Abstract
Background
Neural crest cells are multipotent cells that migrate extensively throughout vertebrate embryos to form diverse lineages. Cell migration requires polarized, organized actin networks that provide the driving force for motility. Actin-binding proteins that regulate neural crest cell migration are just beginning to be defined.
Results
We recently identified a number of actin-associated factors through proteomic profiling of methylated proteins in migratory neural crest cells. Here, we report the previously undocumented expression pattern of three of these proteins in chick early neural crest development: doublecortin (DCX), tropomyosin-1 (TPM-1), and actin depolymerizing factor (ADF). All three genes are expressed with varying degrees of specificity and intensity in premigratory and migratory neural crest cells, and their resulting proteins exhibit distinct subcellular localization in migratory neural crest cells. Morpholino knock down of ADF reveals it is required for Sox10 gene expression, but minimally important during neural crest migration.
Conclusions
Neural crest cells express DCX, TPM-1 and ADF. ADF is necessary during neural crest specification, but largely dispensable for migration.
Keywords: ADF, DCX, TPM-1, neural crest, migration
Introduction
The neural crest is a mulitpotent, migratory cell type that arises from the dorsal neural tube early in vertebrate development. Neural crest cells migrate extensively throughout the embryo to form a diverse set of lineages including the craniofacial skeleton, peripheral nervous system, melanocytes and aorticopulmonary septum of the heart (LeDouarin and Kalcheim, 1999). Progress has been made in deciphering the mechanisms these cells use to migrate away from the neural tube (Theveneau and Mayor, 2012). Nevertheless, much remains to be learned about neural crest motility in comparison to other migratory cell types (Petrie et al., 2009).
We recently performed a mass spectrometry-based screen and identified a number of cytoskeleton-associated factors that are putatively methylated in migratory neural crest cells (Vermillion et al., in press). Of the proteins we identified, three affect actin remodeling and regulate motility in other migratory cell types and were of particular interest to us. However, all three of these proteins are poorly characterized in the neural crest. Thus, we aimed to determine the expression patterns of these three factors in order to assess their relevance to neural crest development.
Doublecortin (DCX) is a member of a family of microtubule-associated proteins that are required for neuronal migration during cortical development (des Portes et al., 1998). DCX binds microtubules and actin filaments to link the actin and microtubule cytoskeletons (Gleeson et al., 1999; Horesh et al., 1999; Tsukada et al., 2005). In DCX mutants, F-actin is redistributed and actin associated proteins are dysregulated within neurons, leading to axon guidance defects (Fu et al., 2013). Although DCX is highly expressed in dorsal root ganglia, a neural crest derivative (Francis et al., 1999; Gleeson et al., 1999), and despite its intriguing neuronal migration-related functions, DCX expression in early embryogenesis, and particularly in early neural crest development, is unknown.
Tropomyosins (TPM) are a large family of actin-binding proteins with over 40 different isoforms (Lees et al., 2011). TPM-1, which we identified, is one of four tropomyosin genes and produces five differentially spliced transcripts. Tropomyosins form head to tail polymers in the actin filament major groove (Smillie, 1979), and generally stabilize actin filaments, competing for binding with proteins that destabilize actin filaments like ADF/cofilin and Arp2/3 (Bernstein and Bamburg, 1982; Blanchoin et al., 2001; Ono and Ono, 2002). Accordingly, tropomyosin regulates actin stress fiber formation, focal adhesion assembly, and leading edge lamella stiffness and contractility (Gupton et al., 2005; Bach et al., 2009; Tojkander et al., 2011). However, the expression of TPM-1 in neural crest cells has not been determined.
Actin depolymerizing factor (ADF) and highly related cofilin-1 (non-muscle) and cofilin-2 (muscle) promote actin filament disassembly and are essential for actin driven motility (reviewed by (Moon and Drubin, 1995; Bamburg and Wiggan, 2002). ADF/cofilin proteins exhibit tissue-specific expression patterns throughout development, suggesting they serve non-overlapping roles (Nishida et al., 1984; Abe et al., 1989). Accordingly, cofilin activity is required for neural crest directional migration, and cofilin-1 mutant mouse embryos exhibit neural crest migration defects despite 3- to 4-fold upregulation of ADF in these embryos (Gurniak et al., 2005). Although ADF cannot compensate for cofilin-1 function during neural crest migration, it could have other roles. While ADF mutant mice only have corneal defects (Ikeda et al., 2003), early neural crest development is highly regulative (Barembaum and Bronner-Fraser, 2005), and the requirement for ADF in specific developmental events must be evaluated before a role for ADF in neural crest cells is ruled out. Moreover, a careful analysis of ADF expression in the neural crest is lacking.
In this report we determine the expression patterns of DCX, TPM-1, and ADF, and evaluate the necessity for ADF in chick early neural crest development. Our results indicate that DCX, TPM-1, and ADF each exhibit a distinct subcellular localization in neural crest cells, suggesting they may regulate actin dynamics during neural crest migration as they do in other motile cell types. Moreover, ADF has a previously unrecognized, essential role in neural crest gene expression, although it is largely dispensable for migration.
Results
DCX is expressed early in neural crest development, continuing through migration
Although DCX regulates neuronal migration late in development (des Portes et al., 1998; Hannan et al., 1999; Fu et al., 2013), DCX expression in early embryos has not been evaluated. At 5 somites (s) DCX mRNA is apparent in chick neural folds (Fig. 1A, C, black arrowheads). Subsequently, DCX expression was abundant cranially and in rostral trunk, but was expressed at lower levels caudally (Fig. 1B). In cross section, midbrain migratory neural crest cells positive for HNK-1 (Fig. 1D′, red, white arrowheads) expressed DCX mRNA throughout their migration (Fig. 1D, black arrowheads). Non-neural ectoderm was also DCX-positive (Fig. 1D, black arrow), while head mesenchyme expressed low levels of DCX mRNA. Like its transcript, DCX protein (Fig. 1E, green; E′, white arrowhead) was abundant in migratory neural crest cells immunostained with HNK-1 (Fig. 1E, red; E″, white arrowhead) compared to surrounding head mesenchyme cells in cross-sections of a 9-somite chick embryo. Because cells in sections overlap one another, to evaluate DCX subcellular localization we assayed DCX immunofluorescence in cultured individual migratory neural crest cells. DCX protein (Fig. 1F, green; F′, white arrowhead) was diffusely cytoplasmic and surrounding the nucleus in HNK-1 positive (Fig. 1F, red; F″, white arrowhead) migratory cranial neural crest cells. Thus, DCX is expressed during early chick development and is particularly abundant in premigratory and migratory neural crest cells.
Figure 1. Doublecortin is expressed throughout neural crest development.
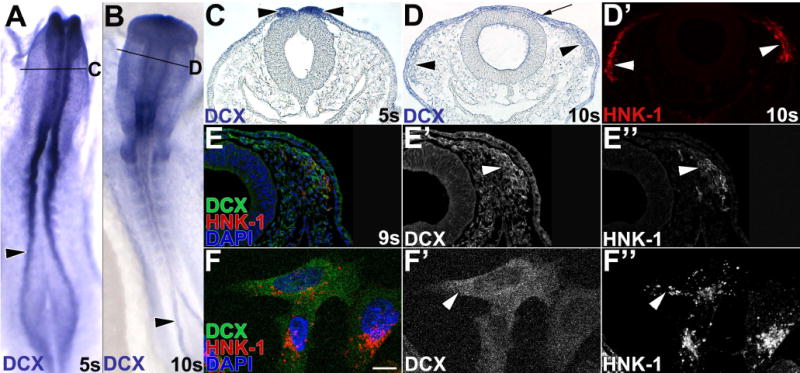
(A–D) Premigratory and migratory neural crest cells express DCX mRNA. DCX expression was visualized in 5 somite (s; A,C) and 10s (B,D) chick embryos by in situ hybridization. Sections (C,D) were taken at the level indicated in the whole mount view (A,B). At 5s, DCX mRNA is broadly expressed, but particularly abundant in neural folds (black arrowheads) in whole mount (A) and cross-section (C). At 10s in whole mount (B), DCX is highest cranially and in developing somites and caudal neural folds. In a midbrain cross-section at 10s (D), DCX mRNA is expressed in non-neural ectoderm (arrow), and in HNK-1-positive (D′, white arrowheads) migratory neural crest cells (black arrowheads). A,B, dorsal view. (E–F) DCX protein is expressed in migratory neural crest cells by immunofluorescence. In a 9 somite (E–E″) chick embryo midbrain section, DCX protein is abundant in cranial migratory neural crest cells (E, green; E′, white arrowhead) positive for HNK-1 (E, red; E″, white arrowhead), and expressed at lower levels in non-neural ectoderm and head mesenchyme. In cultured cranial migratory neural crest cells (F), DCX protein (F, green; F′, white arrowhead) is cytoplasmic and highest around the nucleus.
TPM-1 protein is expressed in migratory neural crest cells
Our mass spectrometry analysis identified two TPM-1 peptides with high confidence (Vermillion et al., in press). However, both of these peptides mapped to domains that are present in all TPM-1 isoforms. Thus, we generated an in situ probe over the entire TPM-1 gene and set out to define TPM-1 expression in neural crest cells. By in situ hybridization, TPM-1 mRNA was broadly expressed in whole mount chicken embryos (Fig. 2A,B), although it was enriched in neural crest precursors in the dorsal neural tube at 5 somites (Fig. 2C, black arrowheads). However, by 8s this expression had declined, and TPM-1 mRNA was most abundant in the non-neural ectoderm (Fig. 2C,D, arrows), with only low levels of TPM-1 transcripts detectable in HNK-1 positive midbrain migratory neural crest cells (Fig. 2D,D′, arrowheads). To determine whether TPM-1 is relevant to neural crest migration, we took advantage of a unique TPM-1 antibody (CG-1) that recognizes a TPM-1 epitope present only in motile cells (Hegmann et al., 1988). Immunofluorescence with this antibody showed strong, specific TPM-1 immunoreactivity (Fig. 2E, green, 2E′, white arrowhead) in leading midbrain migratory neural crest cells that are HNK-1 positive (Fig. 2E″, white arrowhead). Non-neural ectoderm was not detected, confirming the specificity of this antibody for a motility-dependent TPM-1 epitope (Hegmann et al., 1988). Finally, we evaluated TPM-1 subcellular localization in individual cultured cranial migratory neural crest cells (Fig. 2F–G, red; F″, G″). TPM-1 protein was expressed throughout the cell, typically in a filamentous pattern that resembled F-actin (Fig. 2F–G, green; F′, G′). While this expression pattern is similar to TPM localization in other cell types (Bach et al., 2009), it has not previously been documented in neural crest cells. Moreover, although TPM-1 mRNA is expressed fairly ubiquitously, leading migratory neural crest cells contain motility-activated TPM-1, suggesting that TPM-1 may be important for neural crest cell migration.
Figure 2. Tropomyosin 1 expression in the neural crest.
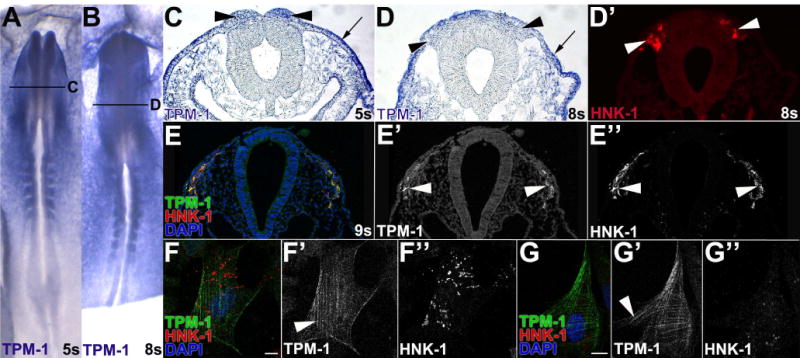
(A–D) Neural crest cells express low-levels of TPM-1 mRNA. TPM-1 expression was visualized in 5 somite (s; A,C) and 8s (B,D) chick embryos by in situ hybridization. Sections (C,D) were taken at the level indicated in the whole mount view (A,B). In whole mount, TPM-1 mRNA is expressed everywhere except the midline and neural plate (A,B). In a midbrain sections, non-neural ectoderm and head mesenchyme strongly express TPM-1, while the neural tube does not (C,D). TPM-1 expression is moderate in neural folds (C, black arrowheads), but declines in HNK-1-positive (D′, white arrowhead) migratory neural crest cells moving away from the neural tube (D, black arrowheads). A,B, dorsal view. (E–G) Leading migratory neural crest cells contain the motility-activated TPM-1 epitope. In a midbrain section of a 9 somite (E–E″) chick embryo, immunofluorescence with the CG-1 antibody (Hegmann et al., 1988) shows motility-dependent TPM-1 immunoreactivity in cranial migratory neural crest cells (E, green, E′, white arrowhead) that are positive for HNK-1 (E″, white arrowhead). HNK-1 positive (F,G, red; F″, G″) cultured cranial migratory neural crest cells exhibit motility-activated TPM-1 immunoreactivity (F,G, green; F′, G′) in striped patterns throughout the cell body.
Neural crest cells express ADF
Finally, we evaluated the expression of ADF, which competes with TPM-1 to bind actin (Kuhn and Bamburg, 2008). ADF mRNA was broadly expressed in early chick embryos (Fig. 3A), and at 5 somites was abundant in the dorsal neural folds, which contain neural crest precursors (Fig. 3C, black arrowheads). At 9 somites, ADF transcripts persisted in HNK-1 positive (Fig. 3D′, red, white arrowheads) cranial migratory neural crest cells (Fig. 3D, black arrowheads). ADF expression was also high in the non-neural ectoderm (Fig. 3C,D, black arrow) and along the basal surface of the dorsal neural tube. Correspondingly, ADF immunoreactivity (Fig. 3E, green; E′) was apparent in the non-neural ectoderm as well as in HNK-1-positive migratory neural crest cells (E′, white arrowheads) at 8 somites (Fig. 3E, red; E″, white arrowheads). At higher magnification, neural crest cells exhibited both cytoplasmic (Fig. 3F′, arrowhead) and nuclear ADF immunoreactivity (Fig. 3F′, arrows). To better evaluate the subcellular localization of ADF protein, we cultured individual cranial migratory neural crest cells. ADF protein was apparent in the nucleus (Fig. 3G′, white arrow) and along the cell periphery and in the tips of protrusions (Fig. 3G, green; G′, white arrowheads) in HNK-1 positive neural crest cells (Fig. 3G, red; G″). ADF protein expression in cranial neural folds and cranial migratory neural crest cell lysates was confirmed by western blot (data not shown). Therefore, ADF is expressed at the right time and place to be involved in chick neural crest cell development.
Figure 3. Premigratory and migratory neural crest cells express ADF.
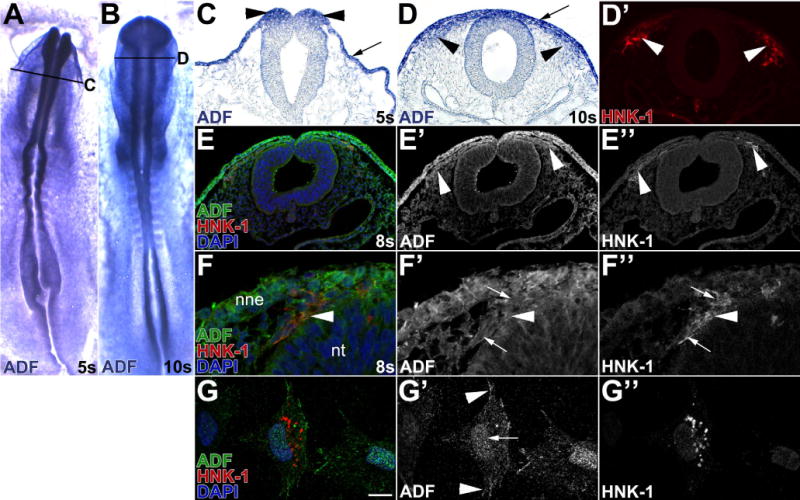
(A–D) Neural crest cells express ADF mRNA. In situ hybridization for ADF in 5 somite (s; A,C) and 10s (B,D) chick embryos. Sections (C–D) were taken at the level indicated in the whole mount view (A,B). In whole mount, ADF mRNA is widely expressed (A–B). In sections, ADF is particularly abundant in premigratory neural crest cells in the neural folds (C, black arrowheads) and in HNK-1-positive (D′, white arrowhead) migratory neural crest cells (D, black arrowheads). Non-neural ectoderm (C,D, arrow) and the basal surface of the neural tube also strongly express ADF. (E–G) Migratory neural crest cells express ADF protein. In 8 somite chick embryo midbrain sections (E–F), immunofluorescence reveals abundant ADF protein (E,F, green; E′,F′) in the non-neural ectoderm (nne) and HNK-1-positive (E,F red; E″,F″ white arrowheads) cranial migratory neural crest cells. ADF is both cytoplasmic (Fig. 3F′, arrowhead) and nuclear (Fig. 3F′ white arrows). In cultured cranial neural neural crest cells (G–G″). ADF protein (G, green; G′, white arrowheads) is nuclear (G′, white arrow), along the cell periphery, and at the tips of protrusions of HNK-1-positive (G, red; G″) migratory neural crest cells (G′, arrowheads).
ADF is required for neural crest specification and migration
While previous work in mouse suggested ADF was not necessary for neural crest development (Ikeda et al., 2003; Gurniak et al., 2005), the identification of ADF as a putatively methylated protein in chick (Vermillion et al., in press), and its expression in the periphery of cultured migratory neural crest cells (Fig. 3G) suggested it could have an overlooked function. To assess the requirement for ADF during chick neural crest specification and migration, we designed an antisense morpholino oligonucleotide (MO) to disrupt ADF mRNA splicing (Fig. 4A). To assess the efficacy of the MO, we unilaterally electroporated ADF MO into neural crest precursors at late gastrula, at the time of neural crest induction (Basch et al., 2006; Gammill and Krull, 2011). After incubation for 8–14 hours to 4–7 somites, neural folds well-targeted with fluorescein-modified ADF MO or control MO (CO MO) were dissected and RT-PCR was performed to visualize ADF mRNA splice products. Introduction of ADF MO caused the expected shorter, mis-spliced product to accumulate (Fig. 4B; full-length product is still apparent because electroporation is mosaic and cells in the dissected neural fold are not necessarily uniformly MO-positive). Meanwhile, electroporation of standard control morpholino (CO MO) only produced full-length ADF (Fig. 4B). To assess subsequent loss of ADF protein, embryos electroporated with ADF MO were sectioned and ADF protein levels visualized by immunofluorescence. ADF immunostaining was depleted (Fig. 4C, red; C″, white arrow) in cells targeted with ADF MO (Fig. 4C, green; C′) which can be seen most clearly in the non-neural ectoderm, which normally expresses high levels of ADF protein (Fig. 4C red, C″, black arrow). Together these results show that ADF MO knocks down ADF protein.
Figure 4. ADF MO elicits ADF-specific knock down.
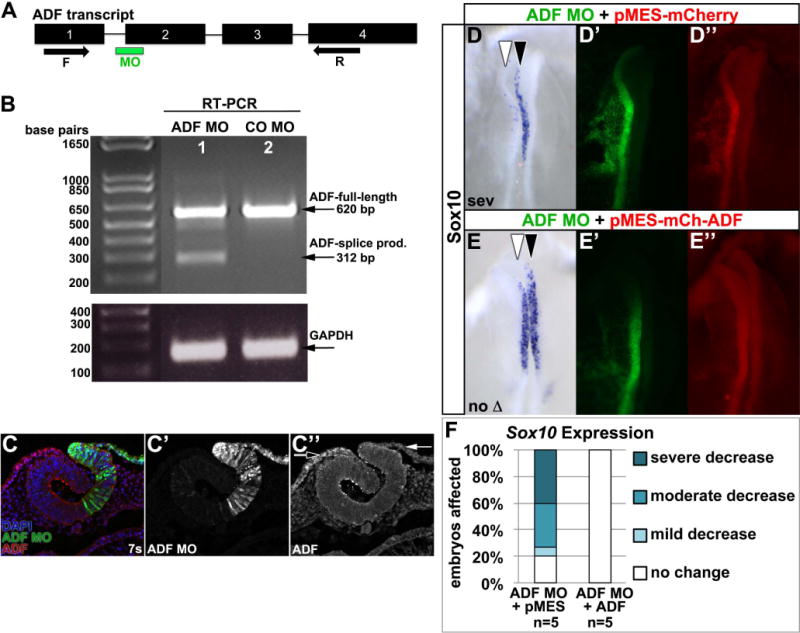
(A) ADF exon (numbered rectangles) and intron (lines) organization, splice-blocking MO (green bar) site, and forward (F) and reverse (R) primer (arrows) locations. (B,C) Embryos were unilaterally electroporated with ADF MO (B, lane 1; C, green; C′) or standard control MO (CO MO; B, lane 2) at late gastrula, reincubated to 7 somites (7s), and RT-PCR (B) or ADF immunofluorescence (C) performed. ADF MO promotes ADF mis-splicing (B, lane 1; 312 bp (base pair) product), while CO MO yields normally spliced ADF (B, lane 2; 620 bp product). GAPDH shows equal input cDNA. Meanwhile, in ADF MO-electroporated embryo sections, ADF immunoreactivity (C, red; C″) is reduced in ADF MO-targeted cells (C, green; C′; C″, white arrow) compared to untargeted cells (C, green; C″, black arrow). (D–F) ADF co-expression rescues ADF knock down. Embryos were unilaterally electroporated with ADF MO (D′,E′, green) mixed with pMES-mCherry (D″, red) or pMES-mCherry-ADF (E″, red) at late gastrula, reincubated to 4–6 somites, and Sox10 visualized by in situ hybridization (D,E, purple; dorsal view). White arrowhead, targeted side; black arrowhead, untargeted side. (F) Stacked bar graph shows that ADF co-expression rescues ADF MO-dependent loss of Sox10 expression.
Next we used the ADF MO to test the requirement for ADF during neural crest specification and migration. Because ADF’s counterpart cofilin shuttles actin to the nucleus for efficient transcription (Gieni and Hendzel, 2009; Visa and Percipalle, 2010; Obrdlik and Percipalle, 2011), we reasoned that knocking down ADF might affect neural crest gene expression. To assess this, we unilaterally electroporated ADF MO at late gastrula and cultured embryos to 4–6 somites, at the onset of neural crest specifier gene expression (Khudyakov and Bronner-Fraser, 2009). We then used in situ hybridization to evaluate the expression of key neural crest transcription factors Sox10, FoxD3, and Snail2 in ADF MO-targeted (Fig. 5, white arrowheads) versus untargeted (Fig. 5, black arrowheads) sides of electroporated embryos. Control (CO) MO electroporation elicits only a slight variation in Sox10, FoxD3, or Snail2 expression on the targeted side in a minority of embryos (Fig. 5A,D,E,H,I,L). In contrast, ADF knockdown had a significant impact on Sox10 expression, with about half of ADF MO-electroporated embryos showing a moderate to severely reduced Sox10 expression domain (Fig. 5B,C, white arrowhead; Fig. 5D, p=0.025). This phenotype could be rescued by co-electroporating wildtype ADF (Fig. 4D–F), indicating the effects of ADF knock down are specific. Meanwhile, FoxD3 (Fig. 5F,G, white arrowhead; Fig. 5H, p=0.297) and Snail2 (Fig. 5J,K, white arrowhead; Fig. 5L, p=0.500) were unaffected by ADF knock down. Thus, ADF is required for Sox10 gene expression during neural crest specification.
Fig. 5. ADF is required for neural crest specification and migration.
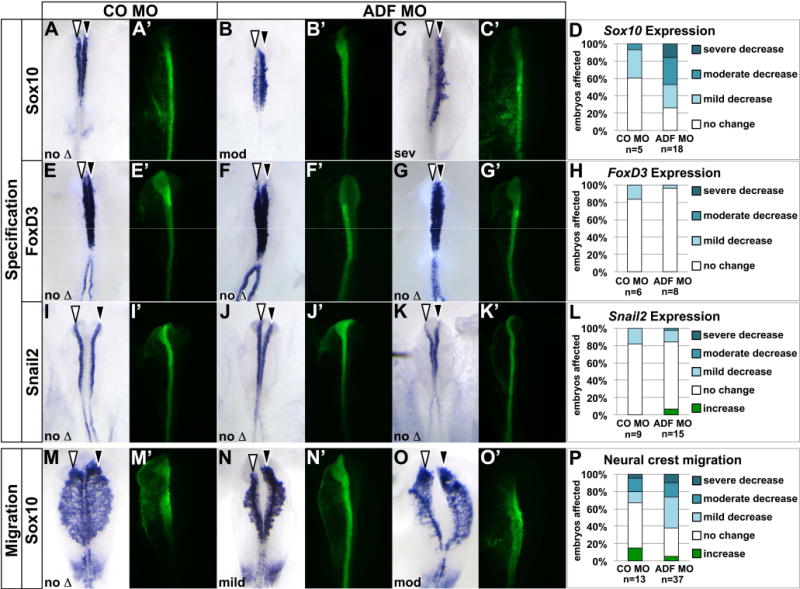
Embryos were unilaterally electroporated with standard control MO (CO MO; A,E,I,M, green) or ADF MO (B,C,F,G,J,K,N,O, green) at late gastrula, reincubated to 4–6 somites (A–C, E–G, I–K) or 8–10 somites (M–O), and processed by in situ hybridization (purple) to visualize expression of Sox10 (A–C, M–O), FoxD3 (E–G), or Snail2 (I–K). Dorsal view of in situ hybridization in left panel, fluorescent MO targeting in right panel. White arrowhead, targeted side of embryo; black arrowhead, untargeted side of embryo. (A–J) ADF is necessary for Sox10, but not FoxD3 or Snail2 expression. (A–C, E–G, I–K) Representative examples of electroporated embryos, showing only Sox10 expression defects on the ADF MO-targeted side. (D,H,L) Stacked bar graph depicting the frequency and severity of gene expression defects in embryos electroporated with CO MO or ADF MO. (M–P) ADF knockdown causes subtle defects in neural crest migration distance. (M–O) Representative examples of electroporated embryos, with the majority of ADF-targeted embryos mildly affected. (N) Stacked bar graph depicting the frequency and severity of migration defects in embryos electroporated with CO MO or ADF MO.
Since ADF also regulates the actin cytoskeleton in migrating cells (Sarmiere and Bamburg, 2004), we evaluated the requirement for ADF during neural crest migration as well. To this end, we unilaterally electroporated ADF MO at late gastrula and cultured embryos to 8 to 10 somites, when we evaluated the distance Sox10-positive cells migrated from ADF MO-targeted versus untargeted sides of the embryo. Although ADF knock down affected Sox10 expression in premigratory neural crest cells (Fig. 5B–D), by migratory stages, expression levels on the targeted side had recovered (see Fig. 5N for example). Relative to control (CO) MO-electroporated embryos (Fig. 5M), ADF MO-targeted neural crest cell migration distance was reduced, and this reduction was significant (Fig. 5N,O, white arrowheads; Fig. 5P, p=0.01). However, most embryos were only mildly affected, suggesting ADF is not a major factor in migratory neural crest cells.
Discussion
Migrating cells use polarized actin networks and actin cytoskeletal remodeling to provide the driving force for cell motility (Lauffenburger and Horwitz, 1996; Ridley et al., 2003). Actin-binding proteins that regulate directional migration of neural crest cells are still poorly defined. Here, we report the previously undocumented expression of three actin-binding proteins, DCX, TPM-1, and ADF, in chick early neural crest development. All three proteins exhibit distinct subcellular localizations consistent with their known roles in other cell types. Moreover, knockdown of ADF led to a significant reduction in the Sox10 expression domain in premigratory neural crest cells, and subsequently caused minor deficiencies in neural crest migration, uncovering a formerly unappreciated role for ADF in neural crest development (Fig. 5). Altogether our work increases the number of cytoskeletal proteins known to be expressed in neural crest cells, and provides new insight into the role of ADF in the neural crest.
The localization of these proteins suggests they serve similar functions in neural crest cells as they do in other motile cell types. ADF alters the helical twist and stability of actin filaments, increasing actin depolymerization (Bobkov et al., 2006) and promoting protrusive activity in motile cells (Condeelis, 1993; Carlier et al., 1997). Consequently, ADF is typically found at the leading edge, as seen in migratory neural crest cells (Fig. 3G′). On the other hand, most TPM isoforms bind and stabilize actin filaments to prevent bending or breaking (Kuhn and Bamburg, 2008; Lees et al., 2011; Tojkander et al., 2011). As a result, most forms of TPM localize to the cell interior, where they regulate focal adhesions and stabilize stress fibers that generate the force required for motility (DesMarais et al., 2002; Bach et al., 2009). The filamentous pattern of TPM-1 immunoreactivity in migratory neural crest cells (Fig. 2F′,G′) is consistent with such a role in the neural crest as well. Interestingly, only leading migratory neural crest cells exhibit motility-activated TPM-1 (Fig. 2E′; (Hegmann et al., 1988). Leading and trailing migratory neural crest cells express different genes (McLennan et al., 2012) and display unique behaviors (Kulesa et al., 2010); the presence of motility activated TPM-1 immunoreactivity in only the leading population defines a novel difference in the leading cranial migratory neural crest population. Finally, DCX, while best known as a microtubule-associated protein, has recently been shown in neurons to bind actin, affect actin localization, and be required for axon guidance (Tsukada et al., 2005; Fu et al., 2013). Our results suggest that DCX is also likely to be involved in neural crest migration, although its relationship to the actin cytoskeleton is less obvious from its pattern of staining by immunofluorescence (Fig. 1). Characterizing the expression of ADF, TPM-1 and DCX in the neural crest is a first step toward a more complete understanding of actin cytoskeletal regulation in neural crest cells.
To directly test the importance of these actin-binding proteins in neural crest development, we focused on ADF in particular and found a previously unappreciated, transient requirement for ADF during neural crest specification. It is well established that transcription by RNA polymerase II requires a constant supply of G-actin monomers in the nucleus (Hofmann et al., 2004; Kukalev et al., 2005). Actin does not have a nuclear localization signal (NLS), however, both ADF and cofilin do (Iida et al., 1992), and cofilin is necessary for nuclear translocation of actin (Dopie et al., 2012). Moreover, in the nucleus, cofilin is required for elongation of nascent transcripts by facilitating association of the transcriptional machinery with actively transcribed genes (Obrdlik and Percipalle, 2011). ADF exhibits nuclear localization in migratory neural crest cells (Fig. 3F′,G′), and is required for Sox10 expression (Fig. 5A–D). Thus, ADF may likewise be involved in actin nuclear transport and transcriptional regulation. However, the effect of ADF knock down on Sox10 expression was not fully penetrant (Fig. 5D) and recovered with time, so that Sox10 was expressed normally at migratory stages (Fig. 5N–P). This is unlikely to be due to diminished knock down efficiency, as ADF MO fluorescence was still abundant in migratory stage embryos (Fig. 5N′,O′) and ADF immunoreactivity was absent in ADF MO-targeted cells at 7 somites as migration initiated (Fig. 4C″). Instead, other factors such as cofilin likely functionally compensate for ADF over time.
It is curious that ADF knock down did not affect all neural crest transcription factors equally (Fig. 5A–L). It could be that ADF is only necessary for the expression of certain genes. Alternatively, the fact that FoxD3 and Snail2 are expressed earlier than Sox10 and are input targets in neural crest gene regulatory module (Prasad et al., 2012; Simoes-Costa et al., 2012) could affect the experimental outcome. One possibility is that insufficient time had elapsed for the MO to knock down ADF protein when FoxD3 and Snail2 expression initiated, but transcription was affected by the time Sox10 was expressed. Another possibility is that compensation for ADF function takes place in a protein complex-dependent, gene-by-gene basis, and does not recover generally. Because FoxD3 and Snail2 are expressed earlier, compensation for loss of ADF might already have taken place by the time we assayed their expression.
Importantly, our results are consistent with and expand results in mutant mice. The recovery of Sox10 expression in chick ADF-deficient cells explains why ADF mutant mice do not exhibit a neural crest phenotype (Ikeda et al., 2003). Meanwhile, the mild effects of ADF knock down on neural crest migration indicate that ADF does have a minor, cofilin-independent function during migration (Fig. 5N–P), although apparently ADF-deficient embryos recover from this slight defect over time as well.
Experimental Procedures
Embryos
Fertilized chicken embryos were obtained from local sources. Eggs were incubated in a humidified incubator at 37°C until the desired stage of development (Hamburger and Hamilton, 1992). Embryos were judged by counting somite pairs.
In situ Hybridization
Primer pairs against the NCBI Reference Sequences for chick ADF (NM_205528; F: 5′-CGAGAATTCGCCACCATGGCATCTGG-3′; R: 5′-GGGCCCAGGCCTCTACACAGGACTT-3′), TPM-1 (NM_205401; F: 5′-CGAGAATTCGCCACCATGGATGCCAT-3′; R: 5′-GGGCCCAGGCCTTCACATGTTGTTTA-3′), and DCX (NM_204335; F: 5′-CGAGAATTCGCCACCATGGAACTTGA-3′; R: 5′-GGGCCCAGGCCTTTACATGGAATCTC-3′). With these primer pairs, full-length coding sequences were PCR amplified from 4–10 somite chick random primed cDNA and cloned into CS107 (Baker et al., 1999). Clone identity was verified by sequencing. Each plasmid was linearized and used as template for RNA probe syntehsis (ADF-EcoRI, T7; TPM-1-EcoRI, T7, DCX-EcoRI, T7; Sox10 (Cheng et al., 2000)). Digoxigenin-labeled probes and chick whole mount chick in situ hybridization were performed as previously described (Wilkinson, 1992). Embryos were imaged in whole mount using a Discovery V8 stereoscope and AxioCam MRc5 (both from Zeiss; Oberkochen, Germany).
Histology
Embryos were infiltrated with 5% and 15% sucrose, embedded in gelatin in 15% sucrose, frozen in liquid nitrogen and sectioned with a CM1900 cryostat (Leica; Buffalo Grove, IL) at 10–20 μm. Gelatin was removed from the sections by incubating for 30 minutes in 42°C PBS.
Immunostaining
Sections and cultures were blocked in PBS + 10% Fetal bovine serum + 0.1% Triton X-100 for 30 minutes at room temperature and stained with anti-HNK-1 (ATCC; Manassas, VA), anti-ADF (Bamburg and Bray, 1987), anti-TPM1 (CG-1) (DHSB, Iowa City, IA), anti-DCX (C-18) (Santa Cruz Biotechnology; Dallas, TX) followed by the appropriate secondary antibody (mouse AF488, rabbit AF568, mouse AF568, Life Technologies; Grand Island, NY; or Cy2 anti-mouse/rabbit IgG, Cy5 anti-mouse/rabbit IgG, RRX anti-mouse IgM, Jackson ImmunoResearch; West Grove, PA) as indicated. For some assays the signal was amplified using a mouse anti-rabbit IgG or mouse anti-goat IgG (Jackson Labs, Inc; West Grove, PA). Slides and cultures were mounted with Permafluor (Thermo Fisher Scientific; Waltham, MA) containing DAPI and viewed on either a Zeiss AxioImager A1 or Zeiss LSM 710 confocal microscope. Images were assembled in Photoshop (Adobe).
Neural Crest Cultures
Cranial neural folds were dissected from 4–7 somite embryos. Cranial explants were then cultured for 14–16 hours at 37°C on 10–100 μg/ml fibronectin coated glass coverslips in neural crest complete media (L15, 1% L-glutamine, 0.1% Pen/Strep (all from Life Technologies; Grand Island, NY), 10% FBS (VWR Scientific; Radnor, PA), and 10% chick embryo extract (Bronner-Fraser and Garcia-Castro, 2008). For evaluation, cultures were fixed in 4% paraformaldehyde and permeabilized in PBS + 1.0% Triton X-100.
Morpholinos
FITC-tagged morpholinos (MOs) were synthesized by GeneTools, LLC (Philmouth, OR) with the following sequences: splice blocking ADF MO AGATGCCTGCAAAGATGAGAACAAA (ADF MO); and standard control MO CCTCTTACCTCAGTTACAATTTATA (CO MO).
Electroporation
Ex ovo early embryo electroporation was performed on late gastrula stage 4–5 embryos with ADF or CO MO at 1.0 mM as previously described (Gammill and Krull, 2011; Roffers-Agarwal et al., 2012).
RT-PCR
Embryos electroporated at late gastrula were incubated until 3–7 somites, and well-targeted head folds were dissected. RNA was isolated with Trizol and cDNA was synthesized with SuperScript III (both from Life Technologies). PCR was performed with Choice Taq Blue (Denville Scientific) using primers (F-GTGACGTCGGCAGCGTTT; R-GCCTCCTAGCTTCTCAGCAA) that spanned the splice acceptor site of ADF exon 2, which is targeted by the MO (Fig. 4A). GAPDH was amplified as a control (F-GGACACTTCAAGGGCACTGT; R-TCTCCATGGTGGTGAAGACA).
Phenotype evaluation
Embryos well-targeted with fluorescent MO in the dorsal neural tube were evaluated at the cranial axial level. Effects on premigratory gene expression were categorized by judging the relative amount of colorimetric in situ hybridization signal on the targeted and untargeted side of whole mount embryos. Effects on migration were scored by comparing the distance migrated on targeted and untargeted sides. All phenotypes were scored blind by three independent judges. The number of embryos in each scoring class was graphed to quantitatively represent the severity of the defects observed in the experimental population, and statistics were performed using two–sided Fisher’s exact test in R (R Development Core Team, 2011).
doublecortin, tropomyosin 1, and actin depolymerizing factor mRNA are expressed in premigratory and migratory neural crest cells.
these proteins exhibit distinct localization in migratory neural crest cells
expression of the neural crest transcription factor Sox10 requires actin depolymerizing factor.
actin depolymerizing factor is minimally important for chick neural crest migration.
Acknowledgments
We are grateful to Yi-Chuan Cheng for the kind gift of Sox10 plasmid. We would like to thank the members of the Gammill lab for experimental suggestions, advice and comments over the progress of this research, particularly Bridget Jacques-Fricke for help with blind scoring. Many thanks to Wuming Gong for assistance with statistical analysis. Special thanks to Paul Letourneau and Jim Bamburg for the kind gift of ADF antibody (Bamburg and Bray, 1987). We gratefully acknowledge the resources of the Minnesota Supercomputing Institute. The monoclonal antibody CG1, developed by J. Jung-Ching Lin, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
This work was supported by National Institutes of Health Individual NRSA F31 DE019755-01 to K.L.V, and National Science Foundation Research Grant IOS-1052101, March of Dimes Basil O’ Connor Starter Scholar Award to L.S.G., and U of MN Academic Health Center Seed Grant awarded to L.S.G.
Grant Sponsors: NSF IOS-1052101, March of Dimes 5-FY09-39, NIH F31 DE019755, U of MN Academic Health Center Seed Grant.
References
- Abe H, Ohshima S, Obinata T. A cofilin-like protein is involved in the regulation of actin assembly in developing skeletal muscle. J Biochem. 1989;106:696–702. doi: 10.1093/oxfordjournals.jbchem.a122919. [DOI] [PubMed] [Google Scholar]
- Bach CT, Creed S, Zhong J, Mahmassani M, Schevzov G, Stehn J, Cowell LN, Naumanen P, Lappalainen P, Gunning PW, O’Neill GM. Tropomyosin isoform expression regulates the transition of adhesions to determine cell speed and direction. Mol Cell Biol. 2009;29:1506–1514. doi: 10.1128/MCB.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JC, Beddington RS, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 1999;13:3149–3159. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Bray D. Distribution and cellular localization of actin depolymerizing factor. J Cell Biol. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- Barembaum M, Bronner-Fraser M. Early steps in neural crest specification. Seminars Cell Dev Biol. 2005;16:642–646. doi: 10.1016/j.semcdb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell motility. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complexnucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Garcia-Castro M. Manipulations of neural crest cells or their migratory pathways. Methods Cell Biol. 2008;87:75–96. doi: 10.1016/S0091-679X(08)00204-5. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res Dev Brain Res. 2000;121:233–241. doi: 10.1016/s0165-3806(00)00049-3. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Life at the leading edge: the formation of cell protrusions. Annu Rev Cell Biol. 1993;9:411–444. doi: 10.1146/annurev.cb.09.110193.002211. [DOI] [PubMed] [Google Scholar]
- des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, Catala M, Kahn A, Beldjord C, Chelly J. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A. 2012;109:E544–552. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis F, Koulakoff A, Boucher D, Chafey P, Schaar B, Vinet MC, Friocourt G, McDonnell N, Reiner O, Kahn A, McConnell SK, Berwald-Netter Y, Denoulet P, Chelly J. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- Fu X, Brown KJ, Yap CC, Winckler B, Jaiswal JK, Liu JS. Doublecortin (Dcx) family proteins regulate filamentous actin structure in developing neurons. J Neurosci. 2013;33:709–721. doi: 10.1523/JNEUROSCI.4603-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammill LS, Krull CE. Embryological and genetic manipulation of chick development. Methods Mol Biol. 2011;770:119–137. doi: 10.1007/978-1-61779-210-6_5. [DOI] [PubMed] [Google Scholar]
- Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, Waterman-Storer CM. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurniak CB, Perlas E, Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev Biol. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Henke RC, Seeto GS, Capes-Davis A, Dunn J, Jeffrey PL. Expression of doublecortin correlates with neuronal migration and pattern formation in diverse regions of the developing chick brain. J Neurosci. 1999;55:650–657. doi: 10.1002/(SICI)1097-4547(19990301)55:5<650::AID-JNR12>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Hegmann TE, Lin JL, Lin JJ. Motility-dependence of the heterogenous staining of culture cells by a monoclonal anti-tropomyosin antibody. J Cell Biol. 1988;106:385–393. doi: 10.1083/jcb.106.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, Hozak P, de Lanerolle P. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- Horesh D, Sapir T, Francis F, Wolf SG, Caspi M, Elbaum M, Chelly J, Reiner O. Doublecortin, a stabilizer of microtubules. Hum Mol Genet. 1999;8:1599–1610. doi: 10.1093/hmg/8.9.1599. [DOI] [PubMed] [Google Scholar]
- Iida K, Matsumoto S, Yahara I. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct Funct. 1992;17:39–46. doi: 10.1247/csf.17.39. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Cunningham LA, Boggess D, Hawes N, Hobson CD, Sundberg JP, Naggert JK, Smith RS, Nishina PM. Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor) Hum Mol Genet. 2003;12:1029–1037. doi: 10.1093/hmg/ddg112. [DOI] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev Dyn. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TB, Bamburg JR. Tropomyosin and ADF/cofilin as collaborators and competitors. Adv Exp Med Biol. 2008;644:232–249. doi: 10.1007/978-0-387-85766-4_18. [DOI] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol. 2005;12:238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Bailey CM, Kasemeier-Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Dev Biol. 2010;344:543–554. doi: 10.1016/j.ydbio.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- LeDouarin N, Kalcheim C. The Neural Crest. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Lees JG, Bach CT, O’Neill GM. Interior decoration: tropomyosin in actin dynamics and cell migration. Cell Adh Migr. 2011;5:181–186. doi: 10.4161/cam.5.2.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan R, Dyson L, Prather KW, Morrison JA, Baker RE, Maini PK, Kulesa PM. Multiscale mechanisms of cell migration during development: theory and experiment. Development. 2012;139:2935–2944. doi: 10.1242/dev.081471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A, Drubin DG. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E, Maekawa S, Sakai H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry. 1984;23:5307–5313. doi: 10.1021/bi00317a032. [DOI] [PubMed] [Google Scholar]
- Obrdlik A, Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus. 2011;2:72–79. doi: 10.4161/nucl.2.1.14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev Biol. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. Retrieved from http://www.R-project.org. [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Roffers-Agarwal J, Hutt KJ, Gammill LS. Paladin is an antiphosphatase that regulates neural crest cell formation and migration. Dev Biol. 2012;371:180–190. doi: 10.1016/j.ydbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol. 2004;58:103–117. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genetics. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie LB. Structure and function of tropomyosins from muscle and non-muscle sources. Trends Biochem Sci. 1979;4:151–155. [Google Scholar]
- Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev Biol. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21:539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Prokscha A, Ungewickell E, Eichele G. Doublecortin association with actin filaments is regulated by neurabin II. J Biol Chem. 2005;280:11361–11368. doi: 10.1074/jbc.M405525200. [DOI] [PubMed] [Google Scholar]
- Vermillion KL, Lidberg KA, Gammill LS. Cytoplasmic protein methylation is essential for neural crest migration. J Cell Biol. doi: 10.1083/jcb.201306071. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harbor Persp Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. In Situ Hybridization: A Practical Approach. Oxford: IRL Press; 1992. [Google Scholar]


