Abstract
RNAi based strategies to induce gene silencing are commonly employed in numerous model organisms but have not been extensively used in zebrafish. We found that introduction of transgenes containing convergent transcription units in zebrafish embryos induced stable transcriptional gene silencing (TGS) in cis and trans for reporter (mCherry) and endogenous (One-Eyed Pinhead (OEP) and miR-27a/b) genes. Convergent transcription enabled detection of both sense and antisense transcripts and silencing was suppressed upon Dicer knockdown, indicating processing of double stranded RNA. By ChIP analyses, increased silencing was accompanied by enrichment of the heterochromatin mark H3K9me3 in the two convergently arranged promoters and in the intervening reading frame. Our work demonstrates that convergent transcription can induce gene silencing in zebrafish providing another tool to create specific temporal and spatial control of gene expression.
Zebrafish is a powerful model vertebrate organism to elucidate mechanisms regulating development and disease1. The availability of large numbers of external, optically transparent developing embryos, combined with genetics and imaging, have provided exceptionally useful tools to study development and as powerful animal models of numerous human diseases. However, a critical missing tool is a simple, straightforward way to post-transcriptionally knock down genes in a sequence specific manner, especially at later stages of development. Recent reports have described the possibility that shRNA approaches might be useful for temporal and spatial knockdown of genes in zebrafish2,3. More commonly, antisense morpholinos have been used to knockdown gene expression during early development and have proven quite useful but with important caveats related to toxicity, limited duration, and potential off-target effects4. Here, we describe a novel RNA interference (RNAi) mediated mechanism to silence gene expression in zebrafish. First discovered in C. elegans, the discovery of RNAi ushered in a new era of reverse genetics allowing sequence specific knockdown of genes in multiple model organisms5. As an umbrella term, RNAi encompasses a number of gene regulatory mechanisms that ultimately depend on the production of small dsRNAs. Post-transcriptional gene silencing (PTGS) methods utilize small RNAs derived either from endogenous genes (miRNAs) or from exogenously delivered small interfering RNAs (siRNAs)6. Primary transcripts encoding miRNAs are initially processed to precursor miRNAs (pre-miRNAs) in the nucleus by Drosha7 followed by a second processing step in the cytoplasm by the enzyme Dicer that produces mature 21–23 nucleotide dsRNAs8. Following Dicer processing, one of the strands is incorporated into a multi-component complex called the RNA Induced Silencing Complex (RISC) containing Argonaute proteins9. Complementary base pairing then determines the fate of targeted mRNAs. Perfect pairing (siRNAs) results in mRNA cleavage whereas imperfect pairing (miRNAs) results in translational repression, deadenylation, and subsequent degradation of target mRNAs10.
Despite the fact that the zebrafish genome encodes Dicer, Drosha, and RISC components11, siRNA-mediated gene knockdown remains controversial with the limited number of reports claiming successful knockdown countered by results suggesting that the effects are entirely nonspecific12,13,14,15,16,17,18,19. These conflicting reports account for the fact that morpholino-mediated knockdown is prevalent in zebrafish, especially during early development. Given the limitations of morpholinos, we sought to determine whether RNAi-mediated chromatin silencing could silence genes in zebrafish. Transcriptional gene silencing (TGS) directed by small RNAs has been widely reported and recent work has raised the possibility that similar mechanisms may apply in higher eukaryotes20,21,22. While examining transcription termination in Schizosaccharomyces pombe, the Proudfoot lab discovered that overlapping transcripts derived from genes organized in a convergent manner can generate dsRNAs that activate nuclear RNAi leading to histone methylation, recruitment of cohesin, and silencing of the convergent genes23. The methylation marks that regulate chromatin are removed during G2 enabling transient production of overlapping RNAs that then silence the genes during G1/S. A number of genes encoding RNAi components in the S. pombe genome are arranged in a convergent manner enabling autoregulation24. Interestingly, the same mechanism can silence genes in mammalian cells25,26. Based on the presence of functional RNAi genes in the zebrafish genome, we hypothesized that convergent transcription could be used to specifically silence genes via a comparable mechanism. Our data demonstrate that transgenic zebrafish harboring convergently arranged genes generate dsRNAs that direct chromatin silencing of both reporter constructs and endogenous genes. Silencing leads to H3K9me3 deposition and can be suppressed by Dicer knockdown, consistent with the S. pombe data. This paper is the first paper to demonstrate that RNAi-mediated chromatin silencing is possible in zebrafish with the ability to precisely control spatial and temporal gene expression in embryos and adult zebrafish.
Results
Convergent transcription induces gene silencing in zebrafish
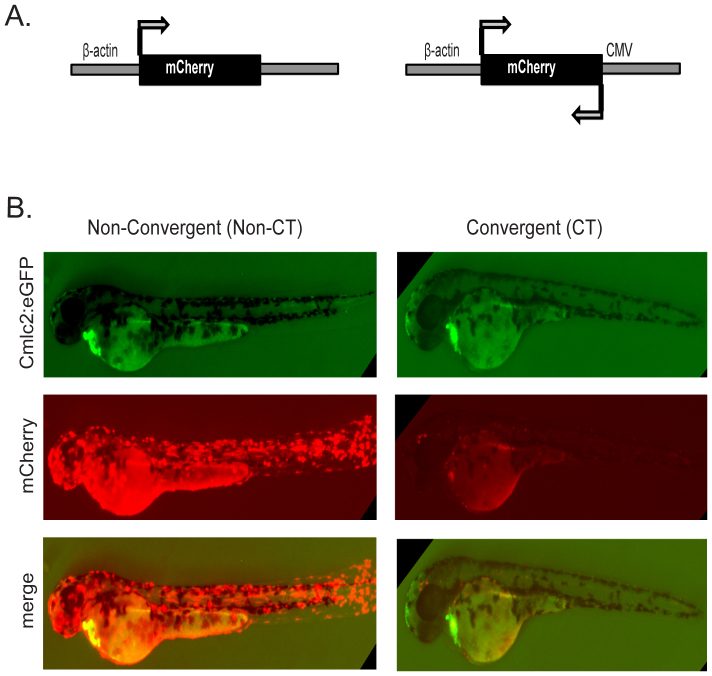
To determine if we could trigger RNAi-mediated gene silencing in zebrafish via convergent transcription, we created plasmids with transcriptional promoters arranged in either a convergent (CT) or nonconvergent (non-CT) manner. The experimental setup included insertion of open reading frame (ORF) sequences between the two promoters with no 5′ or 3′ UTR sequences. For proof of principle, we first targeted mCherry for rapid visualization of silencing. For the non-CT mCherry construct, the full length zebrafish β-actin promoter was inserted upstream of the mCherry open reading frame (ORF). The CT mCherry construct harbored a β-actin promoter at the 5′ end and an inverted CMV promoter at the 3′ end (Fig. 1A). Transient transgenic animals were created by co-injecting embryos at the 1–2 cell stage with the non-CT mCherry construct and Tol2 transposase mRNA27. As a marker of transgenesis, the cardiac myosin light chain promoter (cmlc2) was fused to GFP driving fluorescent expression in the heart28. As expected, transgenic animals containing the non-CT mCherry construct displayed robust expression of mCherry throughout the entire developing embryo (Fig. 1B). In contrast, there was a striking absence of mCherry in transgenic animals created by injection of the CT mCherry construct (Fig. 1B). Except for a few small puncta and some yolk autofluorescence, the levels of mCherry were dramatically reduced to near zero. The lack of mCherry was not due to the absence of the transgene as readily detectable levels of heart GFP were observed (Fig. 1B). Silencing was robust across multiple injections with undetectable levels of mCherry in greater than 92% of the transgenic CT-mCherry embryos. Nearly ~100% of non-CT mCherry transgenic embryos broadly expressed mCherry. Importantly, crossing of founders to generate F1 and F2 generations showed that silencing of mCherry is stable and continues to be maintained.
Figure 1. Convergent silencing of mCherry.
(A) Non-CT and CT constructs were designed using the Tol2 transgenesis system with the β-actin promoter driving sense transcription and an inverted CMV promoter driving antisense transcription of the mCherry open reading frame (see Supplement for maps and sequence information). All constructs harbored heart-specific GFP (cmlc2-GFP) which enabled identification of transgenic embryos. (B) Widespread expression of mCherry was observed in non-CT embryos (386 out of 386 transgenic embryos), whereas near complete loss of mCherry was observed in CT-mCherry embryos at 2 dpf (370 out of 402 transgenic embryos).
Convergent transcription produces dsRNA and silencing is suppressed upon knockdown of dicer
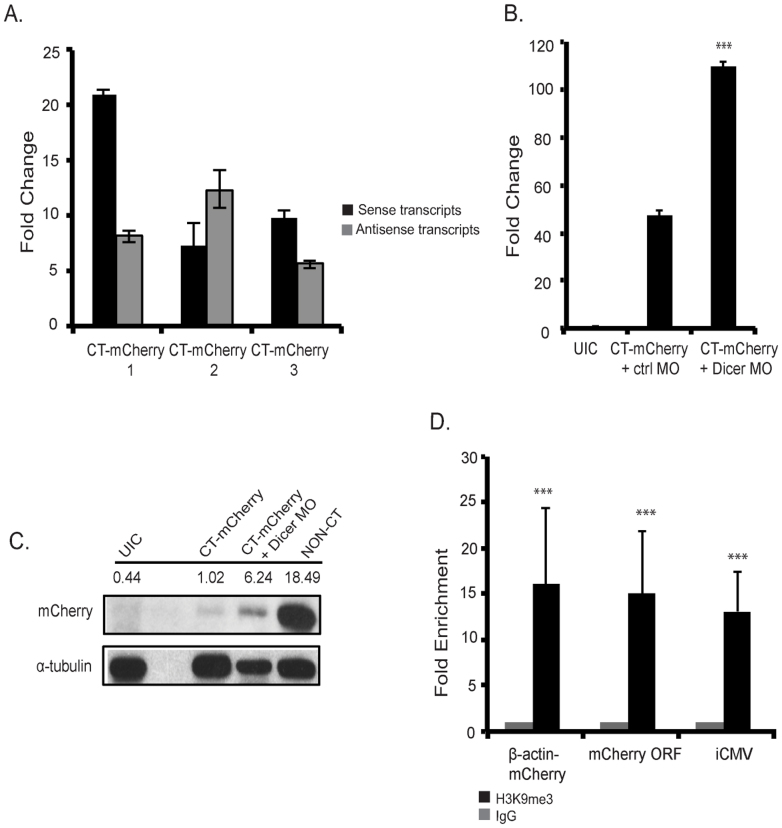
In order to determine whether the mechanism of mCherry silencing was as predicted based on convergent silencing in S. pombe25, we designed primers to amplify sense and antisense mCherry transcripts. We reasoned that convergent transcription should produce both sense and antisense transcripts. As expected, the levels of sense mCherry transcripts in non-CT F2 embryos were abundant, 30-fold more than the levels detected in CT-mCherry animals, whereas the levels of antisense transcripts were at or just barely above background levels. In contrast, RNA isolation and qPCR analyses from F2 embryos derived from multiple F1s confirmed elevated levels of both sense and antisense mCherry transcripts with slight variation between lines (Fig. 2A). These data argue against the idea that the loss of mCherry is due to simple steric interference from colliding convergent polymerases.
Figure 2. mCherry silencing is Dicer dependent and results in increased levels of H3K9me3 chromatin modification.
(A) Detection of sense and antisense mCherry transcripts via qPCR analysis in F2 embryos from 3 different CT-mCherry F1 lines. (B) Increased mCherry sense mRNA levels upon co-injection of a Dicer MO compared to control (ctrl) MO injected transient transgenics at 54 hpf. (C) Rescue of mCherry protein levels upon co-injection of Dicer MO. Protein lysates were prepared from embryos as in (B) and Western blots were prepared with antibodies against mCherry and α-tubulin. The full gel is shown in Supplemental Fig. 1. For comparison, protein levels in a non-CT embryo are as shown. (D) ChIP-qPCR showing significant enrichment of H3K9me3 levels on convergent chromatin from CT-mCherry F2 embryos (F1 line 9119). Enrichment was determined compared to negative IgG control at 5 dpf. ChIP values and standard deviations are shown from three independent biological experiments. ***p < .001 based on unpaired, two-tailed distribution Student's t-test.
If the mechanism of silencing that we observe is an RNAi-mediated event, as suggested by the presence of sense and antisense transcripts, it should require Dicer activity to cleave dsRNA for packaging into RNA Induced Silencing Complexes29. To test whether decreased levels of Dicer would suppress CT-mCherry silencing, we used antisense morpholinos to knockdown Dicer. Compared to scrambled control morpholinos, Dicer morphants displayed an increase in mCherry levels (Fig. 2B). Not only could we detect increased mCherry transcript levels after Dicer knockdown, we also observed increased mCherry protein levels using Western blots (Fig. 2C; full gel in Supp. Fig. 1). Taken together, these results indicate that silencing of mCherry is suppressed upon Dicer knockdown and consistent with the formation of dsRNA from sense and antisense transcripts. The fact that the levels of suppression are not complete is expected and is likely due to the efficiency of Dicer knockdown but could also indicate a requirement for other components involved in nuclear RNAi mediated silencing.
RNAi-mediated chromatin modification
To further elucidate the mechanism of sustained mCherry gene suppression, we tested whether increased levels of H3K9me3 could be detected in the convergent promoters and within the mCherry open reading frame. Increased levels of H3K9me3 would be indicative of heterochromatin formation in vivo and would be further confirmation that convergent silencing in zebrafish utilizes a similar mechanism as proposed in S. pombe. To test for histone modification, we utilized formaldehyde-based in vivo cross-linking/immunoprecipitation (Chromatin immunoprecipitation; ChIP) with antibodies against H3K9me3. Chromatin was isolated from non-CT and CT mCherry injected embryos and specific primers were then used to amplify the border region between the β-actin promoter and the mCherry ORF, within the mCherry ORF, and in the inverted CMV promoter. We performed ChIP on F1 and F2 embryos (Supp. Fig. 2,3). As shown in Fig. 2D in F2 embryos, compared to the IgG control, we detected a 16-fold enrichment for H3K9me3 modified DNA on the β-actin promoter-mCherry border, a 15 fold enrichment within the mCherry ORF, and a 13 fold enrichment within the inverted CMV promoter. As an additional control, we examined the levels of H3K9me3 on an unrelated, active zebrafish gene, the flk-1 promoter30. We were not able to detect any enrichment for H3K9me3 in the flk-1 promoter (data not shown). Combining experiments to detect sense and antisense transcripts, suppression upon Dicer knockdown, and H3K9me3 marks, our data are consistent with a model of RNAi-mediated heterochromatin silencing in zebrafish.
Convergent silencing in trans: One Eyed Pinhead
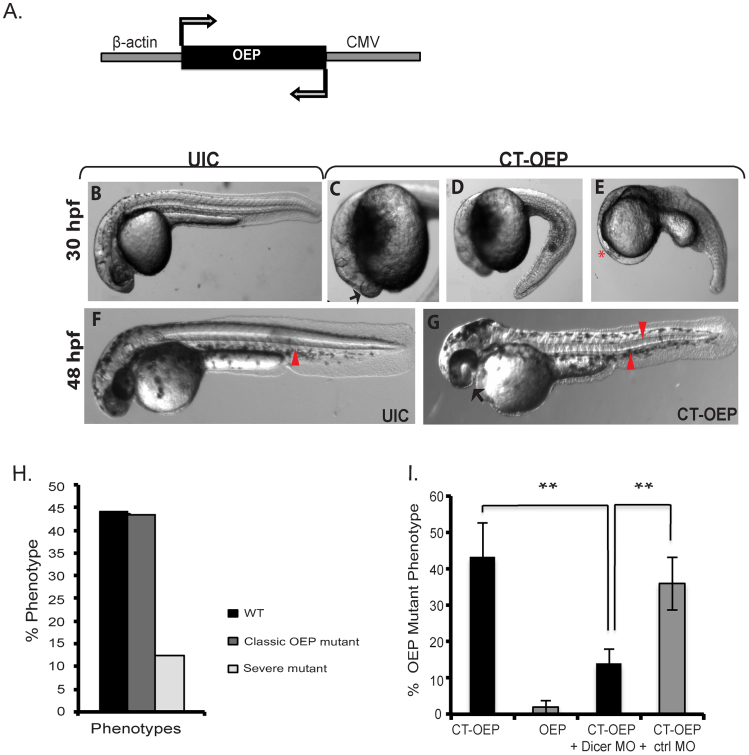
To address the ability of convergent transcription to silence genes in trans, we generated a convergent construct targeting the endogenous One-Eyed Pinhead (OEP) gene31 (Fig. 3A). We chose this gene because the OEP phenotype is well characterized and easy to score. As shown in Fig. 3B–G and quantified in Fig. 3H, 43% of transient transgenic animals injected with CT-OEP displayed cyclopia, strong ventral curvature of the body axis, and a reduced/misshapen notochord, consistent with OEP mutants and morpholino knockdown of OEP31,32. More severe defects were observed in 12% of the embryos with eyeless, headless, or severely reduced body axis. Interestingly, the more severe phenotypes resemble those observed with high concentrations of OEP MOs which were proposed to be due to the loss of both maternal and zygotic OEP expression32. In our experiments, the more severe effects would not be due to destruction of maternal transcripts but are consistent with increased silencing of the endogenous OEP gene.
Figure 3. Silencing of One Eyed Pinhead.
Convergent silencing of the zebrafish One Eyed Pinhead (OEP) gene with the β-actin promoter driving sense transcription and an inverted CMV promoter driving antisense transcription. The OEP open reading frame was directly cloned between the two promoters. (B–G) Compared to uninjected control embryos (UICs), CT-OEP injected embryos phenocopied OEP mutants with curved body axis, cyclopia (black arrow in C), and reduced notochord (red arrows in F compared to G). In 12% of the CT-OEP embryos, we observed a more severe phenotype with complete loss of eye development (red asterisk in E). (H) Graph showing % transient transgenics showing WT, classic OEP, or more severe phenotypes. n = 167. (I). Silencing of OEP is Dicer dependent. Embryos were injected as indicated in the absence or presence of Dicer MO or a control mismatch morpholino. **p < 0.005.
To test whether silencing of OEP in trans could be suppressed by knockdown of Dicer, embryos were co-injected with the Dicer MO. As shown in Fig. 3I, knockdown of Dicer resulted in a substantial decrease in the number of embryos displaying the OEP phenotype from 43% to 15%. The OEP experiments suggest that convergent silencing can be used to silence endogenous zebrafish genes in trans.
Convergent silencing of miR-27a/b
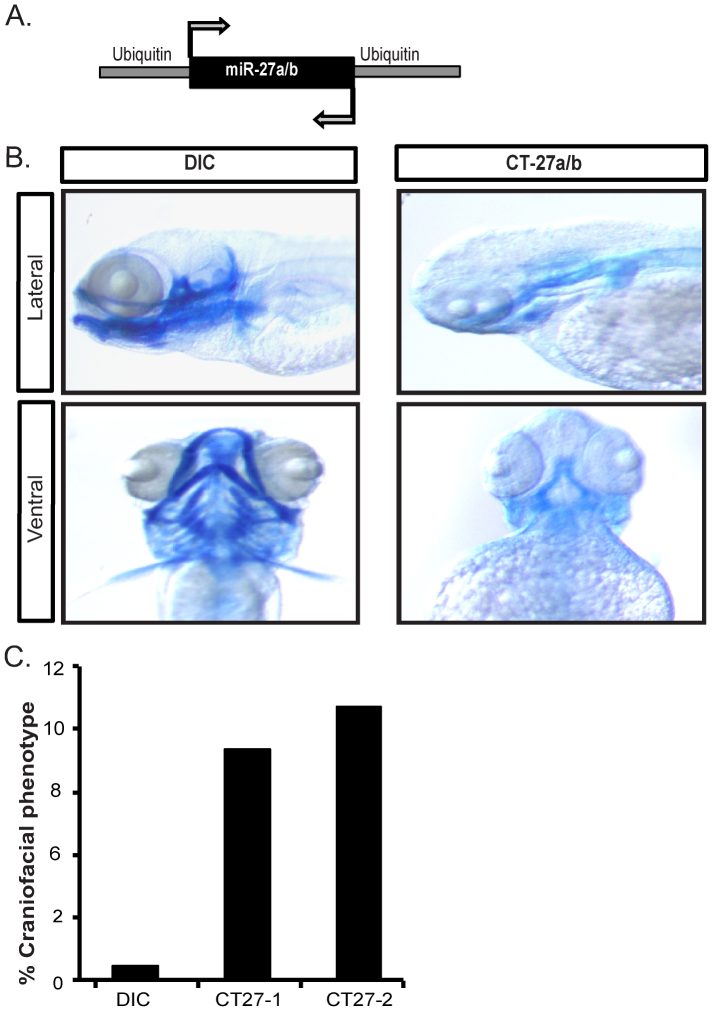
To further demonstrate the utility of targeting endogenous genes with convergent transcription, we decided to target a miRNA gene. We chose miR-27a/b because morpholino and CRISPR knockdown of miR-27a/b results in dramatic craniofacial defects and impairment of pectoral fin outgrowth that can be readily detected both visually, and by staining extracellular matrix (ECM) and cartilage with alcian blue (Kara et al, manuscript in preparation). Tol2 based CT and non-CT constructs were generated with identical convergently arranged zebrafish Ubiquitin promoters flanking a fusion of DNA sequences encoding the precursors of both miR-27a and miR-27b (Fig. 4A). We chose to use the Ubiquitin promoter as a means to test the utility of multiple promoters and to begin to address questions related to differential promoter strength. Although the frequency of the defect was lower than what we observed with mCherry and OEP convergent silencing, we found that transgenic animals containing the CT construct showed a nearly identical phenotype to that produced upon morpholino knockdown (Supp Fig. 4) of miR-27a/b (Fig. 4B). This included loss of craniofacial structures, loss of upper and lower jaws, and impaired pectoral fin outgrowth. Only ~8–10% of transgenic animals displayed the phenotype but this was significantly higher than that compared to DICs where much less than 1% of the embryos showed some form of jaw defect or slight changes in alcian blue staining patterns. While the reasons for less efficient silencing of the miR-27a/b genes are not clear, the results indicate that convergent transcription can be used to silence noncoding RNAs in trans.
Figure 4. Silencing of miR-27a/b.
(A) Convergent silencing of the miR-27a/b genes with ubiquitin promoters driving both sense and antisense transcription. A synthetic DNA sequence encompassing precursor sequences for both miR-27a and miR-27b were directly cloned between the two promoters. (B) Compared to control embryos injected with dye only (DIC), lateral and ventral views show that CT-miR-27a/b injected embryos phenocopy miR-27a/b morphants (see Supplemental Figure 4) with defects in pharyngeal arch morphogenesis, craniofacial defects, and inhibition of pectoral fin outgrowth as shown by decreased staining of ECM (cartilage) with alcian blue. (C) Compared to DICs, craniofacial defects observed with alcian blue staining were observed in 8–9.5% of CT-miR-27a/b embryos using two different plasmid clones of the construct in (A). n = 191 for DIC, n = 458 for CT-miR-27a/b clone 1, and n = 272 for CT-miR-27a/b clone 2. All images are at 4 dpf.
Discussion
Using convergent transcription to induce nuclear RNAi-mediated gene silencing, we achieved targeted silencing of exogenous mCherry and endogenous mRNA (OEP) and miRNA (miR-27a/b) encoding genes. This paper is the first to show that nuclear RNAi mediated gene silencing can be used in zebrafish to trigger heterochromatin formation. The mechanism of silencing in zebrafish is hypothesized to follow that described in fission yeast, which is supported by two sets of experiments. First, Dicer knockdown suppressed transcriptional gene suppression of both mCherry and OEP, suggesting that long dsRNAs derived from the convergent promoters fail to be efficiently processed and thereby unable to elicit TGS. Second, our ChIP analyses showed increased H3K9me3 levels coincident with silencing. Enrichment of H3K9me3 was maintained and became even more pronounced from generation to generation, with greatest H3K9me3 occupancy observed in F2 generations. These novel findings demonstrate that convergent transcription can trigger nuclear RNAi pathways to allow reverse genetics in zebrafish. With multiple tools available to regulate transcription in zebrafish, precise spatial and temporal control of gene expression is possible using convergent transcription33,34.
Currently, the most common method to knockdown genes in zebrafish utilizes antisense morpholinos to block mRNA translation, inhibit pre-mRNA splicing, or interfere with miRNA function32,35. Despite ongoing work, the use of RNAi in zebrafish remains elusive36. Reports of successful knockdown via RNAi have been countered by reports of broad nonspecificity12,13,14,15,16,17,18,19. Recently, shRNA expression vectors have been generated to produce siRNAs mimicking miRNA pathways2,3,37 but it remains to be determined how useful or efficient these methods will be in generating stable knockdown lines. For mCherry knockdown, we observed stable, ongoing silencing. However, we also observed gene dependent differences in efficiency. Silencing of mCherry was greater than 92%, silencing of OEP was approximately 50%, and silencing of miR-27a,b was the least efficient at ~9%. Differences in efficiency can be due to numerous causes with obvious challenges for silencing high copy genes with stable mRNAs. Additionally, as more is learned about how small RNAs direct sequence specific silencing, we will likely learn more about how chromatin is altered and how chromosome location and positioning might affect silencing. Nevertheless, the ability to generate stable convergent lines creates an advantage over standard morpholinos that can lack precise spatial and temporal control, not to mention possible off target and nonspecific effects. As we develop this strategy further, we will examine the expansion of heterochromatin marks on different promoters and ORFs as well as discern the exact requirements for the size and abundance of the convergent transcripts to gain a better understanding of the mechanism and functional relevance of convergent silencing in zebrafish.
In addition to variable efficiencies of silencing in mCherry, OEP and miR-27a,b, we also found that some genes were not able to be targeted for silencing. Convergent constructs targeting different pigment genes (tyrosinase, golden, sandy) showed a delay or only very limited silencing of pigment expression, provided the embryos survived, especially for tyrosinase. Despite changes in the use of different strength promoters or different length ORF sequences in the convergent constructs, we were never able to generate non-pigmented embryos. A potential explanation for the lack of silencing with the CT-pigment constructs could be due to limiting levels of Dicer since Dicer over-expression was shown to promote primary siRNA generation in a genome wide dependent manner in S. pombe38. Further, sequence context and chromosomal position can also inhibit siRNA-mediated heterochromatin formation38. Lastly, even though we have focused extensively on the role of Dicer, other components of the RNAi pathway, such as Argonaute proteins39 are likely required and might be limiting for nuclear RNAi. Future work will be necessary to answer such questions as well as positional and sequence-dependent effects on convergent gene silencing. Nevertheless, our results demonstrate that RNAi-mediated heterochromatin formation can be used to silence genes in zebrafish.
Tools to efficiently knockdown gene expression or generate gene knockouts in zebrafish are rapidly evolving with CRISPR mediated gene editing rapidly becoming a common technique in zebrafish45. In our hands, knockdown of miR-27a/b is most efficient with antisense morpholinos, followed by CRISPR knockout, followed by convergent silencing. As discussed above, morpholino based knockdowns suffer from temporal and spatial limitations. Although spatial restrictions can be overcome using CRISPR technology, temporal and reversible strategies are still lacking and it may not always be desirable to create irreversible genetic mutations. For some applications, convergent silencing could prove useful, especially with tissue specific and inducible promoters that can be further used to control temporal and spatial production of dsRNA.
Methods
Ethics statement
All zebrafish experiments and methods were carried out in accordance with the approved guidelines from the Vanderbilt Institutional Animal Care and Use Committee (IACUC) under protocol M-09-398.
Plasmid constructs
Convergent vectors were created by inserting ORF sequences directly between convergent promoters with no 5′ or 3′ UTR sequences. For miR-27a/b, a region encompassing the precursor sequence for both miRNAs was inserted directly between convergent Ubiquitin promoters (see below). All plasmid constructs were created using Gateway technology28. Destination vectors were created in the pDEST2G2 backbone by Gateway LR Clonase II reactions (Invitrogen) using the following vectors:
For non-CT-mCherry: p5E-β-Actin, pME-mCherry, p3E-pA. CT-mCherry: p5E-β-Actin, pME-mCherry, p3E-iCMV. non-CT OEP: p5E-β-Actin, pME-OEP, p3E-pA. CT-OEP: p5E-β-actin or p5E-CMV, pME-OEP, p3E-iCMV or p3E-iβ-Actin.
Inverted promoters were inserted into p3E (3′-entry vectors) to transcribe the antisense strand of target genes. P3E-iCMV, p3E-iβActin and p3E-Ubi promoter sequences were PCR amplified with Phusion polymerase (NEB) from p5E-CMV/SP6, p5E-β-Actin (Tol2 Kit) and p5E-Ubi (a kind gift from Dr. Josh Gamse), respectively. Forward and reverse primers were flanked by attB2 and attB3 sites (lowercase, respectively:
iCMV: F- 5′ggggacagctttcttgtacaaagtggaTCACCTAAATCAAGCTTGCTC; R- 5′ ggggacaactttgtataataaagttgCCAAGGCCTCTTCGC 3′
iβ-Actin: F- 5′ ggggacagctttcttgtacaaagtggaGGATCCGGCTGAACTGTAAAAGAAAG; R- 5′ ggggacaactttgtataataaagttgCCGGTACCAATTCCAGTTTGAAG
iUbi: F- 5′ ggggacagctttcttgtacaaagtggaGGATCCCTGTAAACAAATTCAAAG; R- 5′ ggggacaactttgtataataaagttgCCCTCGAGACCAGCAAAGTTCTAG
pME-OEP: OEP cDNAs were generated using 500 ng of total RNA from sphere stage wild type embryos using Superscript II reverse transcriptase (Invitrogen). Forward and reverse primers flanked by attB1 and attB2 sites (lowercase) were then used for PCR amplification:
F- 5′ ggggacaagtttgtacaaaaaagcaggctTGCCACCATGACGAGTCAACTGTTCG 3′
R- 5′ ggggaccactttgtacaagaaagctgggtGCATACGGAGCGTTACAGTACA 3′
pME-miR27a/b: The following sequence was ordered from GeneArt (Life Technologies) containing a fusion of precursor sequences for miR-27a and miR-27b:
GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGGGCCAATAGCTAATATGGCCAATGATTTggagagcaTCTGGATATGATGTCTGCTGAAGTTTCGTGAGGTGCAGGACTTAGCTCACTCTGTGAACAGATCTCGGATATCCTATGTTCACAGTGGCTAAGTTCCGCTCCTCTGAGGCCCACACTCGAAATCAGCCAGGaggtgagaacacaaacatgacGCGGCCGCTCTTTTCTAGCAGGTGCAGAGCTTAGCTGATTGGTGAACAGTGATTGAACTCTTTGTTCACAGTGGCTAAGTTCTGCATCTGAGGAGAGGACAGTGTACCCAGCTTTCTTGTACAAAGTGGTCCCC
BP reactions (Invitrogen) were performed to recombine the PCR amplicons into entry vectors following the manufacturers protocol.
All entry vectors were sequence verified with M13 F and M13 (-21) R primers. The exact sequences of the different promoters and maps of the various vectors are contained in the Supplement.
Transgenic animals
Transgenic animals were created using Gateway vectors and the Tol2 transgenesis system28,40 (http://chien.neuro.utah.edu/tol2kitwiki/index.php/Main_Page). For injections, embryos from wild type AB fish were collected from a 15-minute mating period and microinjections of pDEST-based plasmids were performed41. Briefly, plasmid DNA and Tol2 transposase mRNAs were injected into 1–2 cell stage embryos. For non-CT-mCherry and CT-mCherry, 22.5 pg plasmid DNA and 7.5 pg transposase mRNAs were injected. For CT-OEP, 11.25 pg DNA and 7.5 pg transposase mRNAs was injected. For CT-miR-27a/b, 30 pg DNA and 7.5 pg transposase mRNAs were injected.
RNA isolation and sense/antisense transcript detection
Total RNA from GFP+ 4 dpf embryos was isolated using Tri-Reagent (MRC). 20 embryos were used per 1 ml Tri-reagent. 800 ng of RNA was reversed transcribed with M-MLV RT (Promega) using Oligo(dT)16 (Applied Bioscience) or strand-specific primers. Sense and antisense transcripts were generated and quantified by SYBR green (BioRad) in real-time PCR (BioRad). Transcripts were normalized to GAPDH and relative fold changes were normalized to values of WT embryos, set at 1, and determined by the ΔΔCt method42.
Strand-specific primers:
mCherry anti-sense detection: 5′ CGACATCCCCGACTACTTGAAGC
mCherry sense detection: 5′ TCTTGGCCTTGTAGGTGGTCTT
qRT-PCR primers:
GAPDH: F- 5′ GGCAGAAGGCGGCAAACT; R- 5′ CTGGGTCCCTCTCGCTATAGA
mCherry: F- 5′ CCCCGTAATGCAGAAGAAGA; R- 5′ TCTTGGCCTTGTAGGTGGTC
Dicer knock-down
Fluorescein-tagged antisense morpholinos (Gene Tools) against the 5′UTR of Dicer (dicerMO1)43 or the translational start (dicerstart)44 were co-injected with the CT construct and transposase mRNAs into 1–2 cell stage wild-type embryos of the AB strain. As a control, a mismatched Dicer MO (dicermm2)43 was used. Injection of either dicerMO1 and dicerstart produced the same phenotype and comparable levels of rescue. Embryos were examined at 54 hpf.
Western blots
At 72 hpf, GFP+ embryos were de-yolked and protein lysates were prepared from 30 embryos in lysis buffer as described35. 25 μg of total protein was loaded per lane as determined by Bradford assays (Biorad). Blots were probed with anti-mCherry1C51 (Novus Biologicals) and anti-α-Tubulin (Abcam). For detection, ECL Mouse- and ECL Rabbit-IgG-HRP-linked secondary antibodies (GE Sciences-NA931) were used followed by visualization with ECL (Perkin Elmer).
ChIP and qPCR
Approximately 25–60 embryos were washed in 1× PBS. Embryos were incubated with 2.22% formaldehyde for 10 minutes at 25°C with gentle rotation. 150 mM of glycine was used to quench the reaction for 10 minutes at 25°C with gentle rotation. Embryos were then washed three times with 1× PBS for 5 minutes each and then dissociated in ChIP Whole Cell Lysis Buffer (10 mM Tris-HCl, pH 8.1, 10 mM NaCl, 3 mM MgCl2, 1% NP40, 1% SDS, 0.5% DOC, and 1× protease inhibitor cocktail). The protease cocktail was from Sigma (P8340). DNA was fragmented using a probe sonicator (Heat Systems Ultrasonics) followed by water bath sonication (Diagenode) at the high setting (3 rounds of 5 minutes each). After centrifugation for 20 minutes, supernatants were collected. Reverse crosslinking of chromatin aliquots, DNA isolation, and fragmentation checks were performed by incubation at 65°C overnight, phenol/chloroform extraction, and agarose gel electrophoresis, respectively. Immunoprecipitation was conducted using 10 ug of chromatin with 1 ug of either negative control rabbit IgG (Cell Signaling) or H3K9me3 antibody (Diagenode pAb-056-050) incubated overnight at 4°C. Magnetic Protein A beads (Millipore) were pre-washed with chip dilution buffer (16 mM Tris-HCl, pH 8.1,162 mM NaCl, .0096 mg SDS, 2% Triton-X 100) and then blocked by incubation overnight with 1.5% BSA, 0.03% protease inhibitor cocktail, and 0.006% of Herring sperm ssDNA at 4°C. An equal volume of blocked bead solution was then added to each chromatin-antibody bound sample for 1 hour at 4°C. Samples were then washed for 5 minutes each with three different buffers. Wash Buffer 1 (20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100), Wash Buffer 2 (20.5 mM Tris-HCl, pH 8.1, 493 mM NaCl, 2 mM EDTA, 0.103% SDS, and 1% Triton X-100) and Wash Buffer 3 (10 mM Tris-HCl, pH 8.1, 1 mM EDTA, 250 mM LiCl, 1% NP-40, and 1% DOC). Following two washes in TE buffer, antibody bound chromatin was eluted in 1% SDS and 10 mM NaHCO3. Tubes were then incubated at 65°C overnight after addition of 200 mM NaCl and then placed at 45°C after addition of 40 mM Tris-HCl (pH 7.5), 10 mM of EDTA (500 mM) and 20 ug of PK (20 mg/mL) for 1–2 hours. Following reverse crosslinking, DNA was isolated (Qiagen) and dissolved in H20. ChIP-qPCR was performed and quantified using Sybr green (Bio-Rad). Fold enrichment was calculated by normalization of signal (H3K9me3 CT values) to background (IgG CT values) using the ΔΔCT method.
Primers for ChIP-qPCR:
Inverted CMV promoter
F_CMV6879 – 5′ GAC CCC TCC TCA AAC TTG GCA C 3′ Tm = 60.3°C
R_CMV7059 – 5′ GGG ACT CAT GCC AAT TCA ATA TGG TGG ATC 3′ Tm = 60.8°C
Sense β-actin promoter with reverse mCherry ORF
F_5201 bactin qpcr 5′ GGC CAG ACT GTA GAA TGC AGA GC 3′ Tm = 59.8°C
R_5446 mcherry qpcr 5′ CTG AAC TCG TGG CCG TTC ACG 3′ Tm = 60.8°C
mCherry ORF
mCherry_F_414 5′ CCC CGT AAT GCA GAA GAA GA Tm = 54.5°C
mCherry_R_568 5′TCT TGG CCT TGT AGG TGG TC 3 Tm = 56.8°C
Alcian blue staining
Embryos were fixed in 4% PFA and rinsed with PBS-0.1% Tween (PBT) twice for 5 minutes. After briefly rinsing in water, embryos were bleached for 30 minutes in 1.5% H202 in 1% KOH. Following several rinses in PBT, embryos were stained in 0.1% alcian blue 8GX (Sigma) overnight on a shaker at 25°C. Embryos were then washed three times for 1 hour in 70% ethanol in 5% HCL and afterwards washed twice for 1 hour in 0.25% KOH in 20% glycerol. Next, embryos were washed overnight in 0.25% KOH in 50% glycerol and stored at 4°C in 0.1% KOH in 50% glycerol solution. Before viewing stained embryos under the stereomicroscope, 80–90% glycerol was added to embryos and rotated overnight at 4°C.
Author Contributions
All authors contributed to the design of the experiments. O.E.A. and D.J.C. performed experiments, developed methods, analyzed data. C.W. contributed Supplemental Figure 4. O.E.A. and J.G.P. wrote the manuscript.
Supplementary Material
Supplemental Information
Acknowledgments
We thank Qiang Guan for excellent zebrafish care at the Vanderbilt University Stevenson Center Fish Facility. This study was supported by a grant from the National Eye Institute–National Institutes of Health to J.G.P. (R21 EY 019759). O.E.A. was supported in part by grants from the NIH (IMSD GM062459) and the Microenvironmental Influences in Cancer training grant (T32 CA009592). O.E.A., D.J.C. and C.W. were supported in part by the Department of Biological Sciences and the Gisela Mosig Fund. The authors wish to thank Dr. Josh Gamse and Sataree Khuansuwan for Tol2 based reagents and constructs. We also thank Brian McKenna and Dr. Roland Stein for help and advice with ChIP experiments and Daniel Levic for alcian blue staining.
References
- Lieschke G. J. & Currie P. D. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367, 10.1038/nrg2091 (2007). [DOI] [PubMed] [Google Scholar]
- Dong Z., Peng J. & Guo S. Stable gene silencing in zebrafish with spatiotemporally targetable RNA interference. Genetics 193, 1065–1071, 10.1534/genetics.112.147892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rienzo G., Gutzman J. H. & Sive H. Efficient shRNA-mediated inhibition of gene expression in zebrafish. Zebrafish 9, 97–107, 10.1089/zeb.2012.0770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill B. R., Petzold A. M., Clark K. J., Schimmenti L. A. & Ekker S. C. A primer for morpholino use in zebrafish. Zebrafish 6, 69–77, 10.1089/zeb.2008.0555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu, Mary K., Kostas A., Driver E. & Mello C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- Hammond S., Caudy A. & Hannon G. J. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2, 110–119 (2001). [DOI] [PubMed] [Google Scholar]
- Lee Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A., Hammond S. & Hannon G. Role for a bidentate ribonuclease in the intiation step of RNA interference. Nature 409, 363–366 (2001). [DOI] [PubMed] [Google Scholar]
- Cenik E. S. & Zamore P. D. Argonaute proteins. Curr. Biol. 21, R446–449, 10.1016/j.cub.2011.05.020 (2011). [DOI] [PubMed] [Google Scholar]
- Djuranovic S., Nahvi A. & Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240, 10.1126/science.1215691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A. S., Thatcher E. J. & Patton J. G. RNA Interference and miRNAs in Zebrafish. Regulation of Gene Expression by Small RNAs (eds Gaur, R. K. & Rossi, J. J.) 149–172 (CRC Press Boca Raton, 2009). [Google Scholar]
- Wargelius A., Ellingsen S. & Fjose A. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Comm. 263, 156–161, 10.1006/bbrc.1999.1343 (1999). [DOI] [PubMed] [Google Scholar]
- Acosta J., Carpio Y., Borroto I., Gonzalez O. & Estrada M. P. Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J Biotechnol 119, 324–331, 10.1016/j.jbiotec.2005.04.023 (2005). [DOI] [PubMed] [Google Scholar]
- Zhao X. F., Fjose A., Larsen N., Helvik J. V. & Drivenes O. Treatment with small interfering RNA affects the microRNA pathway and causes unspecific defects in zebrafish embryos. FEBS J 275, 2177–2184, 10.1111/j.1742-4658.2008.06371.x (2008). [DOI] [PubMed] [Google Scholar]
- Li Y.-X., Farrell M. J., Liu R., Mohanty N. & Kirby M. L. Double-Stranded RNA Injection Produces Null Phenotypes in Zebrafish. Dev Biol 217, 394 (2000). [DOI] [PubMed] [Google Scholar]
- Oates A. C., Bruce A. E. & Ho R. K. Too much interference: injection of double-stranded RNA has nonspecific effects in the zebrafish embryo. Dev Biol 224, 20–28 (2000). [DOI] [PubMed] [Google Scholar]
- Mangos S., Vanderbeld B., Krawetz R., Sudol K. & Kelly G. M. Ran binding protein RanBP1 in zebrafish embryonic development. Mol Reprod Dev 59, 235–248, 10.1002/mrd.1028 (2001). [DOI] [PubMed] [Google Scholar]
- Gruber J., Manninga H., Tuschl T., Osborn M. & Weber K. Specific RNAi mediated gene knockdown in zebrafish cell lines. RNA Biol. 2, 101–105 (2005). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. U6 promoter-driven siRNA injection has nonspecific effects in zebrafish. Biochem. Biophys. Res. Comm. 391, 1363–1368, 10.1016/j.bbrc.2009.12.065 (2010). [DOI] [PubMed] [Google Scholar]
- Martienssen R. A., Zaratiegui M. & Goto D. B. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 21, 450–456, 10.1016/j.tig.2005.06.005 (2005). [DOI] [PubMed] [Google Scholar]
- Sabin L. R., Delas M. J. & Hannon G. J. Dogma derailed: the many influences of RNA on the genome. Mol. Cell. 49, 783–794, 10.1016/j.molcel.2013.02.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420, 10.1038/nature07756 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M. & Proudfoot N. J. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132, 983–995, 10.1016/j.cell.2008.02.040 (2008). [DOI] [PubMed] [Google Scholar]
- Gullerova M., Moazed D. & Proudfoot N. J. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 25, 556–568, 10.1101/gad.618611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M. & Proudfoot N. J. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat Struct Mol Biol 19, 1193–1201, 10.1038/nsmb.2392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Nieto F. J., Bert A. G. & Cockerill P. N. Transcription-dependent silencing of inducible convergent transgenes in transgenic mice. Epigenet. & Chromatin 3, 3, 10.1186/1756-8935-3-3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. The transgenesis and gene and enhancer trap: Methods in zebrafish by using the Tol2 transposable element. Essential Zebrafish Methods (ed Detrich, H. M., Westerfield, M. & Zon, L.) Ch. 8, 153–174 (Academic Press, Oxford 2009). [Google Scholar]
- Kwan K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
- Castel S. E. & Martienssen R. A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112, 10.1038/nrg3355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. W., Beis D., Mitchell T., Chen J. N. & Stainier D. Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209, 10.1242/dev.02087 (2005). [DOI] [PubMed] [Google Scholar]
- Schier A. F., Neuhauss S. C., Helde K. A., Talbot W. S. & Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development 124, 327–342 (1997). [DOI] [PubMed] [Google Scholar]
- Nasevicius A. & Ekker S. C. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet 26, 216 (2000). [DOI] [PubMed] [Google Scholar]
- Halpern M. E. et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Dong J. & Stuart G. W. Transgene manipulation in zebrafish by using recombinases. Essential Zebrafish Methods Genetics and Genomics (eds Detrich, H. W., Westterfield, M. & Zon, L. I.) Ch. 12, 233–254 (Academic Press, Oxford, 2009). [Google Scholar]
- Flynt A., Li N., Thatcher E., Solnica-Krezel L. & Patton J. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39, 259–263, 10.1038/ng1953 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. & Hurlstone A. F. The use of RNAi technologies for gene knockdown in zebrafish. Brief Funct Genomics 10, 189–196, 10.1093/bfgp/elr014 (2011). [DOI] [PubMed] [Google Scholar]
- Dong M. et al. Heritable and lineage-specific gene knockdown in zebrafish embryo. PloS One 4, e6125, 10.1371/journal.pone.0006125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R., Jih G., Iglesias N. & Moazed D. Determinants of heterochromatic siRNA biogenesis and function. Mol Cell 53, 262–276, 10.1016/j.molcel.2013.11.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Sheets M. D., Imboden S. B. & Dahlberg J. E. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 25, 1121–1131, 10.1101/gad.2038811 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc J. A., Amigo J. & Lawson N. D. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077–3087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster M. L., Kikuta H., Urasaki A., Asakawa K. & Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol 561, 41–63, 10.1007/978-1-60327-019-9_3 (2009). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- Wienholds E., Koudijs M. J., van Eeden F. J., Cuppen E. & Plasterk R. H. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet 35, 217–218 (2003). [DOI] [PubMed] [Google Scholar]
- Thatcher E., Paydar I., Anderson K. K. & Patton J. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci USA 105, 18384–18389, 10.1073/pnas.0803713105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L.-E., Wente S. R. & Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110, 13904–13909, 10.1073/pnas.1308335110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information