Abstract
Rhesus (rh) but not human (hu) TRIM5α potently restricts human immunodeficiency virus (HIV)-1 infection. It is not clear why huTRIM5α fails to effectively block HIV infection, but it is thought to have a lower affinity for the viral core. Using primary human CD4 T cells, we investigated the ability of huTRIM5α, rhTRIM5α, and the huTRIM5αR323–332 B30.2/SPRY patch-mutant to form cytoplasmic bodies, postulated as key components of the HIV-1 restriction apparatus. Both rhTRIM5α and huTRIM5αR323–332 formed pronounced cytoplasmic bodies, whereas cytoplasmic bodies in T cells overexpressing huTRIM5α were present but more difficult to detect. As expression of all three TRIM5α orthologs was similar at the RNA level, we next investigated the role of protein stability in conferring TRIM5α-mediated HIV-1 restriction. Both steady-state and pulse-chase experiments revealed that the huTRIM5α protein was much less stable than rhTRIM5α, and this difference correlated with higher self-ubiquitination activity. Using a stabilized form of huTRIM5α in which the steady-state expression level was more similar to rhTRIM5α, we observed comparable HIV-1 restriction activity in multi-round HIV-1 challenge assays. Lastly, primary human CD4 T cells expressing a stabilized huTRIM5α were protected from HIV-1-mediated destruction in vivo, indicating that efforts to stabilize huTRIM5α should have significant long-term therapeutic value.
Introduction
TRIM5α serves as both an innate cytoplasmic receptor for retroviral capsid to alert the cell of infection via the type I interferon (IFN) response, as well as a host restriction factor that blocks retroviral infection in the cytoplasm postentry.1,2 Rhesus (rh) TRIM5α is capable of restricting human immunodeficiency virus (HIV) infection at multiple stages in the cytoplasm by binding and inducing the rapid disassembly and degradation of incoming mature cytosolic capsid, disrupting reverse transcription and preventing the viral preintegration complex from accessing the nucleus.3,4 The exact mechanism by which rhTRIM5α restricts HIV is not clear, but capsid binding is essential. TRIM5α is thought to bind symmetrical interfaces of incoming mature viral capsid in the cytoplasm by forming a complementary lattice via higher order multimerization that increases its affinity for capsid and ability to restrict HIV.5,6 Despite the potency by which rhTRIM5α restricts HIV-1 infection, understanding specific interactions between TRIM5α and the HIV-1 capsid has been difficult, as TRIM5α self-associates into dimers7 and higher order structures,8 and recognizes capsid only when it is part of a mature viral core.5 These structures have proven difficult to mimic in a cell-free system, although the TRIM5-21R fusion variant appears more amenable to traditional structure-function studies.5 Thus, while some indirect measurements such as capsid sedimentation assays suggest that rhTRIM5α has a considerably higher affinity for HIV-1 capsid than huTRIM5α,9,10,11 it is not clear how great of a difference this is or the extent to which it explains the vastly different HIV-1 restriction activities of these isoforms.
Human TRIM5α, on the other hand, is thought to have at best modest anti-HIV-1 activity,1,9,12,13,14 and is often used as a negative control in HIV-1 restriction studies.15,16,17 Nonetheless, cohort studies indicate that selected huTRIM5α alleles are associated with altered HIV-1 disease progression,18,19,20,21 suggesting that huTRIM5α does play a role in ameliorating HIV-1 pathogenesis. Importantly, selected mutations within huTRIM5α that mirror rhTRIM5α dramatically augment restriction activity, with only a single substitution (R332P) required within the B30.2/SPRY domain to confer measureable restriction activity in single-round assays,22 and as few as five substitutions within the B30.2/SPRY domain conferring potent restriction activity approaching that of rhTRIM5α.14,17 Surprisingly, we have found that these mutations augment TRIM5α protein stability and localization in cytoplasmic bodies in primary human T cells. By linking TRIM5α to mCherry, we enhanced the stability of wild-type huTRIM5α protein and this resulted in potent restriction of HIV-1 in primary human CD4 T cells approaching that of rhTRIM5α. Lastly, using a humanized murine model of HIV-1 infection, we observed that overexpression of mCherry-huTRIM5α in vivo protects T cells from infection long term. Thus, approaches that stabilize huTRIM5α in infected individuals may be highly effective in controlling HIV-1 replication.
Results
The ability to form distinct and pronounced cytoplasmic bodies correlates with TRIM5α restriction activity
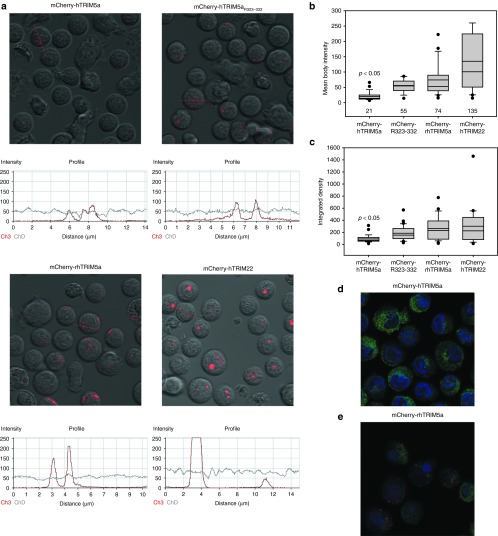
Previously, it has been noted that the ability of TRIM5α-CypA fusions to restrict HIV-1 infection correlated with their ability to form cytoplasmic bodies.23,24 We wished to see whether the ability of TRIM5α orthologs to form cytoplasmic bodies correlated with restriction activity in primary human T cells. To do this, we first fused mCherry to the N-terminus of huTRIM5α, rhTRIM5α, and huTRIM5αR323–332, a modified huTRIM5α bearing 5 rhesus substitutions (P323R, K324N, I328M, G330Q, and R332P) in the B30.2/SPRY viral recognition motif.14 We next transduced primary human T cells with lentiviral vectors encoding these mCherry-TRIM5α fusions along with mCherry-huTRIM22 control, and then measured their ability to form cytoplasmic or nuclear bodies by confocal microscopy (Figure 1a). By measuring the mean intensity (Figure 1b) and integrated density (Figure 1c) of each cytoplasmic or nuclear body in transduced T cells, as well as the number of bodies, we determined that when imaged with the same exposure time, huTRIM5α forms much less pronounced cytoplasmic bodies than either huTRIM5αR323–332 or rhTRIM5α (P < 0.05). These data indicate that huTRIM5α is less able to form larger, brighter cytoplasmic bodies, and this correlates with its inability to restrict HIV-1 infection as effectively. The more restrictive huTRIM5αR323–332 patch mutant formed cytoplasmic bodies with mean intensity and density not significantly different than those observed with rhTRIM5α, indicating the ability to form pronounced cytoplasmic bodies in primary human T cells is determined at least in part by the sequence of this small region of the B30.2/SPRY domain of TRIM5α. Although they are fewer and fainter, huTRIM5α still forms and is localized to cytoplasmic bodies (Figure 1d), like rhTRIM5α (Figure 1e), as evident using fixed cells and optimized exposure times.
Figure 1.
The ability to form distinct and pronounced cytoplasmic bodies correlates with TRIM5α restriction activity. (a) Primary human CD4 T cells were activated with CD3/28 coated beads and transduced with the indicated mCherry-TRIM5α fusion or mCherry-TRIM22. Confocal microscopy was used to visualize and quantify the intensity of mCherry-TRIM5α cytoplasmic bodies in live human CD4 T cells on day 8 post-transduction. (b) Summary and quantification of the mean body intensity of cytoplasmic bodies in all cells shown in Figure 1a, as determined using the Zeiss ZLM Image Browser software (Carl Zeiss Microscopy LLC, Thornwood, NY). (c) Summary and quantification of the integrated density of cytoplasmic bodies in all cells shown in Figure 1a via ImageJ (http://fiji.sc/). For each condition, 15–20 cells and over 100 cytoplasmic bodies were analyzed per panel. Statistical testing to determine whether huTRIM5α formed less pronounced cytoplasmic bodies than other TRIM5α isoforms was performed using the Kruskal–Wallis one-way analysis of variance on Ranks test with Dunn's Method to adjust for multiple comparisons, utilizing Sigma Plot 11.0 software (Systat Software, Chicago, IL). (d) Imaging mCherry-huTRIM5α bodies in the cytoplasm (green) of fixed CD4 T cells with optimized exposure time; nucleus (blue) stained with DAPI. (e) Fixed cells expressing mCherry-rhTRIM5α bodies in the cytoplasm (green) with DAPI nuclear stain. DAPI, 4‘,6-diamidino-2-phenylindole.
rhTRIM5α accumulates much more readily than huTRIM5α in primary human T cells
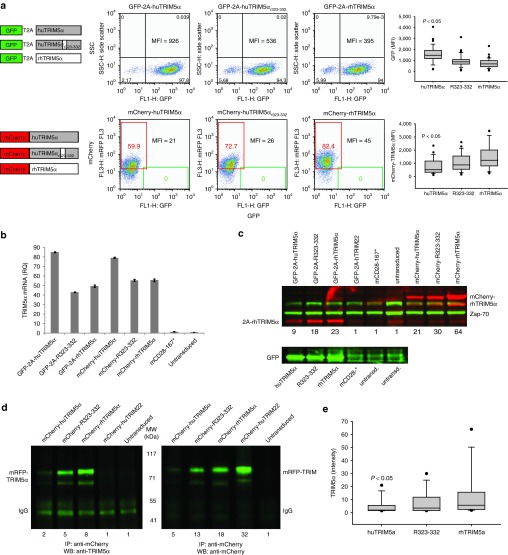
We speculated that protein stability might play a role in the inability of huTRIM5α to form pronounced cytoplasmic bodies in CD4 T cells. To investigate this, we took advantage of the fact that the TRIM5α and green fluorescent proteins are expressed distinctly and in an equimolar manner from the same transcript when linked by the T2A sequence.17 Thus, the relative ratio of TRIM5α to green fluorescent protein provides insight into TRIM5α stability. We consistently observed that the highest level of GFP was seen in T cells transduced with huTRIM5α, followed by cells transduced with huTRIM5αR323–332, and the lowest GFP expression was seen in T cells transduced with rhTRIM5α (Figure 2a). Since GFP and huTRIM5α are expressed as a single transcript via the T2A motif, it was not surprising to see that T cells transduced with GFP-2A-huTRIM5α, which showed the brightest GFP expression via fluorescence-activated cell sorting (Figure 2a), also had the highestTRIM5α expression at the RNA level by revese transcription polymerase chain reaction (Figure 2b). However, when we examined the amount of cytoplasmic TRIM5α protein by western blotting, we observed that cells expressing the most TRIM5α mRNA and GFP protein, GFP-T2A-huTRIMα, displayed the lowest steady-state levels of TRIM5α protein (Figure 2c). That is, despite higher huTRIM5α mRNA levels, huTRIM5α was the least abundant cytoplasmic TRIM5α protein, suggesting it was much less stable than rhTRIM5α.
Figure 2.
RhTRIM5α accumulates much more readily than huTRIM5α in primary human T cells. (a) CD3/28-activated CD4 T cells were transduced with lentiviral vectors encoding TRIM5α isoforms linked to green fluorescent protein (GFP) via a 2A sequence (top) or fused to mCherry (bottom). GFP and mCherry expression was measured by flow cytometry after 8 days of culture (shown), and at other timepoints in replicate experiments (box-plot). (b) The relative abundance of each TRIM5α isoform mCherry was measured by quantitative revese transcription polymerase chain reaction. (c) Protein lysates were made from the cells shown in Figure 2a and blotted for TRIM5α (red) or ZAP-70 (green); relative expression of each TRIM5α isoform compared to untransduced T cells is shown below each sample (upper panel). Lysates from GFP-2A-TRIM5α transduced cells and controls were blotted separately for GFP (lower panel). (d) Immunoprecipitation of mCherry-TRIM proteins from transduced primary human CD4 T cells via monoclonal antibody to mCherry, followed by western blotting with anti-TRIM5α (left panel) and anti-mCherry (right panel) polyclonal antibodies. (e) Densitometry data are representative of at least four independent experiments.
Fluorescent proteins like GFP and mCherry have been shown to be very stable and often augment the stability of molecules to which they are fused;25 so, we wanted to determine whether mCherry could stabilize TRIM5α. We observed the same disconnect between TRIM5α mRNA and protein expression using the mCherry-TRIM5α fusions as seen with GFP-T2A-TRIM5α configurations: the mCherry-huTRIM5α vector expressed the most TRIM5α RNA but had the dimmest mCherry expression as measured by fluorescence-activated cell sorting (Figure 2a,b) and the least TRIM5α protein by western (Figure 2c). In order to improve our sensitivity in measuring differences in cytoplasmic protein expression levels between the three TRIM5α isoforms, we performed immunoprecipitations of mCherry-TRIM5α fusion proteins with anti-mCherry mAb followed by western blotting for TRIM5α or mCherry and observed approximately three- to fourfold more rhTRIM5α than huTRIM5α (Figure 2d), in spite of the lower amount of rhTRIM5α at the RNA level (Figure 2b). The lower relative expression of huTRIM5α protein compared to rhTRIM5α was a consistent observation in densitometry experiments (Figure 2e) and was ameliorated by addition of the rhesus B30.2/SPRY patch in huTRIM5αR323–332. Importantly, we also observed a two- to threefold increase in huTRIM5α expression in T cells transduced with mCherry fused to the N-terminus of TRIM5α relative to the expression of TRIM5α in T cells transduced with GFP-T2A-huTRIM5α vector, despite comparable expression at the RNA level (Figure 2b). These data suggest that huTRIM5α is a much less stable protein in primary human T cells than rhTRIM5α, and that huTRIM5α can be stabilized by fusion to mCherry as well as by incorporating select rhesus substitutions in the B30.2/SPRY viral recognition motif.
HuTRIM5α is less stable than rhTRIM5α in primary human CD4 T cells
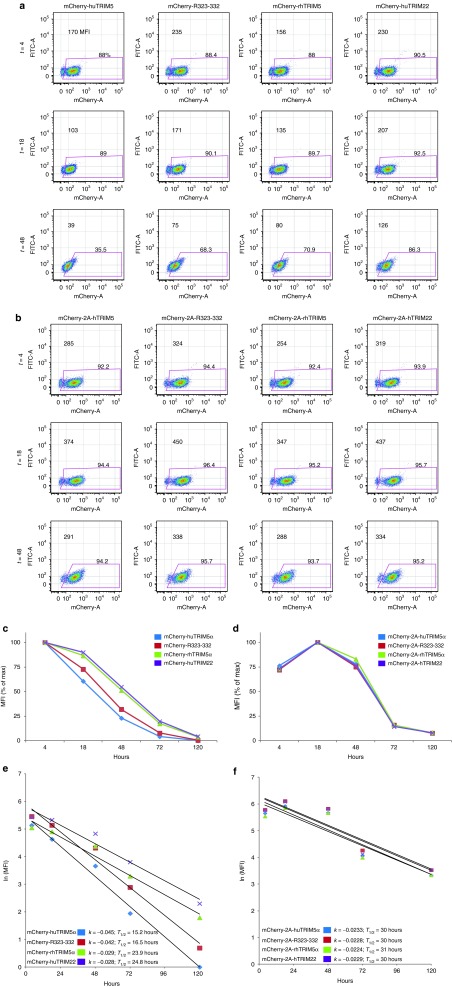
Previous studies have measured TRIM5α stability using pulse/chase approaches and found no remarkable differences between huTRIM5α and rhTRIM5α.13,26 We wanted to determine whether the differences observed between our results and previous findings were the result of the assay (steady-state versus pulse/chase) or the cell type (primary human T cell versus HeLa cells). Since mCherry-TRIM5α fusions demonstrated the same hierarchy of stability as nonfused TRIM5α, we decided to use these constructs to evaluate relative stability in primary human T cells, given the ability to detect mCherry is far more sensitive than the ability to detect TRIM5α. Moreover, since cycloheximide blocks all protein synthesis, and use of cycloheximide to study protein stability may not reflect protein stability under physiologic conditions,27 we decided to use RNA transfection as means to specifically measure the decay of TRIM5α isoforms. Differences in stability and decay of mRNA transcripts were controlled for within the assay by comparing each mCherry-TRIM fusions with its mCherry-T2A-TRIM fusion counterpart, which is also encoded on a single (separate) mRNA with identical 5′ cap and 3′ polyadenylation sequence. Four hours after transfection with in vitro transcribed RNA encoding mCherry-TRIM5α fusions (Figure 3a,c,e) or mCherry-T2A-TRIM5α (Figure 3b,d,f), a baseline TRIM5α expression level is measured, with additional timepoints taken thereafter. For graphical presentation, the decay of mCherry mean fluorescence intensity was normalized to input values at T = 4 hours (Figure 3c,d). Using an approach described previously,28 the natural log of mean fluorescence intensity was plotted versus time, and the rates of decay (k) were solved via the slope of the line, with the half-life (T1/2) then calculated using the equation T1/2 = ln(2)/k (Figure 3e,f). After 120 hours of culture, we calculated that rhTRIM5α had a half-life of 23.9 hours whereas huTRIM5α had a half-life of only 15.2 hours (Figure 3e). In contrast, we observed no differences in mCherry stability when expressed with TRIM5α isoforms via the T2A motif (Figure 3f). Together, these data indicate that the huTRIM5α protein is much less stable than rhTRIM5α.
Figure 3.
HuTRIM5α is less stable than rhTRIM5α in primary human CD4 T cells. Resting primary human CD4 T cells were electroporated with 20 µg of in vitro transcribed RNA encoding mCherry-TRIM5α fusion proteins (a,c,e) or an mCherry-T2A-TRIM5α cassette (b,d,f), which expresses mCherry and TRIM in tandem but separately. (a,b) The mean fluorescence intensity (MFI) of mCherry was monitored by flow cytometry at T = 4, 18, 48, 72, and 120 hours after electroporation; CD4 T cells were CD3/28 costimulated at T = 18 hours. (c,d). Line graphs showing the decay of mCherry normalized to input values at T = 4 hours. (e,f) By taking the natural log of the MFI values and plotting them versus time, the slope or rate of decay (k), and half-life (T1/2) were calculated for each mCherry-TRIM5α fusion protein (e), and mCherry expressed separately but in an equimolar manner with TRIM5α or TRIM22 vis T2A motif (f), using the equation (T1/2 = ln(2)/k) (ref. 28). Results are representative of four independent experiments.
HuTRIM5α displays more self-ubiquitination activity than rhTRIM5α
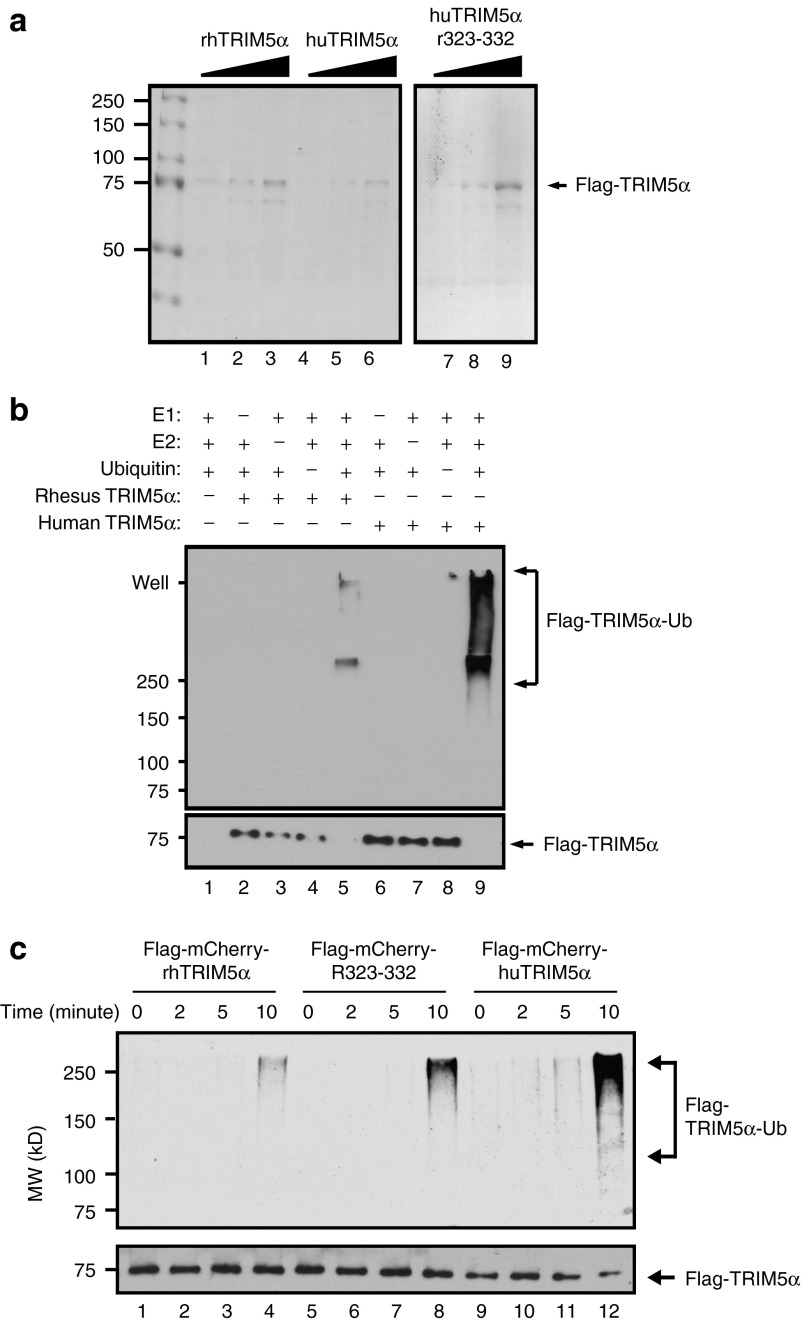
Previous studies have indicated that TRIM5α functions as a RING-finger-type E3 ubiquitin ligase able to add ubiquitin groups to itself or other proteins.26,29,30,31 We postulated that differences in the relative stability of huTRIM5α and rhTRIM5α would correlate with self-ubiquitination activity. To test this, we purified all three TRIM5α isoforms to near homogeneity (Figure 4a). To confirm the specificity of the autoubiquitination assay, it was performed in the presence of all assay components or in the absence of one or more components (Figure 4b). Having confirmed the specificity of the autoubiquitination assay, we incubated the purified TRIM5α proteins with ubiquitin components for up to 10 minutes and observed that huTRIM5α had substantially more self-ubiquitination activity than rhTRIM5α and the huTRIM5αR323–332 construct (Figure 4c). While the relationship between ubiquitination and protein stability is complicated, our data would suggest that huTRIM5α's propensity to self-ubiquitinate leads to a loss of stability, which limits its ability to inhibit HIV-1 infection.
Figure 4.
HuTRIM5α has a higher propensity for self-ubiquitination than rhTRIM5α or huTRIM5αR323–332. (a) Flag-mCherry-TRIM5α proteins (rhTRIM5α, huTRIM5αR323–332, and huTRIM5α) were purified from lysates of transfected HEK293T cells by anti-Flag affinity chromatography. One, three, and ten microliters of purified lysate were analyzed on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel: lanes 1–3 rhTRIM5α, lanes 4–6 huTRIM5α, lanes 7–9 hTRIM5αR323–332. (b) The specificity of the autoubiquitination assay was then confirmed by removing one or more reaction components (indicated by the + and – signs) from the assay, using purified huTRIM5α and rhTRIM5α separately as the E3 ligase. After 10 minutes incubation, samples were boiled in SDS–PAGE sample buffer and analyzed by western blot, using anti-ubiquitin and anti-Flag antibodies. (c) To compare the propensity of the three TRIM5α isoforms to self-ubiquitinate, purified Flag-mCherry-TRIM5α proteins (rhTRIM5α, huTRIM5αR323–332, and huTRIM5α) were incubated with ubquitination reaction components for the indicated time, and subjected to SDS–PAGE and western blotting as above. The amount of ubiquitinated Flag-TRIM5α proteins (top panel) and unmodified Flag-mCherry-TRIM5α proteins (lower panel) are shown and the reaction times, antibodies, and molecular weight standards are indicated on the top, right, and left, respectively.
Stabilized version of huTRIM5α has potent HIV-1 restriction activity
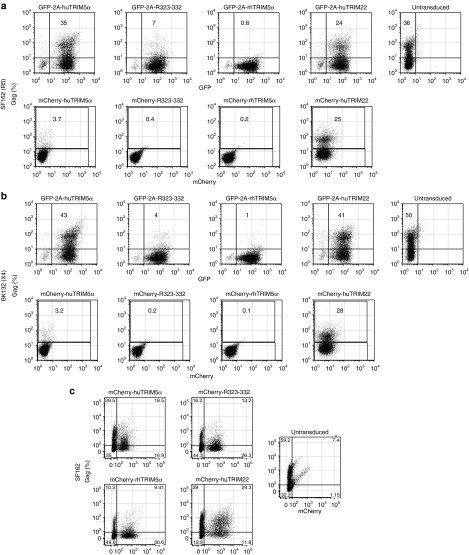
We and others have previously demonstrated that overexpressed huTRIM5α in primary human T cells has modest antiviral activity, whereas, under the same conditions, rhTRIM5α has potent activity.17,32,33,34 Most have interpreted these data to mean that huTRIM5α has a reduced ability to recognize and restrict HIV-1. Since the five rhesus B30.2/SPRY-domain substitutions in the huTRIM5αR323–332 construct enable potent HIV-1 restriction, it has been postulated that these changes to huTRIM5α augment its affinity for capsid. However, our data show that these changes also augment TRIM5α stability. To better understand the extent increased stability was contributing to the ability of huTRIM5αR323–332, we wanted to determine whether a stabilized but unmutated version of huTRIM5α would have potent HIV-1 restriction activity as well. To test this, we compared the huTRIM5α delivery system used previously which coexpressed huTRIM5α and GFP using a T2A sequence, and the system above in which huTRIM5α expression was stabilized by fusion to mCherry. Primary human CD4 T cells were transduced with each TRIM5α ortholog and later infected with HIV-1SF162 (R5 tropic) and HIV-1BK132 (X4 tropic) separately. Consistent with our and others' previous reports,17,32,33,34 T cells transduced with GFP-T2A-huTRIM5α were equally susceptible to HIV-1SF162 infection as untransduced T cells (36 versus 35% HIV-1GAG-positive cells), whereas T cells expressing either GFP-T2A-huTRIM5αR323–332 (7%) or GFP-T2A-rhTRIM5α (<1%) were highly resistant to HIV-1 infection (Figure 5a). In contrast, T cells transduced with the mCherry-huTRIM5α fusion displayed robust antiviral activity relative to the untransduced control (3.7 versus 36%), as well as GFP-T2A-hTRIM5α (3.7 versus 35%). Similar results were observed with regard to susceptibility to X4 isolate HIV-1BK132 infection (Figure 5b). Although huTRIM22 is closely related to TRIM5α and an important component of the type I IFN response to HIV-1 infection,35 it is a nuclear protein not thought to bind the HIV-1 capsid. Unlike huTRIM5α, however, its overexpression conferred little or no resistance to HIV-1 in our assay systems, even when fused to mCherry. These data suggest that introduction of stabilized TRIM5α is having a direct effect on HIV-1 through capsid interactions rather than an effect of overexpressing a member of the type I interferon response. Similar experiments using HIV-1BaL, HIV-1US1, and HIV-1JRFL (data not shown) were performed, and in all cases, T cells transduced with the mCherry-huTRIM5α fusion were more potent in resisting HIV infection than either GFP-T2A-huTRIM5α or untransduced controls, and there was no difference in potency between mCherry-huTRIM5α, mCherry-huTRIM5αR323–332, and mCherry-rhTRIM5α, supporting our hypothesis that stabilized huTRIM5α has potent antiviral activity. In mixed cocultures with untransduced CD4 T cells, mCherry-huTRIM5α, mCherry-huTRIM5αR323–332, and mCherry-rhTRIM5α were all saturable factors following HIV-1 challenge (Figure 5c), presumably due to the high effective multiplicity of infection achieved in these conditions by cell-to-cell transmission from HIV-infected untransduced cells to mCherry-TRIM5α-expressing cells.17 These data indicate that a stabilized version of huTRIM5α has potent yet saturable HIV-1 restriction activity, and in fact, huTRIM5α has not lost the ability to recognize HIV-1.
Figure 5.
Stabilized huTRIM5α has potent human immunodeficiency virus (HIV)-1 restriction activity. (a,b) CD4 T cells expressing TRIM5α constructs fused to mCherry (mCherry-TRIM5α) or expressing TRIM5α as a distinct protein in an equimolar manner with GFP (GFP-T2A-TRIM5α) were infected with cell-free HIV-1SF162 (a) or HIV-1BK132 (b). After 8 days of culture, HIV-1 viral replication was measured by intracellular HIV-1GAG staining. (c) Mixed cultures where initially half the CD4 T cells are untransduced and half express mCherry-TRIM5α were infected with cell-free HIV-1SF162 and assayed after 8 days of culture for intracellular HIV-1GAG staining. Results are representative of those observed in three separate experiments performed in duplicate or larger replicates.
Enhanced overexpression of huTRIM5α results in potent antiviral activity in vivo
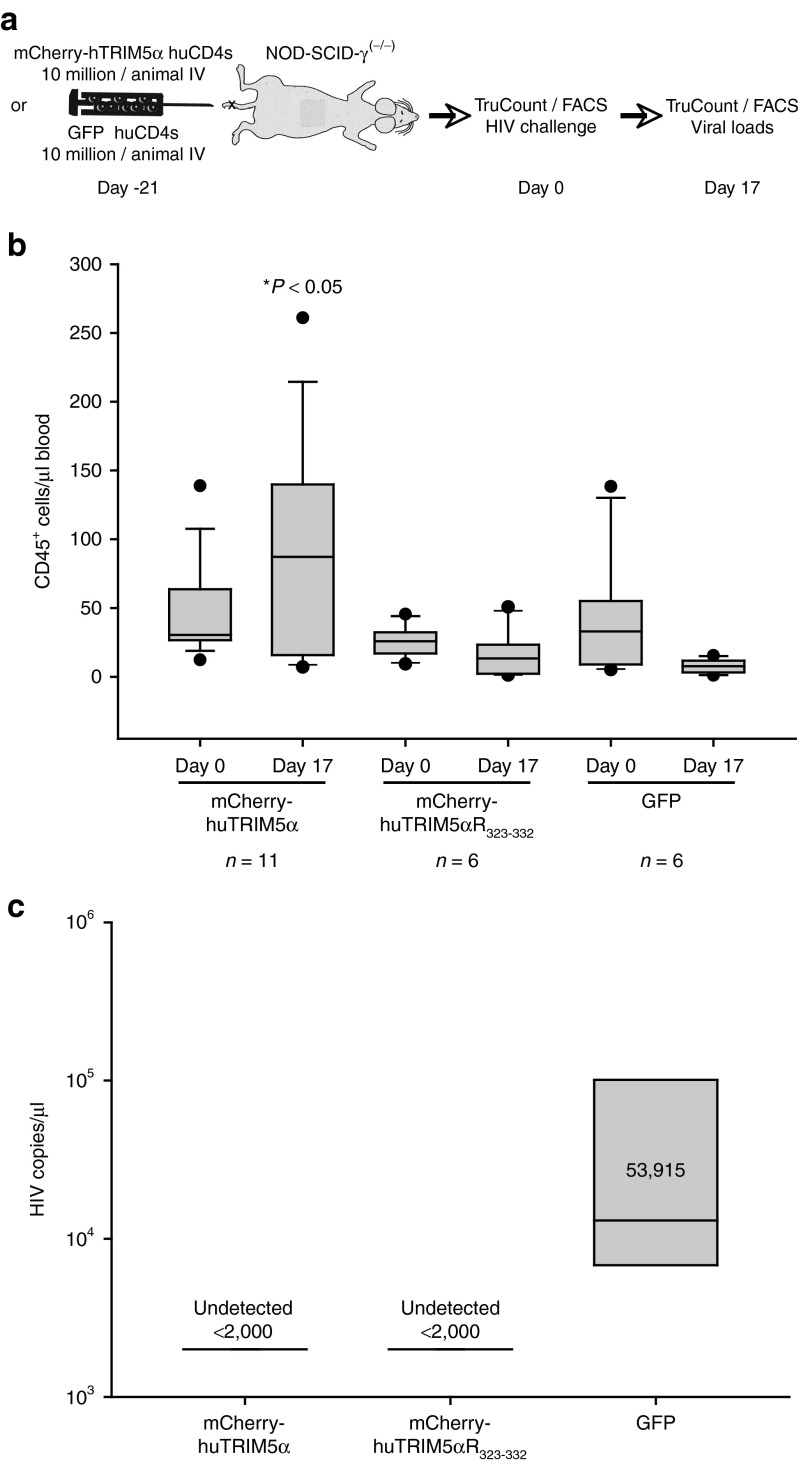
To date, no therapeutic strategy to treat HIV-1 infection employing TRIM5α isoforms has been tested in the clinic. To determine whether stabilized huTRIM5α could protect T cells in vivo, we modified a previous humanized mouse model established to study the ability of CCR5 deficiency to protect T cells in vivo36 and to study the ability of TRIM5α to protect human T cells. A schematic of the experimental design is shown in Figure 6a. NOD-SCID-γ(−/−) mice were engrafted with 107 human CD4 T cells transduced with either mCherry-huTRIM5α, huTRIM5αR323–332, or GFP. After 21 days, engraftment in peripheral blood was measured and groups of mice were selected so that each would have approximately the same number of CD4 T cells engrafted (Figure 6b, day 0). Mice were then infected with HIV-1BaL and both CD45+ CD4 T cell count and HIV-1 viral load were determined 17 days later. Mice engrafted with mCherry-huTRIM5α (n = 11) had higher levels of CD45+ T cells (Figure 6b, day 17) in the absence of detectable viral load (Figure 6c). After HIV-1 infection, mice engrafted with GFP expressing T cells (n = 6) had a substantial reduction of the number of CD45+ T cells present and a high viral load. Mice engrafted with T cells expressing huTRIM5αR323–332 (n = 6) were also protected from HIV-1 infection but did not expand as well in this experiment. Downregulation of CD4 expression in CD45+ T cells was observed mostly in the GFP group, where a small population was visible at later timepoints (data not shown). These studies indicate that a stabilized version of huTRIM5α can protect primary human CD4 T cells in vivo and suggest approaches that stabilize huTRIM5α will be effective HIV-1 therapies.
Figure 6.
Enhanced overexpression of huTRIM5α results in potent antiviral activity in vivo. (a) Schematic for the experiment; 10 million TRIM5α-expressing (mCherry-hTRIM5α or mCherry-hTRIM5αR323–332) or GFP-expressing CD4 T cells were injected intravenously in NOD-SCID-γ(−/−) mice and monitored for engraftment by TruCount for CD45+ cells. Animals in each group—mCherry-hTRIM5α (n = 11), mCherry-hTRIM5αR323–332 (n = 6), or GFP (n = 6)—engrafted similarly. After 3 weeks, intravenous cell-free HIV-1BaL challenge was performed on day 0, following baseline prechallenge TruCount and normalization. TruCount and fluorescence-activated cell sorting (FACS) analysis for cell survival were then performed at day 17 postchallenge. (b) TruCounts for the number of human CD45+ cells at day 0 prechallenge in the three groups, and those remaining on day 17 postchallenge as enumerated by TruCount. (c) Mean HIV-1 viral loads on day 17 postchallenge in TRIM5α-transduced (mCherry-hTRIM5α or mCherry-hTRIM5α R323–332) or GFP groups. This experiment was performed once in this format, but the ability of stabilized huTRIM5α to protect CD4 T cells in vivo was observed in similar experiments. Kruskal–Wallis one-way analysis of variance on Ranks with Dunn's method for multiple comparisons was performed to evaluate differences in CD45+ TruCounts between the three groups.
Discussion
Multiple viral adaptions occurred as SIVcpz evolved into HIV-1.37 Key drivers of viral adaptions are human-encoded restriction factors, which have the potential to severely limit the replication of transmitted simian immunodeficiency virus strains in humans.38 While there is evidence that B-MLV, but not N-MLV, is capable of escaping human TRIM5α restriction,39,40 there are sparse definitive data supporting HIV-1 escape from huTRIM5α. Several studies have provided evidence that rhTRIM5α recognizes the HIV-1 capsid better than huTRIM5α,9,10,11 implying HIV-1 escaped huTRIM5α restriction by modifying its capsid. Other studies suggest the ability of primate TRIM5α to hinder Simian immunodeficiency virus replication correlates with the affinity by which TRIM5α binds capsid.41 These capsid-binding studies were performed by incubating crude lysates from 293T cells expressing TRIM5α with in vitro assembled capsid-nucleocapsid complexes, and centrifuging them over a sucrose cushion, followed by western blotting of input and pellet fractions to quantify TRIM5α bound to and cosedimenting with HIV-1 capsid. One caveat of these studies is that human and rhTRIM5α must have the same stability throughout the experiment to make this comparison valid. Because huTRIM5α is activated and signals via ubiquitin following contact with capsid as part of the type I IFN pathway,2 protein instability may be a direct consequence of encountering the viral core. Therefore, the absence of huTRIM5α in the pellet with capsid could reflect protein degradation due to inherent instability as well as affinity for capsid. Moreover, these studies have most commonly employed HeLa cell lines, which may not reflect the protein turnover and antigen presentation dynamics in primary human CD4 T cells. Thus, although reverse engineering can render HIV-1 capsid more sensitive to restriction by huTRIM5α,16 strong biochemical evidence supporting HIV-1 escape from huTRIM5α recognition is lacking.
Given the uncertainty of whether differential affinity for HIV-1 capsid is sufficient to explain the differences in restriction activity between huTRIM5α and rhTRIM5α, we offer an alternative explanation based on the data presented in this study regarding how HIV-1 avoided huTRIM5α restriction. Our data argue that due to its lack of stability and inability to accumulate in pronounced cytoplasmic bodies likely due to its high self-ubiquitination activity, huTRIM5α was simply ill-equipped to block HIV-1 infection and played little to no role in preventing the HIV-1 epidemic. In contrast, rhTRIM5α is much more stable and is able to accumulate in cytoplasmic bodies, as well as to potently block HIV-1 infection. Our data show that when huTRIM5α is stabilized, it restricts HIV-1 efficiently, suggesting that huTRIM5α and rhTRIM5α when expressed more equivalently have more comparable restriction activities than previously thought, albeit rhTRIM5α is still the most potent likely due to a higher affinity for capsid.9,10,11,15 Moreover, we show that selected mutations in the B30.2/SPRY domain that make huTRIM5α restrict HIV-1 more like rhTRIM5α also augment huTRIM5α's stability, further supporting the concept that TRIM5α stability is another key to HIV-1 restriction. We also show that mCherry-huTRIM5α is a saturable factor when HIV-1 is transmitted by cell-to-cell contact with HIV-1-infected untransduced CD4 T cells, suggesting that human TRIM5α restricts HIV-1 using the same mechanism by which rhTRIM5α restricts HIV-1. While our studies do not rule out or minimize differences in capsid binding affinity, they do suggest that lack of stability and accumulation in cytoplasmic bodies are also significant reasons why huTRIM5α fails to restrict HIV-1 efficiently. Lastly, the concept that huTRIM5α did little as a species-specific restriction factor is further supported by the ease by which Simian immunodeficiency virus is able to infect primary human T cells,42,43 especially when contrasted with the extreme difficulty cell-free HIV-1 has infecting rhesus and other Old World monkey T cells. The evolutionary forces that resulted in TRIM5α being a potent species-specific lentiviral restriction factor in rhesus macaques yet largely ineffectual in humans are unclear, but given the constant onslaught of retroviruses and the need of restriction factors like TRIM5α to adapt to control them,44 it is reasonable to assume that huTRIM5α adapted to control a virus where formation of cytoplasmic bodies and stability were not required for restriction,45 whereas capsid binding and ubiquitin-mediated signal transduction to activate the type I IFN pathway presumably were retained and/or enhanced.
Our studies have important implications for using TRIM5α as a therapeutic. If HIV-1 had escaped huTRIM5α, then the rationale for using potentially immunogenic forms of huTRIM5α that had higher affinity for capsid would be justified. However, our in vitro and in vivo studies indicate that a properly stabilized and overexpressed version of huTRIM5α is highly effective in reducing HIV-1 replication. The recent “Berlin Patient” has done much to increase optimism that an HIV-1 cure is possible,46 and approaches that recapitulate this phenomena in a broader patient population pool have been proposed.36,47 Our data indicate that strategies to augment huTRIM5α expression, stability, and localization to cytoplasmic bodies would be highly effective and an attractive way to promote a cure for HIV-1-infected individuals. Previous studies have shown that introduction of rhTRIM5α into stem cells protected humanized mice from HIV-1 infection.48 Our data would suggest that overexpressed and optimally localized huTRIM5α would be just as effective and much less immunogenic. A key question to move this approach forward is how much TRIM5α expression is required to have a dramatic effect on HIV-1 replication in humans. Our studies show that a while a 5–10-fold improvement of huTRIM5α is not sufficient to protect T cells, 20–30-fold improvement is sufficient. Well-tolerated agents such as IFNα are known to upregulate and stabilize huTRIM5α,32 and this may explain why IFNα and TRIM5α work synergistically.49 Our previous studies demonstrate that for TRIM5α restriction to work efficiently both the infected and target cells must express TRIM5α.17 Therefore, adoptive T cell therapies that only infuse a small number of T cells expressing high, stable levels of huTRIM5α are unlikely to have a benefit, unless they are able to expand and enrich dramatically as a percentage of total T cells and/or selectively repopulate damaged lymphoid organs. Rather, approaches that are able to elevate huTRIM5α in large majority of the T cells within an HIV-1-infected individual by either ingestion of a small molecule stabilizer of huTRIM5α or through stem cell gene therapy would be predicted to lead to functional control of HIV-1 replication.
Materials and Methods
Introduction of TRIM5α into primary human T cells. Deidentified, primary human CD4 T cells obtained from the University of Pennsylvania Human Immunology Core after Declaration of Helsinki protocols were followed and written informed consent and approval by the University of Pennsylvania's institutional review board (Institutional Review Board protocol 705906). Primary human T cells cultures and production of lentiviral vectors expressing TRIM5α isoforms have been previously described.17 The cDNA for huTRIM22 was obtained from Dr. Frederick Bushman's group50 and placed into pELNS as 2A fusion with either GFP or mCherry.17 TRIM5α proteins were also expressed as a fusion protein on the C-terminus of FLAG and/or mCherry. All TRIM5α expression cassettes were transferred into pVAX (Invitrogen, Life Science Technologies, Carlsbad, CA) using standard procedures and in vitro transcribed RNA was generated using mMACHINE Ultra kit (Invitrogen). Electroporation was performed using a prechilled 0.2 cm cuvette (BioRad, Herculues, CA), and an ECM 830 Electro Square Porator (BTX/Harvard Apparatus, Holliston, MA), with 20 µg of in vitro transcribed mRNA added to 10 million cells. Cells were harvested at various timepoints after electroporation for fluorescence-activated cell sorting analysis.
Calculating TRIM protein half-life. The half-lives of mCherry-TRIM fusion proteins, and of mCherry expressed separately but in tandem with TRIM5α on the same transcript via mCherry-2A-TRIM5α cassettes, were calculated using the mean fluorescence intensity of mCherry at time points T = 4, 18, 48, 72, and 120 hours following electroporation. The rate of decay (k) was first calculated by graphing the natural log of the mean fluorescence intensity on the y-axis versus time postelectroporation on the x-axis and fitting this data to a first order decay function; the half-life of each protein was then calculated using the degradation constant k in the equation T1/2 = ln(2)/k.28
Microscopy and flow cytometry. Confocal microscopy was performed using a Zeiss LSM 510 NLO/METAsystem with a Zeiss Axiovert 200M inverted microscope. Images were processed using Zeiss AIM software (Zeiss), and the profile function of the Zeiss AIM analysis software was used to quantify the intensity of TRIM5α cytoplasmic bodies in individual cells. For confocal microscopy, living cells were imaged in inverted chamber slides, and fixed cells were imaged on cover slips. Flow cytometry was performed using a FACSCalibur (BD Biosciences, San Jose, CA) or a LSRII (BD Biosciences).
Western blotting, immunoprecipitation, and quantitative revese transcription polymerase chain reaction. Cytoplasmic extracts were isolated from primary human CD4 and western blotting were performed as previously described17 using a 1:2,000 dilution of anti-mCherry rabbit polyclonal antibody (BioVision, Mountain View, CA), a 1:1,000 dilution of anti-huTRIM5α rabbit polyclonal antibody carboxy terminus (Imgenex, San Diego, CA) or, as a loading control, a 1:2,000 dilution of anti-Zap-70 monoclonal antibody (Clone LIE5; Cell Signaling Technology, Beverly, MA). Secondary antibodies were obtained from LI-COR (LI-COR, Lincoln, Nebraska). Immunoprecipitations from cytoplasmic extracts of TRIM5α -expressing primary human CD4 T cells were performed using the Living Colors anti-mCherry mouse monoclonal antibody (Clontech, Mountain View, CA), and the Immunoprecipitation Kit-Protein G (Invitrogen). Immunoprecipitations from HEK293T cells transfected to express pVAX-FLAG-mCherry-TRIM5α constructs were performed using the anti-FLAG M2 antibody (Sigma-Aldrich, St Louis, MO). Quantitative revese transcription polymerase chain reaction TaqMan assays were performed using primers and probes predesigned by ABI/Life Technologies (Invitrogen), and were performed on an ABI QPCR TaqMan 7900 platform (Invitrogen). After adjusting for glyceraldehyde-3-phosphate dehydrogenase levels in each sample, relative expression levels of huTRIM5α and rhTRIM5α in transduced primary human CD4 T cells were normalized to endogenous huTRIM5α levels in untransduced cells.
TRIM5α protein purification and in vitro ubiquitination assays. Flag-tagged TRIM5α proteins (rhTRIM5α, huTRIM5αR323–332, and huTRIM5α) were transfected in HEK293T cells and purified using anti-Flag M2 beads as per manufacturer recommendations (Sigma-Aldrich). In vitro ubiquitination assays were performed at room temperature (20–25 °C) for 10 minutes in 10 µl reaction buffer (40 mmol/l Tris, pH 7.6, 2 mmol/l Mg2+-ATP, and 2 mmol/l dithiothreitol) containing purified TRIM5α protein (10 ng), E1 EZ (100 nmol/l), UbcH5c (500 nmol/l), and ubiquitin (2.5 µg). The reaction mixtures were heated at 95 °C in 2% sodium dodecyl sulfate sample buffer for 10 minutes and analyzed by western blotting using MAb anti-Ubiquitin clone P4D1 (Santa Cruz Biotechnology, Santa Cruz, CA) and MAb anti-Flag antibody clone M2 (Sigma-Alrdich); secondary antibody was polyclonal goat anti-mouse IgG-HRP (Santa Cruz Biotechnology).
HIV-1 Challenge in vitro and in vivo. Infection of primary human CD4 T cells was performed as previously described.17All humanized mouse experiments were approved by the University of Pennsylvania's Institutional Animal Care and Use Committee (Protocol 802717) and were carried out in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. NSG were engrafted with 107 primary human CD4 T cells expressing mCherry-huTRIM5α, mCherry-huTRIM5αR323–332, or GFP as a control; 15 animals were engrafted initially for the mCherry-huTRIM5α group, while 10 animals were engrafted per group with mCherry-huTRIM5αR323–332, or GFP control. The number of engrafted human CD4 T cells in peripheral blood was enumerated by TruCount (BD Biosciences) as the number of huCD45+ cells/µl, following staining with PerCP-Cy5.5-conjugated anti-human CD45 antibody (eBioscience, San Diego, CA), and FITC-conjugated anti-CD4 antibody (eBioscience). Three weeks after engraftment, mice with lower levels of engrafted human T cells were excluded from this study, and the remaining mice were challenged with 50 µl of cell-free HIV-1BaL; 11 mice were challenged in the mCherry-huTRIM5α group, with 6 mice challenged in each of the mCherry-huTRIM5αR323–332 and GFP control groups. Persistence of engrafted CD45+ CD4 T cells was monitored each week for 3 weeks, and serum was saved for viral load analysis. Animals were housed at the University of Pennsylvania. Serum viral loads were assayed commercially (Bioqual, Rockville, MD), using 10 µl of serum.
Acknowledgments
We gratefully acknowledge Julie Jadlowsky and Caitlin Baiduc for helpful suggestions; CFAR/Cancer Center Immunology Core for primary human T cells; the Xenograft and Stem Cell Core for their expert care and handling of mice, and Jasmine Zhou of the Microscopy Core at the Perelman School of Medicine for skilled assistance with Confocal microscopy. This work was supported in part by National Institutes of Health grants R01-CA-147795 and U19AI082628. M.W.R designed research, performed research, analyzed data, and wrote the paper. L.G. and F.X. designed research, performed research, and analyzed data. X.Y. designed research and analyzed data. J.L.R. designed research, analyzed data, and wrote the paper. None of the authors report any conflict of interest.
References
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Züger S, Guerra J, Lascano J, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Ke D, Vu T, Ahn J, Shah VB, Yang R, et al. Rhesus TRIM5α disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 2011;7:e1002009. doi: 10.1371/journal.ppat.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LR, Aiken C. TRIM5alpha disrupts the structure of assembled HIV-1 capsid complexes in vitro. J Virol. 2010;84:6564–6569. doi: 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353:396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Li X, Gold B, O'hUigin C, Diaz-Griffero F, Song B, Si Z, et al. Unique features of TRIM5alpha among closely related human TRIM family members. Virology. 2007;360:419–433. doi: 10.1016/j.virol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji U, Bobardt MD, Gaskill P, Sheeter D, Fox H, Gallay PA. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J Biol Chem. 2006;281:37025–37033. doi: 10.1074/jbc.M606066200. [DOI] [PubMed] [Google Scholar]
- Maillard PV, Zoete V, Michielin O, Trono D. Homology-based identification of capsid determinants that protect HIV1 from human TRIM5α restriction. J Biol Chem. 2011;286:8128–8140. doi: 10.1074/jbc.M110.187609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson MW, Carroll RG, Stremlau M, Korokhov N, Humeau LM, Silvestri G, et al. Mode of transmission affects the sensitivity of human immunodeficiency virus type 1 to restriction by rhesus TRIM5alpha. J Virol. 2008;82:11117–11128. doi: 10.1128/JVI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Ishida Y, Oleksyk TK, Winkler CA, Roca AL. Evidence for selection at HIV host susceptibility genes in a West Central African human population. BMC Evol Biol. 2012;12:237. doi: 10.1186/1471-2148-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price H, Lacap P, Tuff J, Wachihi C, Kimani J, Ball TB, et al. A TRIM5alpha exon 2 polymorphism is associated with protection from HIV-1 infection in the Pumwani sex worker cohort. AIDS. 2010;24:1813–1821. doi: 10.1097/QAD.0b013e32833b5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt V, Bleiber G, May M, Martinez R, Ortiz M, Telenti A. Swiss HIV Cohort Study Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology. 2006;3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, An P, Gold B, Petersen DC, O'Huigin C, Nelson GW, et al. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grütter C, Martinetti G, et al. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Invest. 2009;119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastri J, O'Connor C, Danielson CM, McRaven M, Perez P, Diaz-Griffero F, et al. Identification of residues within the L2 region of rhesus TRIM5alpha that are required for retroviral restriction and cytoplasmic body localization. Virology. 2010;405:259–266. doi: 10.1016/j.virol.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanenko OV, Verkhusha VV, Kazakov VI, Shavlovsky MM, Kuznetsova IM, Uversky VN, et al. Comparative studies on the structure and stability of fluorescent proteins EGFP, zFP506, mRFP1, “dimer2”, and DsRed1. Biochemistry. 2004;43:14913–14923. doi: 10.1021/bi048725t. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Kornitzer D. Monitoring protein degradation. Methods Enzymol. 2002;351:639–647. doi: 10.1016/s0076-6879(02)51874-7. [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi K, Wada K, Tanji K, Tanaka M, Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5 alpha and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Tipper C, Sodroski J. Role of TRIM5α RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J Virol. 2011;85:8116–8132. doi: 10.1128/JVI.00341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienlaf M, Hayashi F, Di Nunzio F, Tochio N, Kigawa T, Yokoyama S, et al. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha. J Virol. 2011;85:8725–8737. doi: 10.1128/JVI.00497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battivelli E, Lecossier D, Matsuoka S, Migraine J, Clavel F, Hance AJ. Strain-specific differences in the impact of human TRIM5alpha, different TRIM5alpha alleles, and the inhibition of capsid-cyclophilin A interactions on the infectivity of HIV-1. J Virol. 2010;84:11010–11019. doi: 10.1128/JVI.00758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battivelli E, Migraine J, Lecossier D, Matsuoka S, Perez-Bercoff D, Saragosti S, et al. Modulation of TRIM5alpha activity in human cells by alternatively spliced TRIM5 isoforms. J Virol. 2011;85:7828–7835. doi: 10.1128/JVI.00648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battivelli E, Migraine J, Lecossier D, Yeni P, Clavel F, Hance AJ. Gag cytotoxic T lymphocyte escape mutations can increase sensitivity of HIV-1 to human TRIM5alpha, linking intrinsic and acquired immunity. J Virol. 2011;85:11846–11854. doi: 10.1128/JVI.05201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SY, Rogers T, Chan T, Whitney JB, Kim J, Sodroski J, et al. TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys. PLoS Pathog. 2010;6:e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chackerian B, Long EM, Luciw PA, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, et al. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Sawyer SL. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics. 2009;61:163–176. doi: 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- Allers K, Hütter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Akkina R. TRIM5alpharh expression restricts HIV-1 infection in lentiviral vector-transduced CD34+-cell-derived macrophages. Mol Ther. 2005;12:687–696. doi: 10.1016/j.ymthe.2005.07.291. [DOI] [PubMed] [Google Scholar]
- Sakuma R, Mael AA, Ikeda Y. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J Virol. 2007;81:10201–10206. doi: 10.1128/JVI.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]