Abstract
Oncolytic viruses (OVs) and bacteria share the property of tumor-selective replication following systemic administration. In the case of nonpathogenic bacteria, tumor selectivity relates to their ability to grow extracellularly within tumor stroma and is therefore ideally suited to restricting the production of bacterially produced therapeutic agents to tumors. We have previously shown the ability of the type 1 interferon antagonist B18R to enhance the replication and spread of vesicular stomatitis virus (VSV) by overcoming related cellular innate immunity. In this study, we utilized nonpathogenic bacteria (E. coli) expressing B18R to facilitate tumor-specific production of B18R, resulting in a microenvironment depleted of bioactive antiviral cytokine, thus “preconditioning” the tumor to enhance subsequent tumor destruction by the OV. Both in vitro and in vivo infection by VSVΔ51 was greatly enhanced by B18R produced from E. coli. Moreover, a significant increase in therapeutic efficacy resulted from intravenous (IV) injection of bacteria to tumor-bearing mice 5 days prior to IV VSVΔ51 administration, as evidenced by a significant reduction in tumor growth and increased survival in mice. Our strategy is the first example where two such diverse microorganisms are rationally combined and demonstrates the feasibility of combining complementary microorganisms to improve therapeutic outcome.
Introduction
Oncolytic viruses (OVs) promise to improve cancer patient outcomes through their tumor-selective replication and multi-modality attack against cancers.1,2 Vesicular stomatitis virus (VSV) has a broad cancer cell tropism and is effective when administrated intravenously in murine tumor models. The wild-type strain of VSV expresses a protein, the M or matrix protein, that upon infection acts as an intracellular antagonist of interferon (IFN) production by blocking the transport of IFN mRNAs from the nucleus.3 Wild-type VSV is neurotoxic in certain mouse strains, while an attenuated version of the virus (VSVΔ51) retains oncolytic activity but is harmless when administered intravenously as the virus can now only productively infect tumor cells that have a defective IFN response.4 However, one of the major problems with oncolytic virotherapy, is that that some tumors, or regions of tumors, have intact or upregulated antiviral responses, and related intra- and intertumor heterogeneity can result in incomplete oncolysis following VSV therapy. Some human-derived tumor cell lines such as HT29 colon carcinoma retain at least partial responsiveness to IFN and only poorly support the spread of VSVΔ51. We have previously determined the IFN responsiveness of HT29 cells and demonstrated that they could be protected from infection by VSVΔ51 through addition of exogenous IFN-α.1 Previous studies from our lab have identified that localized expression of B18R, a gene from Vaccinia virus which encodes a secreted decoy receptor with a broad antagonizing effect against type 1 IFNs, significantly improved the efficacy of the attenuated VSVΔ51 to grow and kill tumors.1,5,6 A number of studies have explored combination viral strategies,7,8,9 but toxicity and safety remain a concern, as knockdown of type 1 IFN in tissues other than tumor presents the risk of susceptibility to unrestricted replication of other infecting viruses. Therefore, a tightly controlled mechanism for tumor-specific B18R production is required to facilitate this strategy.
The phenomenon of tumor-specific bacterial replication has been known for a century. The specificity is believed to be as a result of the uniqueness of tumor physiology resulting from a combination of factors such as local immune suppression, irregular vasculature, relevant nutrient presence in necrotic tissue and the anaerobic nature of hypoxic/necrotic regions within tumors promoting growth of anaerobic and facultatively anaerobic bacteria.10,11 These regions of tumors are vital to target therapeutically, being a major source of cells responsible for tumor regrowth posttreatment. Many bacteria are readily genetically engineered to produce heterologous proteins. Unlike typical gene delivery vectors, the property of tumor cell invasion is not required, as bacteria themselves can express the gene of interest. As a means of local in situ production therefore, noninvasive bacteria (lacking the ability to mediate disease) present an ideal opportunity to act as a “protein factory” within the tumor. Many species of bacteria are health-promoting or probiotic, including certain strains of Escherichia coli.12,13,14 We have previously demonstrated tumor-specific replication of E. coli MG1655 in mice.15
We hypothesized that delivery of E. coli producing B18R, prior to systemic VSV administration, would safely facilitate tumor-specific knockdown of type 1 IFN and locally enhanced VSV-mediated tumor oncolysis. This study examines the replication and spread of VSVΔ51 at in vitro and in vivo levels in murine tumor models and the effects of E. coli–produced B18R.
Results
E. coli expresses functional B18R
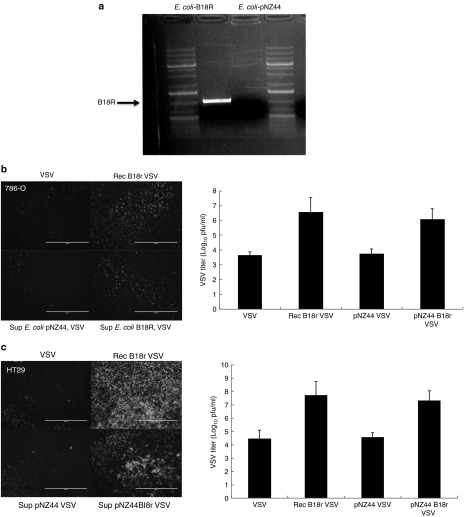
E. coli was engineered to stably express B18R, in addition to a control bacterial construct, containing the plasmid backbone lacking the B18R DNA sequence (E. coli–pNZ44). The new constructs' abilities to express the introduced heterologous gene were demonstrated by reverse transcription polymerase chain reaction (Figure 1a). The activity of the protein produced was examined in an established bioassay for B18R. Supernatant from E. coli cultures expressing or not B18R were compared with purified recombinant B18R protein for their ability to increase VSV IFN sensitive virus replication in two different cell lines (786-O renal cell carcinoma or HT29 colorectal carcinoma) (Figure 1b). In both tumor cell lines, a dramatic increase in VSV titres afforded by E. coli–B18R supernatant was observed, indicating the production of biologically active B18R protein.
Figure 1.
E. coli expresses functional B18R. (a) E. coli pNZ44 and E. coli–B18R were grown for 24 hours in LB-Chloramphenicol, before extraction total RNA. Reverse transcription polymerase chain reaction (RT-PCR) specific for B18R was performed on resultant cDNA. (b) B18R bioassay. 786-O and HT29 tumor cell lines were pretreated or not with recombinant B18R or with supernatant from bacteria expressing or not B18R. Pretreatment was performed 4 hours prior addition of VSVd51-eGFP at MOI = 0.05 for 786-O and 0.001 for HT29. GFP images were taken 36 hours after addition of virus. GFP, green fluorescent protein; VSV, vesicular stomatitis virus.
E. coli–B18R reduces IFN levels in vitro
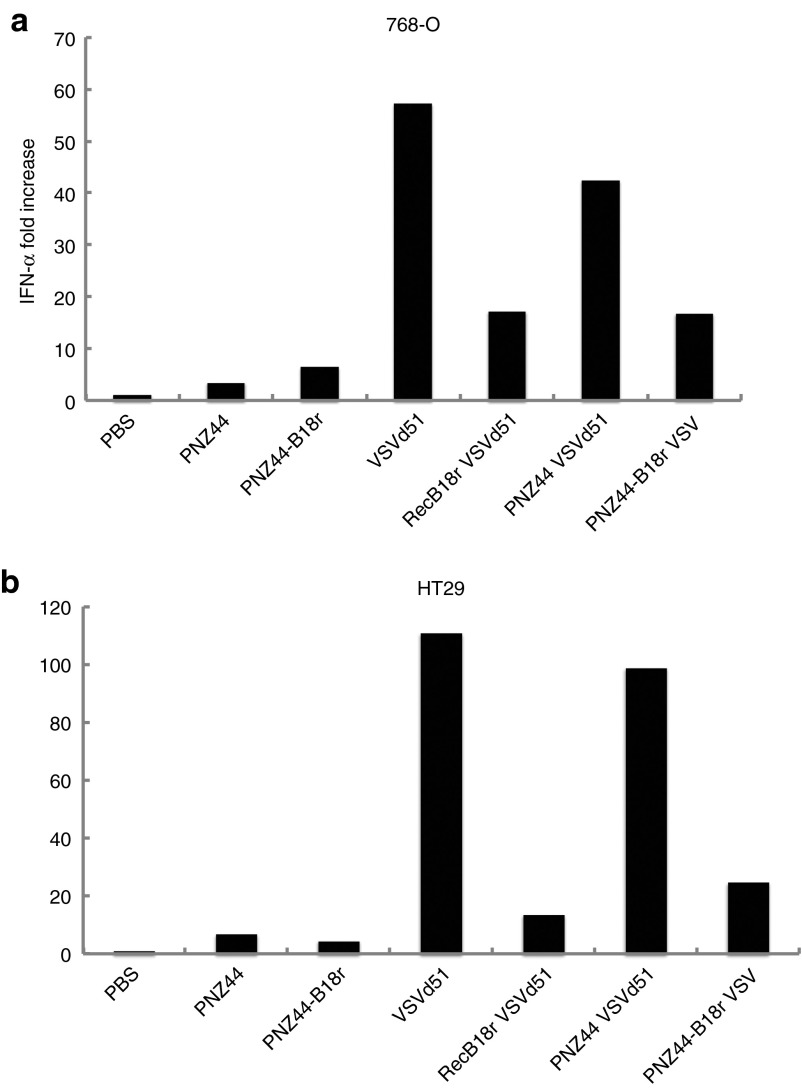
To validate that the effect on VSV replication was related to type 1 IFN inhibition by B18R expression, the effects of bacterial supernatants on IFN-α levels in VSV-infected cell lines were examined by enzyme-linked immunosorbent assay (ELISA). Supernatant from E. coli cultures were compared with purified recombinant B18R protein. Figure 2 shows reductions in IFN-α levels in both 786-O and HT29 cell cultures afforded by E. coli–B18R supernatant, in line with the positive control recombinant B18R protein.
Figure 2.
E. coli–B18R reduces interferon (IFN) levels in cancer cells. The effect of bacterial supernatant on IFN-α levels in vesicular stomatitis virus (VSV) infected cell lines was examined by enzyme-linked immunosorbent assay (ELISA). Reductions in IFN-α levels in both (a) 786-O and (b) HT29 cell cultures was evident in E. coli–B18R supernatant and recombinant B18R protein groups. IFN-α–fold increase is shown.
E. coli–B18R enhances VSV infection in vitro
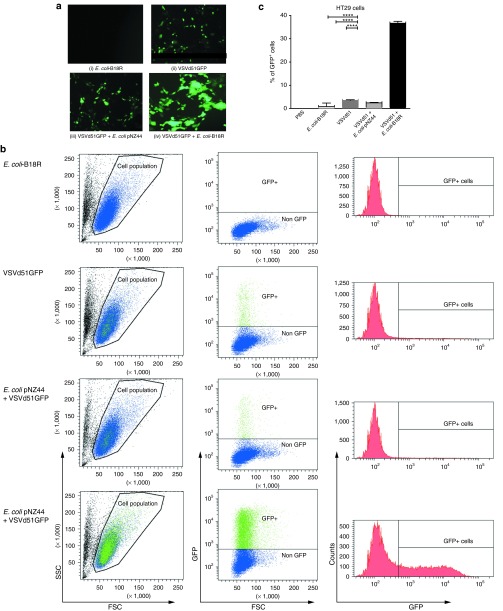
The effects of E. coli–produced B18R on VSVΔ51 replication and spread in cancer cells was examined in HT29 cells in vitro, initially using VSVΔ51-GFP (green fluorescent protein) to permit monitoring of viral spread within the treated culture by fluorescence microscopy. HT29 cell monolayers were unaffected by incubation with the supernatant of E. coli ± B18R in the absence of VSV and no evidence of significant cell killing was detected (P > 0.388) (Supplementary Figure S1b). When E. coli–B18R supernatant incubation was followed by infection with VSVΔ51GFP, a rapid spread of VSVΔ51 was readily detected by fluorescence microscopy. The microscopic examination revealed increased oncolysis due to sensitization of neighboring cells to VSVΔ51 infection (Figure 3a). Total GFP levels were quantified by flow cytometry 6 hours after VSVΔ51 incubation and a significant increase in the total GFP-positive cells was observed in combination treated samples (Figure 3b). The combination treatment resulted in a 1048 ± 396% increase in GFP-positive cells in comparison with virus alone treated wells and a 1497 ± 528% increase over the backbone bacterial vector (E. coli pNZ44) plus virus treated wells (Figure 3c).
Figure 3.
Bacterial B18R production increases VSVΔ51 replication in vitro. The effect of E. coli ± B18R supernatant on VSVΔ51GFP replication within HT29 cells. Bacterial supernatants were coincubated with confluent HT29 cells for 2 hours and washed with PBS. VSVΔ51GFP was added at 105 pfu to the appropriate wells. Six hours later, the cells were analyzed for GFP expression by fluorescence microscopy and flow cytometry. (a) Representative fluorescent microscopy images from (i) E. coli–B18R (bacteria alone), (ii) VSVΔ51GFP (virus alone), (iii) E. coli pNZ44 plus VSVΔ51GFP (backbone vector plus virus), (iv) E. coli–B18R plus VSVΔ51GFP (B18R vector plus virus). (b) Representative dot plots and histograms showing the percentage of GFP+/VSV-infected cells in the presence or absence of E. coli–B18R. (c) Data represent the mean percentage of GFP+ cells and are expressed as the mean ± SEM of 2–4 samples per group. Statistical significance was determined by unpaired Student's t-test,*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. FSC, forward scatter; GFP, green fluorescent protein; SSC, side scatter; VSV, vesicular stomatitis virus.
To further quantitatively examine the effects of bacterial-produced B18R on VSVΔ51 replication levels, a firefly luciferase expressing VSVΔ51FLuc was employed. HT29 cells were incubated with supernatant from either E. coli pNZ44 or E. coli–B18R, alone or in combination with VSVΔ51FLuc. Infection of HT29 cells by VSVΔ51 was significantly enhanced by E. coli–B18R (P = 0.032) but not E. coli pNZ44 (P = 0.545), as evidenced by VSVΔ51FLuc luminescence with a 2.33 ± 0.01-fold increase in luminescence (Supplementary Figure S1a). Lewis lung carcinoma (LLC) cells were also examined in a similar assay, but due to the rapid oncolysis induced by VSVΔ51 alone, no improvements due to E. coli–B18R could be measured within the limits of the in vitro assay (data not shown).
Tumor-specific growth of E. coli following intravenous administration in vivo
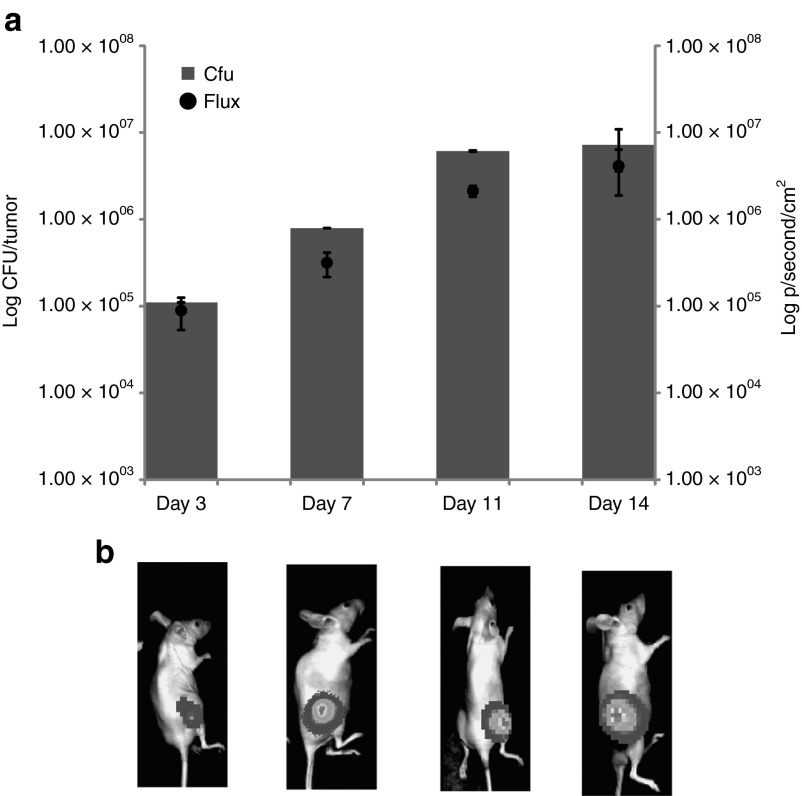
Bacterial homing to, and replication within, subcutaneous tumors following intravenous administration was visualized over time using 2D whole body BioLuminescence Imaging of mice. 1 × 106 E. coli–lux was administered via tail vein injection to athymic mice bearing subcutaneous LLC tumors (n = 6). In vivo BioLuminescence Imaging was performed at various times after bacterial administration. Lux signal was detected specifically in tumors of mice 3–14 days after intravenous administration of E. coli (Figure 4a,b). Bacterial replication in tumors was confirmed by ex vivo bacterial culture (Figure 4a) and viable bacterial numbers correlated with bioluminescence (Figure 4b). Viable bacterial culture indicated E. coli numbers of <100 cfu/g tissue in liver and spleen at the various time points, consistent with our previous findings using this strain,16 confirming tumor specific replication of the bacteria and no off target growth, validating its safety profile.
Figure 4.
IV administered E. coli specifically colonizes tumors in mice. Subcutaneous LLC tumors were induced in MF1 nu/nu mice and bacteria administered upon tumor development (12–14 days; average tumor volume 100 mm3). For intravenous administration, each animal received 106 cells injected directly into the lateral tail vein. (a) Recovery from subcutaneous tumor tissue (bars, y axis) and bacterial lux expression in vivo in live mice (black circles, z axis and images). Time scale (x axis) is time in days after bacterial administration. Increase in bacterial numbers and lux gene expression specifically in tumors was observed over time. (b) There was no detectable luminescence in organs of treated animals LLC, Lewis lung carcinoma.
E. coli–B18R enhances VSVΔ51 replication specifically in tumors
The temporal growth pattern of the E. coli in tumors, based on the above E. coli–lux experiments, was used to design the timing strategy for combination treatments. Nude mice bearing either HT29 or LLC subcutaneous tumors were monitored for tumor development and once tumors reached an average volume of 100 mm3, animals were randomly assigned to the various treatment groups. A nonlux tagged E. coli was used in these experiments, with VSVΔ51-luciferase the sole source of luminescence, used for quantitation of VSVΔ51 replication and biodistribution. A parallel group was run concurrently and injected with the bacterial lux reporter strain (E. coli–lux). Following bioluminescence imaging of the reporter group we observed bacterial tumor colonization as expected at 5 days after injection of the bacteria. At this time, VSVΔ51-luciferase was administered intravenously and expression of the virally encoded luciferase reporter monitored over time.
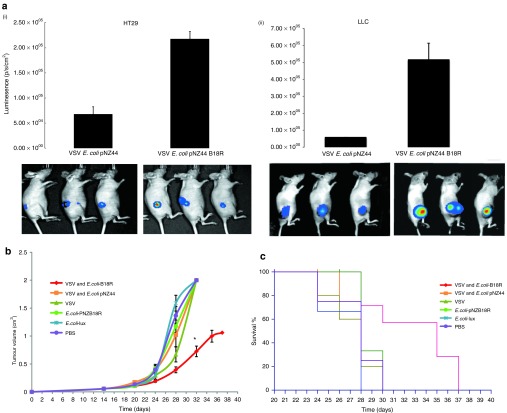
Significant VSVΔ51-associated luciferase expression was observed in tumors only in doubly treated animals (Figure 5a), and was significantly higher than all other groups (P < 0.04). The VSVΔ51-luciferase signal peaked at ~5 days after infection and remained detectable in the tumor for up to 11 days after infection. It should be noted that inevitable cell death of infected cells ultimately reduces VSVΔ51 levels over time, affecting the luminescence read out. Importantly, during daily monitoring of infected animals, no “off-target” VSV infection of healthy tissues was observed.
Figure 5.
E. coli–B18R increases oncolytic virus replication and tumor destruction in vivo. HT29 or LLC subcutaneous tumors were raised in athymic mice. E. colicontaining either an empty vector (pNZ44) or the therapeutic B18R plasmid were administered IV at 106 cfu in 100 µl. One week after bacterial administration, 107 pfu VSVΔ51FLuc were injected IV, and VSVΔ51FLuc luminescence measured by BioLuminescence Imaging (BLI) intermittently, (a) VSV-related luminescence measurements and representative images from (i) HT29 or (ii) LLC xenograft bearing mice in the absence or presence of bacterial B18R expression. Representative data shown relate to 40 hours after VSV administration. (b) LLC tumor growth over time. *Significant difference in tumor volumes at day 32 (P < 0.05). (c) Kaplan–Meier survival curves for LLC-bearing athymic mice treated with (i) E. coli–B18R plus VSVΔ51FLuc (combination treatment, red), (ii) E. coli pNZ44 plus VSVΔ51FLuc (backbone E. coli vector plus VSV, orange), (iii) VSVΔ51FLuc alone (dark green), (iv) E. coli–B18R alone (bright green), (v) E. coli–lux (reporter strain, blue), or (vi) PBS (untreated, purple). Survival was significantly prolonged by E. coli–B18R, with mean survival of the E. coli–B18R plus VSVΔ51FLuc group significantly higher than any of the other groups (P < 0.0211). LLC, Lewis lung carcinoma; VSV, vesicular stomatitis virus.
Effects on tumor growth
The effects of E. coli–B18R on VSVΔ51-mediated tumor destruction was monitored by tumor volume measurements over time. While the virus was capable of replication and LLC cell killing in the absence of B18R, its potency mirrored the in vitro studies described above, with dramatic improvement in antitumor activity in the presence of B18R. Reduced tumor growth and prolonged survival of the combination treatment group in comparison to the backbone vector treatment group (P = 0.002) or the virus alone treatment group (P = 0.006) confirmed the ability of the bacterial B18R to enhance the oncolytic potential of VSVΔ51 in this aggressive tumor model (Figure 5b,c). Survival was significantly prolonged by E. coli–B18R, with mean survival of the E. coli–B18R plus VSVΔ51FLuc group significantly higher than any of the other groups (P < 0.0211).
Tumor specificity of VSVΔ51 replication in combination treated mice
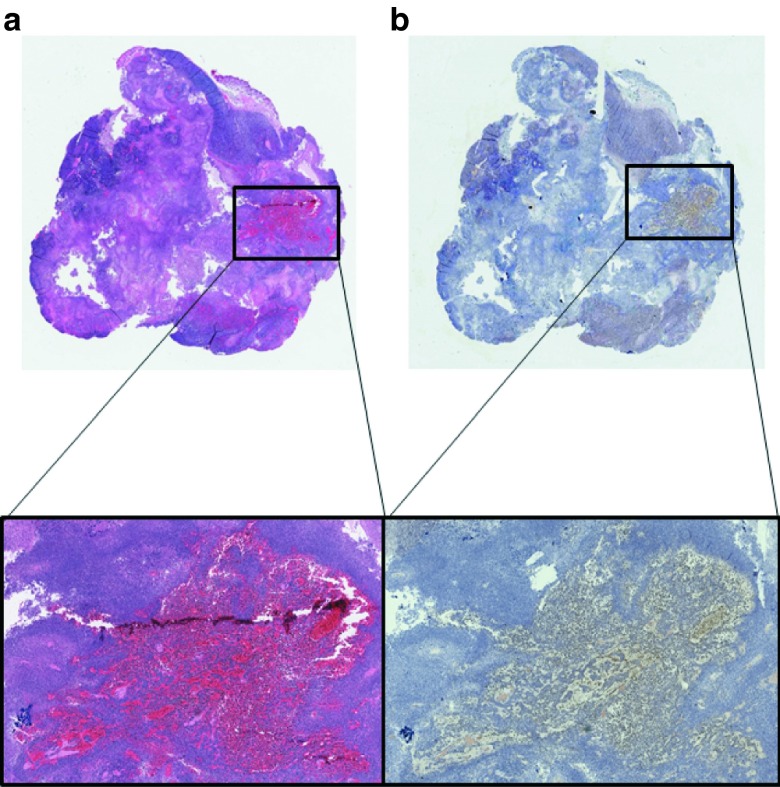
Immunohistochemistry specific for VSV was used to examine the presence of VSV in tumor as well as various organs following treatments. VSV staining was detected by immunohistochemistry 5 days after virus administration only in the tumors of combination treatment animals (Figure 6a). Consecutive sections of all tissues were also stained by H&E, with tumor tissue in combination treatment groups displaying denucleation and evidence of cell death (Figure 6b).
Figure 6.
Histological analyses of treated mouse tissues. Tumors and organs (liver, spleen, kidney, and brain) from the various groups were harvested at necropsy. (a) Immunohistochemistry specific for VSV was performed on sectioned tissues. Positive staining was observed only in combination treated tumors and can be visualized (brown) in the expanded magnification. (b) Consecutive sections were stained with H&E for general morphology and confirm the denucleation and tumor cell destruction caused by the combination therapy.
Immune responses to treatments
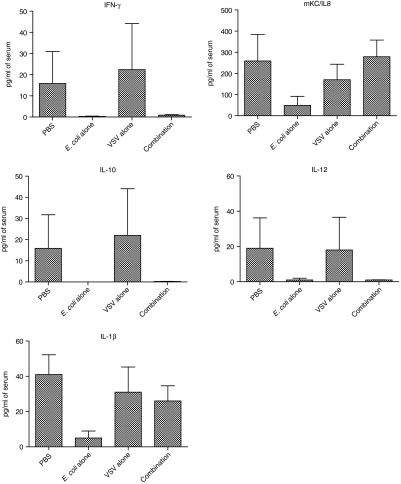
Mindful that this strategy involves the systemic administration of two replication-competent biological agents to the body, we sought to examine potential immune responses to the treatments. Five days after virus administration (=11 days after administration of E. coli), a subset of animals which received either phosphate-buffered saline (PBS), bacteria alone, virus alone or the combination treatment (E. coli–B18R plus VSVΔ51FLuc) were euthanized and serum collected following cardiac puncture. Serum was analyzed using the 7-plex proinflammatory cytokine plate from Meso Scale Discovery. Neither TNF-α nor IL-6 were at detectable levels in any samples. Concentrations of (i) IFN-γ, (ii) mKC, (iii) IL-10, (iv) IL-12p70, and (v) IL-1β were quantified (Figure 7). While increased levels of the neutrophil chemoattractant mKC (the murine homologue of IL-8) was detected in all groups, no treatment group displayed levels statistically significantly higher than the PBS group (P < 0.9024). For the other cytokines profiled IFN-α, IL-10, IL-12, and IL-1β, there was no upregulation (Figure 7). Overall, these data suggest a lack of sustained proinflammatory response to either the bacterial or viral vectors, indicating the safety of this strategy.
Figure 7.
Inflammatory responses to treatments. Serum cytokine profiles. Concentrations of (i) IFN-γ, (ii) mKC, (iii) IL-10, (iv) IL-12p70, and (v) IL-1β in serum at 5 days after virus administration (=11 days after administration of the E. coli) were measured by an ultra-sensitive mouse proinflammatory cytokine kit from Meso Scale Discovery. Whole blood was taken by cardiac puncture and the serum separated from animals which had received either phosphate-buffered saline (PBS), bacteria alone, virus alone or the combination treatment of E. coli–B18R plus VSVΔ51FLuc. All values are represented as pg/ml of serum.
Discussion
Viral-mediated oncolysis is a cancer therapy approach with the potential to be more effective and less toxic than current regimes due to the viruses' selective growth and amplification in tumor cells. To date, OV have proven extremely safe in patients, but have generally fallen short of their expected therapeutic value as monotherapies.17,18 The immune-mediated clearance of the virus is considered to be a significant hindrance to the action of OV and therefore we proposed to develop a new approach combining biotherapeutics to improve efficacy of the OV, and in particular, intratumoral spread. We previously demonstrated that localized expression of the IFN antagonist B18R within a tumor shields the OV from immune-mediated clearance, enhancing the oncolytic potential of the virus to eliminate malignancies.1 However, such a strategy requires an efficient, specific and safe means to localize B18R expression to the desired region of tumors. Nonpathogenic bacterial vectors represent an ideal solution to this problem, since we have demonstrated high-level growth and transgene expression targeted to various tumors.15,19
Various preclinical therapeutic trials have shown the ability of different bacterial strains to traffic to tumor sites, primarily in the context of delivery of DNA for subsequent tumor cell expression.20 In general, invasive or pathogenic species have been exploited for this purpose to date.21,22,23 However, even with safety attenuation, the inherent pathogenicity and immunogenicity of these bacteria has outweighed the therapeutic responses in patients.24 Therefore, we have chosen to exploit a nonpathogenic E. coli strain and validated that this strain colonizes tumors with efficiencies similar to the best-described species in the field, Salmonella Typhimurium.15 The E. coli strain employed here is noninvasive, and therefore does not act as a cell transfection agent, but rather replicates within the tumor stroma, external to tumor cells.
Importantly in our mouse experiments, we found that both virus and bacterial replication, even in coinfected animals, was restricted to tumors with no evidence of infection or colonization of normal tissues detected by imaging (Figures 4b and 5a) and immunohistochemical staining (data not shown). Furthermore, the safety profile of this combination as illustrated by the cytokine profiling demonstrates its potential to be safely used even in immunocompromised hosts. The cytokine profiles showed no overall increase in proinflammatory cytokines, in fact the bacteria for unknown reasons appeared to decrease the cytokine levels; this may be due to its commensal nature.25
At the treatment doses used in these experiments, we found that therapy with either microorganism on its own had little impact on these rapidly growing tumors and did not impact cumulative survival. However, when E. coli–B18R and VSVΔ51 were sequentially administered, substantially slower tumor growth resulted. In addition, the combination treatment could significantly extend survival as compared to all other treatment arms, a considerable improvement given the aggressiveness of the LLC tumor model. We have therefore successfully exploited the local bacterial production of B18R to create a microenvironment depleted of bioactive antiviral cytokines thus permitting robust replication and spread of VSVΔ51. Furthermore, there is potential to enhance this therapeutic approach as both organisms can be further genetically manipulated to improve efficacy. We are cognisant that one of the major factors limiting VSV-mediated oncolysis is that of adaptive immune responses removing virus before it can “finish” the tumor. However, for the purposes of this proof-of-concept study, we opted for athymic mice rather than an immune competent, due to improved growth of the bacteria in xenografts in athymic mice,19 which we believe aids in delineating the mechanism of action of bacterial-produced B18R's involvement in the strategy.
The nature of these organisms' individual actions are confined to the tumor as the bacteria are unable to colonize healthy tissue and consequently, the B18R expression, providing the IFN depleted environment necessary for VSVΔ51 replication and spread, is tumor specific. Combination of these microorganisms, potentially including additional therapeutic genes targeting multiple or sequential pathways, could further enhance efficacy and prevent the development of resistance with no added toxicity. Furthermore, the tumor-specific nature of both of these modalities when administered intravenously permits safe targeting of systemic metastases. Thus, the potential high degree of safety and efficacy predicted for combination therapy of cancer warrants further investigation at both preclinical and clinical levels.
Several cancer therapeutic approaches employing bacteria have been investigated, using different strategies (cancer cell gene delivery, vaccination, prodrug activation, etc.) with varying degrees of success.26,27,28,29 In general, however, hopes for monotherapies employing bacteria have not been fulfilled clinically.21,22,23 The study reported here, represents the first description of such combined biological treatment, with bacteria acting as tumor conditioning agents to support a subsequent directly oncocidal therapeutic. It is possible that combination or neo-adjuvant approaches such as this may represent the most valuable strategy for employment of bacteria in cancer therapy.
Materials and Methods
Bacterial strains. E. coli K12 MG1655 containing the either the control empty pNZ44 plasmid (E. coli pNZ44)30 or pNZ44B18R (B18R polymerase chain reaction amplified from Vaccinia virus) cloned downstream of p44 promoter as NcoI, XbaI insert) plasmid (E. coli–B18R) were grown aerobically at 37 °C in LB medium (Sigma-Aldrich, Co, Wicklow, Ireland) supplemented with 20 μg/ml chloramphenicol (Cm, Sigma-Aldrich). The bioluminescent derivative of MG1655 (E. coli–lux) which was used in a parallel group to monitor bacterial growth through bioluminescence was created using the plasmid p16Slux which contains the constitutive PHELPluxABCDEoperon31 on the backbone of pGh9::ISS1, a thermo-sensitive shuttle vector which integrates randomly into the bacterial chromosome as a consequence of the presence of ISS132 and was grown aerobically at 37 °C in LB medium supplemented with 300 μg/ml erythromycin (Em, Sigma-Aldrich).
Virus preparation. Recombinant AV3 strain of VSV with green fluorescent protein (VSVΔ51-GFP), or luciferase (VSVΔ51-Luc) were propagated in Vero cells. Virions were purified as previously described.33
Tumor cell lines and culture. Murine Lewis Lung Carcinoma and HT29 adenocarcinoma cells were maintained in culture at 37 °C in a humidified atmosphere of 5% CO2, in Dulbecco's modified Eagle's medium (GIBCO, Invitrogen, Paisley, Scotland) supplemented with 10% iron-supplemented donor calf serum (Sigma Aldrich, Ireland), 300 µg/ml L-glutamine.
B18R bioassay. 786-O and HT29 tumor cell lines were pretreated or not with recombinant B18R (0.1 μg/μl, Affymetrix BioScience, San Diego, CA) or with supernatant from bacteria expressing or not B18R. Bacteria from a 24 hours culture (OD600 nm = 2.0 ± 0.2) were pelleted by centrifugation and supernatant filtrated with 0.22 µmol/l filter. Filtrates were used V/V with media on cells. Pretreatment was performed 4 hours prior addition of VSVd51-eGFP at MOI = 0.05 for 786-O and 0.001 for HT29. Fluorescent microscopy (Olympus, Tokyo, Japan) was performed for GFP 36 hours after addition of virus.
In parallel, bacterial supernatant effect on IFN-α levels was investigated by ELISA. Pretreatment were performed 4 hours prior addition or not of VSVd51 at MOI = 0.05 in 786-O and 0.001 in HT29. IFN-α ELISA was performed 24 hours after virus addition on cell supernatants and was carried out using IFN-α (Mabtech, Cincinnati, OH) according to the manufacturer's instructions.
In vitro incubation studies. E. coli containing either pNZ44 (backbone vector) or E. coli–B18R were grown to mid-log OD600 nm of 0.6, harvested by centrifugation and the supernatant removed. This supernatant was filter sterilized and added to confluent six-well plates containing the cell line of interest (HT29) for 2 hours. The cells were then washed in PBS and DMEM ± VSVΔ51-GFP or separate plates with ± VSVΔ51-luc2 at 105 pfu added for 6 hours. Following incubation the cells were imaged for fluorescence or quantified for luminescence using the IVIS imaging system. Subsets of wells were counted for total cell numbers and processed for flow cytometry. The total cell death in virus treated versus untreated cell lines was examined as follows: culture medium was removed from wells; cells were fixed in 96% ethanol for 10 minutes and stained with Prodiff solution C (Braidwood Laboratories, East Sussex, UK). Plates were scanned using the Odyssey IR imaging system (Li-Cor, Cambridge, UK) and viable cells quantified.
FACS analyses. The confluent monolayers of HT29 cells treated with either media alone or media containing filter sterilized bacterial supernatants (E. coli–pNZ44 or E. coli–B18R) and subsequently VSVΔ51GFP were washed, counted and resuspended at 1 × 106 cells/ml for flow cytometric analysis. GFP fluorescence intensity was measured using a LSRII cytometer and BD Diva software (Becton Dickinson, Oxford, UK). For each sample, 30,000 events were recorded. The percentage of GFP+ cells was calculated in wells which were exposed to either virus alone, virus plus backbone vector supernatant (E. coli–pNZ44) or virus plus B18R vector supernatant (E. coli–B18R). The results represent the percentage of positively stained cells in the total cell population exceeding the background staining signal.
Ethics statement. All murine experiments were approved by the Animal Ethics Committee of University College Cork or the Institutional Guidelines Review Board for Animal Care at the University of Ottawa.
Animals and tumor induction. Mice were kept at a constant room temperature (22 °C) with a natural day/night light cycle in a conventional animal colony. Standard laboratory food and water were provided ad libitum. Before experiments, the mice were afforded an adaptation period of at least 7 days. Female mice in good condition, without fungal or other infections, weighing 16–22 g and of 6–8 weeks of age, were included in experiments. For routine tumor induction, the minimum tumorigenic dose of cells (5 × 105 LLC; 3 × 106 HT29) suspended in 200 μl of serum-free culture medium was injected subcutaneously (s.c.) into the flank of athymic MF1-nu/nu mice (Harlan, UK or Charles River Laboratories, Wilmington, MA). The viability of cells used for inoculation was greater than 95% as determined by visual count using a hemocytometer and Trypan Blue Dye Exclusion (Gibco, Thermo Fisher Scientific, Waltham, MA), or the Nucleocounter system (ChemoMetec, Bioimages, Allerød, Denmark). Following tumor establishment, tumors were allowed to grow and develop and were monitored twice weekly. Tumor volume was calculated according to the formula V = (ab2) Π/6, where a is the longest diameter of the tumor and b is the longest diameter perpendicular to diameter a. When tumors reached ~100 mm3 in volume, the mice were randomly divided into experimental groups.
In vivo bacterial administration. Inocula were prepared by growing E. coli pNZ44, E. coli–B18R or the integrated p16Slux aerobically in 100 ml LB broth containing either 20 µg/ml Cm (pNZ derivatives) or 300 µg/ml Em (p16Slux). Cultures were harvested by centrifugation (4,000g for 15 minutes), washed three times with PBS and resuspended in a one-tenth volume of PBS. The viable count of each inoculum was determined by retrospective plating on LB agar containing the appropriate selective antibiotic. For tumor-related studies, mice were randomly divided into experimental groups when tumors reached ~100 mm3 in volume, and administered E. coli or an equal volume of PBS as control. Each animal received 106 E. coli in 100 μl injected directly into the lateral tail vein, as previously described.34
In vivo efficacy studies. Mice were treated with either E. coli–B18R (1 × 106 cfu/ml), E. coli pNZ44 (1 × 106 cfu/ml) or PBS and 5 days later with VSVΔ51 (1 × 108 pfu). Tumors were measured as previously described. At necropsy tumor and healthy tissue were formalin fixed and paraffin embedded for immunohistochemistry. A cardiac bleed was performed and the serum extracted for cytokine profiling using Meso Scale Discovery (Gaithersburg, MD) 7-plex proinflammatory cytokine plate.
Optical image acquisition. 2D in vivo BioLuminescence Imaging was performed using the IVIS100 (Caliper, a Perkin Elmer company, Waltham, MA). At defined time points after bacteria and/or virus administration, animals were anesthetized under 3% isofluorane and whole-body image analysis was performed in the IVIS 100 system for up to 4 minutes at high sensitivity. Regions of interest were identified and quantified using Living Image software (Caliper). To acquire images of the bacterial luciferase signal acquisition times of 4 minutes with bin value of 16 was used to maximize the signal to noise ratio. To acquire images of the firefly luciferase emanating from the virus, d-luciferin (Molecular Imaging Products, Ann Arbor, MI) was injected ~10 minutes prior to imaging using an intraperitoneal injection. For each experiment, images were captured under identical exposure, aperture and pixel binning settings, and bioluminescence is plotted on identical color scales.
Immunohistochemistry. Sectioned tissues were processed as previously described with anti-VSV (1:5,000; 30 minutes) antibody.35 Tumor images were obtained with an Epson Perfection 2450 Photo Scanner (Epson, Toronto, Ontario, Canada) whereas magnifications were captured using a Zeiss Axiocam HRM Inverted fluorescent microscope (Zeiss, Toronto, Ontario, Canada) and analyzed using Axiovision 4.0 software. Consecutive sections were stained with H&E for general morphology.
Cytokine profiling. Concentrations of IL-1β, IL-12p70, IFN-γ, IL-6, mKC, IL-10, and TNF-α in serum at 5 days after virus administration were measured by an ultra-sensitive mouse proinflammatory 7-plex kit from Meso Scale Discovery (MSD, Gaithersburg, MD) according to the manufacturer's instructions. Blood was acquired by cardiac puncture allowed to clot and the serum separated and aliquots frozen at -80 until required. The protocol for the assay was as described by MSD, briefly a spot on the base of each plate was precoated with a capture antibody for each cytokine. The standard and serum samples (50 µl/well) were added to the prepared plates, and allowed to react at room temperature for 2 hours. Afterward, the plates were washed three times with washing buffer (1 × PBS with 0.05% Tween 20). Detection antibody was added and allowed to react at room temperature. After washing the plates three times and adding Read Buffer, the plates were analyzed on the MSD Sector Image 2400 (MSD). Calculation of cytokine concentrations was subsequently determined by 4-parameter logistic nonlinear regression analysis of the standard curve.
Statistical analysis. Two-tailed Student's t-tests were employed to investigate statistical differences. Microsoft Excel 12 (Microsoft) and Prism were used to manage and analyze data.
SUPPLEMENTARY MATERIAL Figure S1. VSV oncolytic activity in vitro.
Acknowledgments
The authors acknowledge support relevant to this manuscript from the Irish Cancer Society (CRF11CRO), the Cork Cancer Research Centre and the Irish Health Research Board (HRA_POR/2010/138) as well as the Ontario Institute for Cancer Research and the CIHR/SNE Research Fellowship. The authors declare no conflict of interest.
Supplementary Material
VSV oncolytic activity in vitro.
References
- Le Boeuf F, Diallo JS, McCart JA, Thorne S, Falls T, Stanford M, et al. Synergistic interaction between oncolytic viruses augments tumor killing. Mol Ther. 2010;18:888–895. doi: 10.1038/mt.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Liu TC, Hwang T, Park BH, Bell J, Kirn DH. The targeted oncolytic poxvirus JX-594 demonstrates antitumoral, antivascular, and anti-HBV activities in patients with hepatocellular carcinoma. Mol Ther. 2008;16:1637–1642. doi: 10.1038/mt.2008.143. [DOI] [PubMed] [Google Scholar]
- Vähä-Koskela MJ, Le Boeuf F, Lemay C, De Silva N, Diallo JS, Cox J, et al. Resistance to two heterologous neurotropic oncolytic viruses, Semliki Forest virus and vaccinia virus, in experimental glioma. J Virol. 2013;87:2363–2366. doi: 10.1128/JVI.01609-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bœuf F, Batenchuk C, Vähä-Koskela M, Breton S, Roy D, Lemay C, et al. Model-based rational design of an oncolytic virus with improved therapeutic potential. Nat Commun. 2013;4:1974. doi: 10.1038/ncomms2974. [DOI] [PubMed] [Google Scholar]
- Le Boeuf F, Bell JC. United virus: the oncolytic tag-team against cancer! Cytokine Growth Factor Rev. 2010;21:205–211. doi: 10.1016/j.cytogfr.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Janelle V, Poliquin L, Lamarre A. [Vesicular stomatitis virus in the fight against cancer] Med Sci (Paris) 2013;29:175–182. doi: 10.1051/medsci/2013292015. [DOI] [PubMed] [Google Scholar]
- Potts KG, Hitt MM, Moore RB. Oncolytic viruses in the treatment of bladder cancer. Adv Urol. 2012;2012:404581. doi: 10.1155/2012/404581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey D, O'Sullivan GC, Tangney M. Tumour targeting with systemically administered bacteria. Curr Gene Ther. 2010;10:3–14. doi: 10.2174/156652310790945575. [DOI] [PubMed] [Google Scholar]
- Baban CK, Cronin M, O'Hanlon D, O'Sullivan GC, Tangney M. Bacteria as vectors for gene therapy of cancer. Bioeng Bugs. 2010;1:385–394. doi: 10.4161/bbug.1.6.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins FS, Silva AA, Vieira AT, Barbosa FH, Arantes RM, Teixeira MM, et al. Comparative study of Bifidobacterium animalis, Escherichia coli, Lactobacillus casei and Saccharomyces boulardii probiotic properties. Arch Microbiol. 2009;191:623–630. doi: 10.1007/s00203-009-0491-x. [DOI] [PubMed] [Google Scholar]
- Schultz M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1012–1018. doi: 10.1002/ibd.20377. [DOI] [PubMed] [Google Scholar]
- Stritzker J, Weibel S, Hill PJ, Oelschlaeger TA, Goebel W, Szalay AA. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol. 2007;297:151–162. doi: 10.1016/j.ijmm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Cronin M, Akin AR, Collins SA, Meganck J, Kim JB, Baban CK, et al. High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS ONE. 2012;7:e30940. doi: 10.1371/journal.pone.0030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin M, Stanton RM, Francis KP, Tangney M. Bacterial vectors for imaging and cancer gene therapy: a review. Cancer Gene Ther. 2012;19:731–740. doi: 10.1038/cgt.2012.59. [DOI] [PubMed] [Google Scholar]
- Bell J, Kirn D. Recent advances in the development of oncolytic viruses as cancer therapeutics. Foreword. Cytokine Growth Factor Rev. 2010;21:83–84. doi: 10.1016/j.cytogfr.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Tedcastle A, Cawood R, Di Y, Fisher KD, Seymour LW. Virotherapy–cancer targeted pharmacology. Drug Discov Today. 2012;17:215–220. doi: 10.1016/j.drudis.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Cronin M, Morrissey D, Rajendran S, El Mashad SM, van Sinderen D, O'Sullivan GC, et al. Orally administered bifidobacteria as vehicles for delivery of agents to systemic tumors. Mol Ther. 2010;18:1397–1407. doi: 10.1038/mt.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants. Infect Agents Cancer. 2013;8:11. doi: 10.1186/1750-9378-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemunaitis J, Cunningham C, Senzer N, Kuhn J, Cramm J, Litz C, et al. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther. 2003;10:737–744. doi: 10.1038/sj.cgt.7700634. [DOI] [PubMed] [Google Scholar]
- Heppner F, Möse JR. The liquefaction (oncolysis) of malignant gliomas by a non pathogenic Clostridium. Acta Neurochir (Wien) 1978;42:123–125. doi: 10.1007/BF01406639. [DOI] [PubMed] [Google Scholar]
- Bermudes D, Low B, Pawelek J. Tumor-targeted Salmonella. Highly selective delivery vectors. Adv Exp Med Biol. 2000;465:57–63. doi: 10.1007/0-306-46817-4_6. [DOI] [PubMed] [Google Scholar]
- Karlsson H, Larsson P, Wold AE, Rudin A. Pattern of cytokine responses to gram-positive and gram-negative commensal bacteria is profoundly changed when monocytes differentiate into dendritic cells. Infect Immun. 2004;72:2671–2678. doi: 10.1128/IAI.72.5.2671-2678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehouritis P, Springer C, Tangney M. Bacterial-directed enzyme prodrug therapy. J Control Release. 2013;170:120–131. doi: 10.1016/j.jconrel.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Ahmad S, Casey G, Cronin M, Rajendran S, Sweeney P, Tangney M, et al. Induction of effective antitumor response after mucosal bacterial vector mediated DNA vaccination with endogenous prostate cancer specific antigen. J Urol. 2011;186:687–693. doi: 10.1016/j.juro.2011.03.139. [DOI] [PubMed] [Google Scholar]
- van Pijkeren JP, Morrissey D, Monk IR, Cronin M, Rajendran S, O'Sullivan GC, et al. A novel Listeria monocytogenes-based DNA delivery system for cancer gene therapy. Hum Gene Ther. 2010;21:405–416. doi: 10.1089/hum.2009.022. [DOI] [PubMed] [Google Scholar]
- Tangney M. Gene therapy for cancer: dairy bacteria as delivery vectors. Discov Med. 2010;10:195–200. [PubMed] [Google Scholar]
- McGrath S, Fitzgerald GF, van Sinderen D. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl Environ Microbiol. 2001;67:608–616. doi: 10.1128/AEM.67.2.608-616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CU, Monk IR, Casey PG, Morrissey D, O'Sullivan GC, Tangney M, et al. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Appl Environ Microbiol. 2007;73:3091–3094. doi: 10.1128/AEM.02940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns-Guydish SM, Olomu IN, Zhao H, Wong RJ, Stevenson DK, Contag CH. Monitoring age-related susceptibility of young mice to oral Salmonella enterica serovar Typhimurium infection using an in vivo murine model. Pediatr Res. 2005;58:153–158. doi: 10.1203/01.PDR.0000157725.44213.C4. [DOI] [PubMed] [Google Scholar]
- Brown CW, Stephenson KB, Hanson S, Kucharczyk M, Duncan R, Bell JC, et al. The p14 FAST protein of reptilian reovirus increases vesicular stomatitis virus neuropathogenesis. J Virol. 2009;83:552–561. doi: 10.1128/JVI.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baban CK, Cronin M, Akin AR, O'Brien A, Gao X, Tabirca S, et al. Bioluminescent bacterial imaging in vivo. J Vis Exp. 2012. p. e4318. [DOI] [PMC free article] [PubMed]
- Clancy EK, Duncan R. Reovirus FAST protein transmembrane domains function in a modular, primary sequence-independent manner to mediate cell-cell membrane fusion. J Virol. 2009;83:2941–2950. doi: 10.1128/JVI.01869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VSV oncolytic activity in vitro.