 29.Arya DP, Willis B. J. Am. Chem. Soc. 2003;125:12398. doi: 10.1021/ja036742k. [DOI] [PubMed] [Google Scholar]
- 30.Willis B, Arya DP. Biochemistry. 2010;49:452. doi: 10.1021/bi9016796. [DOI] [PubMed] [Google Scholar]
- 31.Xue L, Xi H, Kumar S, Gray D, Davis E, Hamilton P, Skriba M, Arya DP. Biochemistry. 2010;49:5540. doi: 10.1021/bi100071j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Xue L, Arya DP. J. Am. Chem. Soc. 2011;133:7361. doi: 10.1021/ja108118v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue L, Ranjan N, Arya DP. Biochemisry. 2011;50:2838. doi: 10.1021/bi1017304. [DOI] [PubMed] [Google Scholar]
- 34.Ranjan N, Davis E, Xue L, Arya DP. Chem. Commun. 2013;49:5796. doi: 10.1039/c3cc42721h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charles I, Xi HJ, Arya DP. Bioconjugate Chem. 2007;18:160. doi: 10.1021/bc060249r. [DOI] [PubMed] [Google Scholar]
- 36.Shaw NN, Xi H, Arya DP. Bioorg. Med. Chem. Lett. 2008;18:4142. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Kellish P, Robinson WE, Wang D, Appella DH, Arya DP. Biochemisry. 2012;51:2331. doi: 10.1021/bi201657k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Arya DP. Bioorg. Med. Chem. Lett. 2011;21:4788. doi: 10.1016/j.bmcl.2011.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailly C, Colson P, Houssier C, Hamy F. Nucleic Acids Res. 1996;24:1460. doi: 10.1093/nar/24.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H. Ph.D thesis. University of California; San Diego: United States: 1998. [Google Scholar]
- 41.Quader S, Boyd SE, Jenkins ID, Houston TA. J. Org. Chem. 2007;72:1962. doi: 10.1021/jo0620967. [DOI] [PubMed] [Google Scholar]
- 42.Kelly DP, Bateman SA, Martin RF, Reum ME, Rose M, Whittaker ARD. Aust. J. Chem. 1994;47:247. [Google Scholar]
- 43.Wei P, Yan X, Li J, Ma Y, Yao Y, Huang F. Tetrahedron. 2012;68:9179. [Google Scholar]
- 44.Deniz E, Ray S, Tomasulo M, Impellizzeri S, Sortino S, Raymo FM. J Phys Chem A. 2010;114:11567. doi: 10.1021/jp107116d. [DOI] [PubMed] [Google Scholar]
- 45.Ji Y, Bur D, Häsler W, Runtz Schmitt V, Dorn A, Bailly C, Waring MJ, Hochstrasser R, Leupin W. Bioorg. Med. Chem. 2001;9:2905. doi: 10.1016/s0968-0896(01)00170-5. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto C, Hamasakim K, Mihara H, Ueno A. Bioorg. Med. Chem. Lett. 2000;10:1857. doi: 10.1016/s0960-894x(00)00359-0. [DOI] [PubMed] [Google Scholar]
- 47.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J. Am. Chem. Soc. 2001;123:5878. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 48.Eriksson M, Norden B. Meth. Enzymol. 2001;340:68. doi: 10.1016/s0076-6879(01)40418-6. [DOI] [PubMed] [Google Scholar]
- 49.Kypr J, Kejnovská I, Renciuk D, Vorlícková M. Nucleic Acids Research. 2009;37:1713. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.