Summary
While a few cancer genes are mutated in a high proportion of tumors of a given type (>20%), most are mutated at intermediate frequencies (2–20%). To explore the feasibility of creating a comprehensive catalog of cancer genes, we analyzed somatic point mutations in exome sequence from 4,742 tumor-normal pairs across 21 cancer types. We found that large-scale genomic analysis can identify nearly all known cancer genes in these tumor types. Our analysis also identified 33 genes not previously known to be significantly mutated, including genes related to proliferation, apoptosis, genome stability, chromatin regulation, immune evasion, RNA processing and protein homeostasis. Down-sampling analysis indicates that larger sample sizes will reveal many more genes, mutated at clinically important frequencies. We estimate that near-saturation may be achieved with 600–5000 samples per tumor type, depending on background mutation rate. The results help guide the next stage of cancer genomics.
Introduction
Comprehensive knowledge of the genes underlying human cancers is a critical foundation for cancer diagnostics, therapeutics, clinical trial design and selection of rational combination therapies. It is now possible to use genomic analysis identify cancer genes in an unbiased fashion, based on the presence of somatic mutations at a rate significantly higher than the expected background level.
Systematic studies have revealed many new cancer genes, as well as new classes of cancer genes1,2. They have also made clear that, while some cancer genes are mutated at high frequencies, most cancer genes in most patients occur at intermediate frequencies (2–20%) or lower. Accordingly, a complete catalog of mutations in this frequency class will be essential for recognizing dysregulated pathways and optimal targets for therapeutic intervention. Yet, recent work suggests major gaps in our knowledge of cancer genes of intermediate frequency. For example, a study of 183 lung adenocarcinomas3 found that 15% of patients lacked even a single mutation affecting any of the ten known hallmarks of cancer, and 38% had three or fewer such mutations.
In this paper, we analyzed somatic point mutations (substitutions and small insertion/deletions) in nearly 5000 TN pairs across 21 tumor types. We ask: (1) Can large-scale genomic analysis across tumor types reliably identify all known cancer genes? (2) Will it reveal many new candidate cancer genes? (3) How far do we stand from having a complete catalog of cancer genes – at least those of intermediate frequency? We used rigorous statistical methods to enumerate candidate cancer genes and then carefully inspected each gene to identify those with strong biological connections to cancer and mutational patterns consistent with the expected function.
The analysis reveals nearly all known cancer genes and revealed 33 novel candidates, including genes related to proliferation, apoptosis, genome stability, chromatin regulation, immune evasion, RNA processing, and protein homeostasis. Importantly, the data show that the catalog of cancer genes is still far from complete – with the number of candidate cancer genes still increasing sharply with sample size. These analyses allow us to estimate the sample sizes that will be needed to approach saturation.
Results
Cancer genome data
We collected and analyzed data from 4,742 samples, consisting primarily of whole-exome sequence from TN pairs. The samples span 21 tumor types, which include 12 from The Cancer Genome Atlas (TCGA) and 14 from non-TCGA projects at the Broad Institute, with some overlapping tumor types (Table 1, Supplementary Table 1). The number of samples per tumor type varied between 35 and 892.
Table 1.
List of the 21 tumor types analyzed. Listed for each tumor type is its code (as used in TCGA projects), number of tumor-normal (TN) pairs, median somatic mutation frequency per megabase, the number of significantly mutated genes detected by the MutSig suite when analyzing the full set of genes, and the number of additional significantly mutated genes detected under restricted hypothesis testing (RHT) on just the set of cancer genes found in all the other tumor types. Supplementary Table 3 lists the cancer genes found in each tumor type and their frequencies (percent of patients mutated).
| Tumor type | Tumr type code |
No. of tumor- normal pairs |
Median somatic mutation frequency (per Mb) |
No. of significantly mutated genes |
No. of additional signif. genes found under RHT |
|---|---|---|---|---|---|
| Acute myeloid leukemia | LAML | 196 | 0.4 | 26 | 1 |
| Bladder | BLCA | 99 | 7.1 | 24 | 10 |
| Breast | BRCA | 892 | 1.2 | 32 | 5 |
| Carcinoid | CARC | 54 | 0.5 | 1 | 0 |
| Chronic lymphocytic leukemia | CLL | 159 | 0.6 | 7 | 8 |
| Colorectal | CRC | 233 | 3.1 | 23 | 12 |
| Diffuse large B-cell lymphoma | DLBCL | 58 | 3.3 | 16 | 7 |
| Endometrial | UCEC | 248 | 2.5 | 58 | 15 |
| Esophageal adenocarcinoma | ESO | 141 | 4.0 | 8 | 7 |
| Glioblastoma multiforme | GBM | 291 | 2.2 | 22 | 4 |
| Head and neck | HNSC | 384 | 3.9 | 25 | 9 |
| Kidney clear cell | KIRC | 417 | 1.9 | 15 | 6 |
| Lung adenocarcinoma | LUAD | 405 | 8.1 | 22 | 10 |
| Lung squamous cell carcinoma | LUSC | 178 | 9.9 | 11 | 13 |
| Medulloblastoma | MED | 92 | 0.3 | 2 | 1 |
| Melanoma | MEL | 118 | 12.9 | 19 | 9 |
| Multiple myeloma | MM | 207 | 1.6 | 11 | 3 |
| Neuroblastoma | NB | 81 | 0.5 | 1 | 0 |
| Ovarian | OV | 316 | 1.7 | 5 | 5 |
| Prostate | PRAD | 138 | 0.7 | 4 | 2 |
| Rhabdoid tumor | RHAB | 35 | 0.1 | 1 | 0 |
Data were all analyzed through the Broad’s stringent filtering and annotation pipeline to obtain a uniform set of mutation calls (Methods). The dataset consists of 3,078,483 somatic single nucleotide variations (SSNVs), 77,270 small insertions and deletions (SINDELs), and 29,837 somatic di-/tri-/oligonucleotide variations (DNVs/TNVs/ONVs), with an average of 672 per TN pair. The mutations include 540,831 missense, 207,144 synonymous, 46,264 nonsense, 33,637 splice-site, and 2,294,935 non-coding mutations (used to improve our background model). The analysis has sensitivity > 90% based on the sequencing depth and tumor purity and ploidy4,5.
Mutation frequencies vary over more than five orders of magnitude (from 0.03/Mb to 7000/Mb) within and across tumor types, consistent with our recent study of mutational heterogeneity6 of ~3,000 samples (of which, 2,502 are included in this dataset) (Supplementary Figure 1). Mutation spectra also vary sharply within and across tumor types6 (Supplementary Figure 2).
Cancer genome analysis
We analyzed these data to identify candidate cancer genes – by which we shall mean genes harboring somatic point mutations (that is, substitutions and small insertion/deletions) at a statistically significant rate or pattern in cancer. (Such genes will ultimately need to be verified by biological experiments to be considered validated cancer genes.) In this paper, we do not seek to implicate genes based on other criteria (such as amplification or deletion, translocations or epigenomic modification; but see7).
In principle, candidate cancer genes can be discovered by sequencing enough tumor-normal pairs – based on the presence of an excess of somatic mutations compared to expectation. However, careful analysis is required to assess statistical significance. The mere presence of somatic mutations is insufficient to implicate a gene in cancer, inasmuch as 93% of genes carried mutations in at least five samples.
We showed recently6 that heterogeneity of mutation rates and patterns in cancer can give rise to false positives and described methods to overcome this problem. We applied these methods to identify candidate cancer genes. We used the most recent version of the MutSig suite of tools (Supplementary Figure 3a, Methods), which looks for three independent signals: (i) high mutational burden relative to background expectation, accounting for heterogeneity; (ii) clustering of mutations within the gene8; and (iii) enrichment of mutations in evolutionarily conserved sites8. We combined the significance levels (p-values) from each test to obtain a single significance level per gene (Methods).
We analyzed each tumor type separately, as well as the entire cohort (‘combined’ set), using the same methodology to ensure that the results can be compared across types. We verified that each analysis accurately calculates the significance level of genes, based on the fact that the vast majority of genes fit the null hypothesis and lie on the diagonal in a Q-Q plot (Supplementary Figure 3b). For each analysis, genes with false discovery rate (FDR) q≤0.1 were declared to be candidate cancer genes (Methods). Using an FDR of q≤0.1 ensures that the expected fraction of false positives in each analysis does not exceed 10%. This well-established statistical procedure allows one to increase statistical power to detect true positives, while controlling the proportion of false positives. We also analyzed the merged set of gene × tumor-type pairs identified from the 22 individual analyses (here we include the combined set as one of the ‘tumor types’), using methods discussed below.
Data and results are posted at http://www.tumorportal.org/. The site includes graphical displays of the mutations in each of the 18,388 genes studied; see examples in Figure 1 and Supplementary Figure 4. The site also includes tables of mutational data for each significant gene) and Q-Q plots for each statistical test.
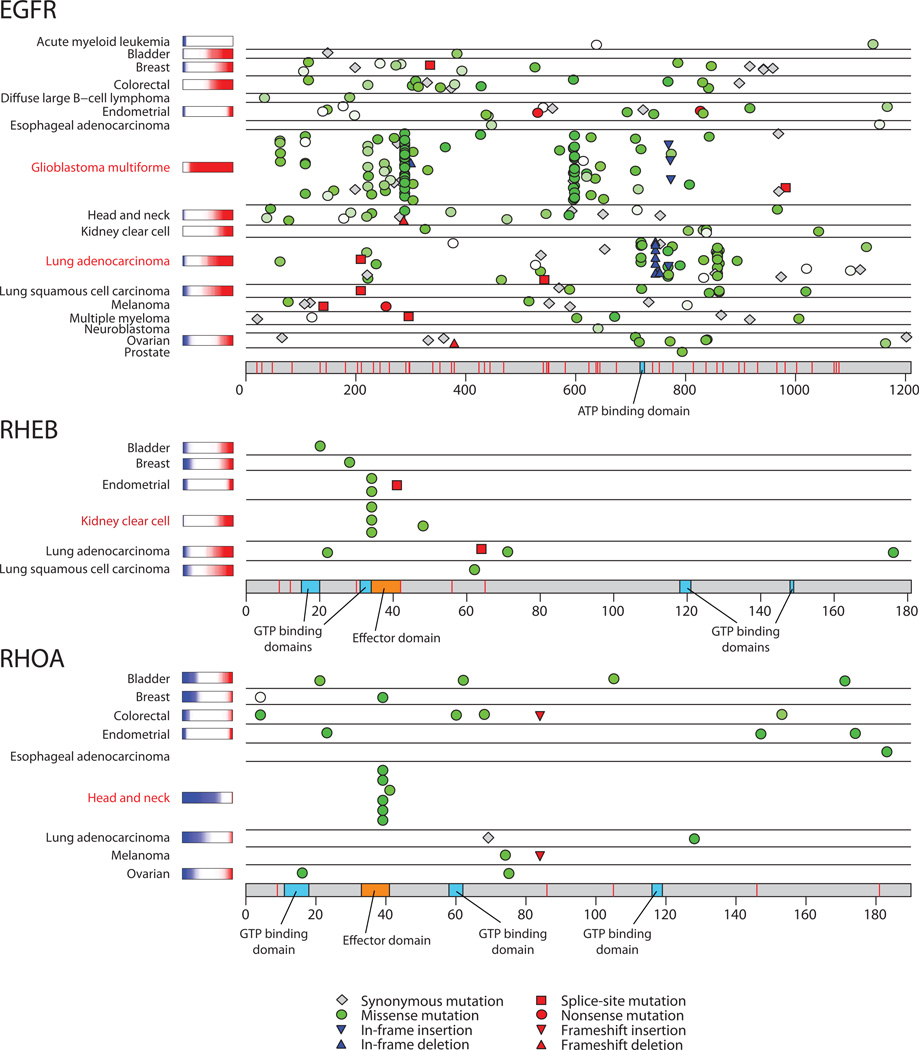
Figure 1.
Mutation patterns for one known and two novel cancer genes. EGFR shows distinctive tumor-type-specific concentrations of mutations in different regions of the gene. RHEB, which encodes a small GTPase in the RAS superfamily, shows a mutational hotspot in the effector domain. RHOA, another a member of the RAS superfamily, also shows a mutational hotspot in the effector domain. Colored bars after tumor type names are copy-ratio distributions for the gene, when available (red=amplified, blue=deleted). See also Supplementary Figure 4. Similar diagrams for all genes are available at http://www.tumorportal.org.
Candidate cancer genes across 21 tumor types
A total of 224 genes were found to be significant in one or more tumor types, and 334 gene × tumor-type pairs were found to be significant. The number of genes detected per tumor type varied considerably (range 1 – 58), with seven types having fewer than 10 genes and two (breast and endometrial) having more than 30 (Figure 2; Supplementary Figure 5; Table 1). The specific genes differed substantially across tumor types, although some pairs of tumor types showed clear similarity, such as lung squamous cancer and head and neck squamous cancer (Methods, Supplementary Figure 6).
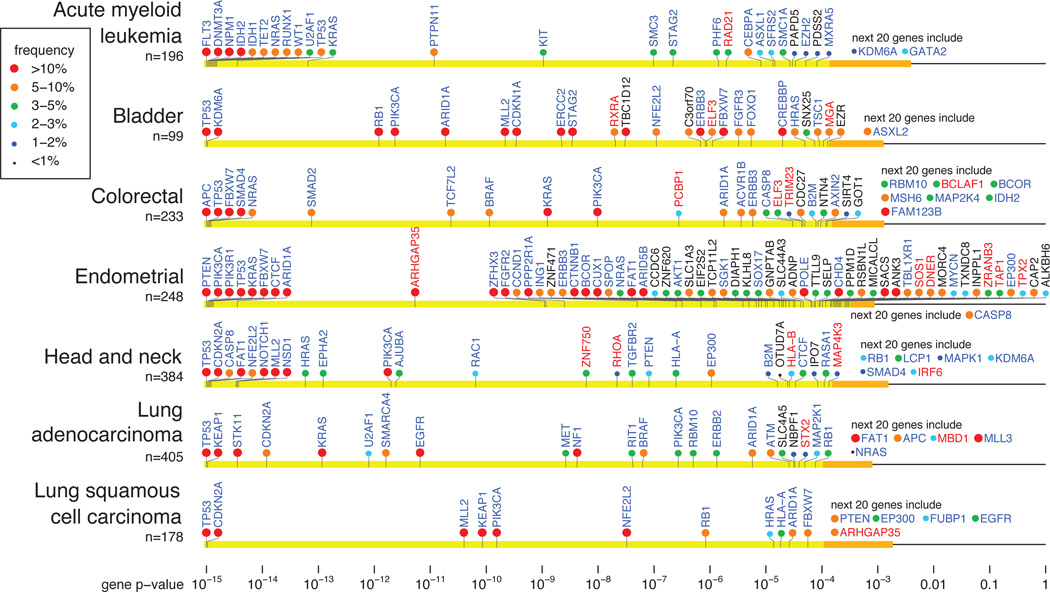
Figure 2.
Cancer genes in selected tumor types. Genes are arranged on the horizontal line according to p-value (combined value for the three tests in MutSig). Yellow region contains genes that achieve FDR q≤0.1. Orange interval contains p-values for the next 20 genes. Gene name color indicates whether the gene is a known cancer gene (blue), a novel gene with clear connection to cancer (red; discussed in text), or an additional novel gene (black). Circle color indicates the frequency (percent of patients carrying non-silent somatic mutations) in that tumor type. See also Supplementary Figure 5.
Strikingly, only 22 genes were declared significant in three or more tumor types. The well-established cancer genes TP53, PIK3CA, PTEN, RB1, KRAS, NRAS, BRAF, CDKN2A, FBXW7, ARID1A and MLL2, as well as STAG2, were significant in four or more tumor types. An additional 10 genes (ATM, CASP8, CTCF, ERBB3, HLA-A, HRAS, IDH1, NF1, NFE2L2, PIK3R1) were significant in three tumor types.
Although the power to detect cancer genes varied across tumor types (based on sample size and background mutation rate), the striking differences across tumor types do not simply reflect differences in detection power. For example, tumor types with low mutation frequency or many samples often show fewer cancer genes despite having greater statistical power to detect them (Table 1). Moreover, many genes that are highly enriched in one (e.g. VHL, WT1) or a few (e.g. HRAS, FBXW7) tumor types fail to show detectable enrichment across the entire data set (Supplementary Table 2). Notably, most of the significant gene × tumor-type pairs involve only a small fraction of patients (with one half of the significant pairs involving ≤ 6.1% of patients, and one quarter involving ≤ 3.1%).
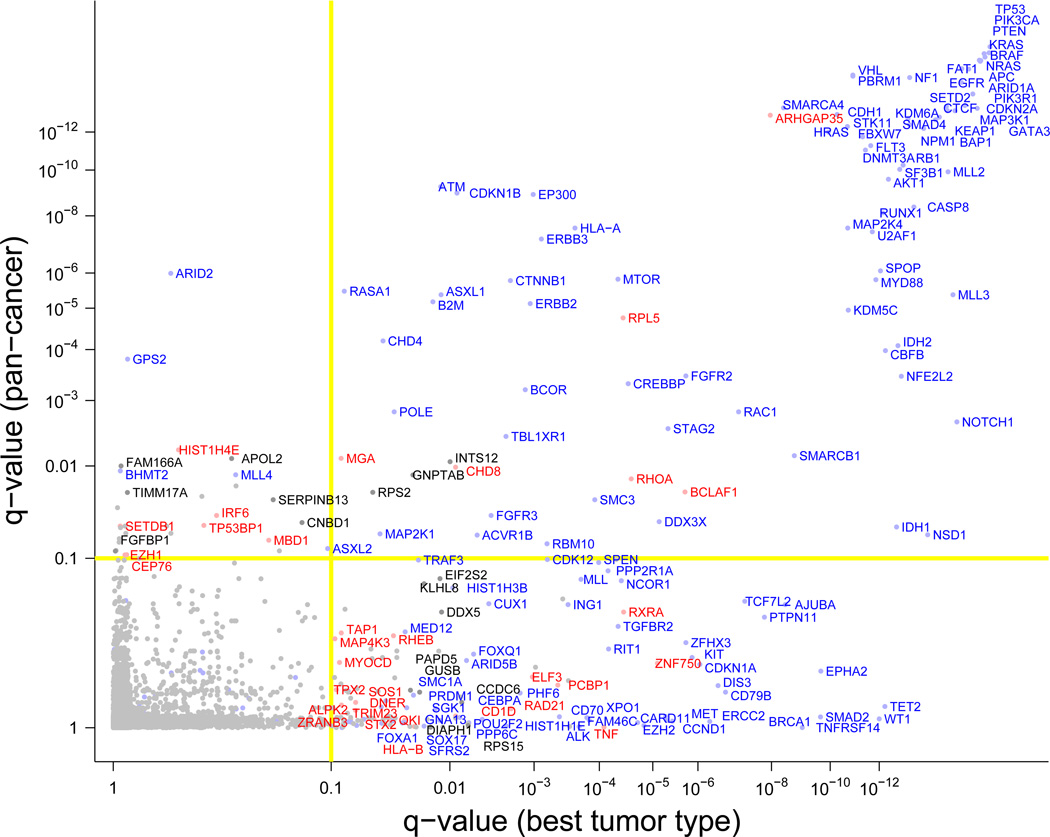
We then analyzed the combined set, which yielded 114 genes (Supplementary Table 2). While 84 of these genes were already identified from analysis of individual tumor types, the remaining 30 achieved significance based only on the frequency of mutations across tumor types – underscoring the value of cross-tumor-type analysis. Conversely, 140 of the 224 genes found in analysis of individual tumor types did not reach significance when analyzing the combined set (Figure 3, lower right quadrant), consistent with the observation that many genes show strong enrichment in only one or a few tumor types.
Figure 3.
Cancer genes identified in 4742-tumor dataset. X-axis indicates the q-value (FDR) in the most significant of the 21 tumor types. Y-axis indicates the q-value when the 4742 tumors are analyzed as a combined cohort. Genes in the upper left quadrant reached significance only in the combined analysis. Genes in the lower right quadrant reached significance only in one or more single-type analyses. Genes in the upper right quadrant were significant in both the combined set and in individual tumor types. Color of gene names is as in Figure 2.
By merging the 22 lists above, we obtained a Cancer5000 set containing 254 genes. Although the expected proportion of false positive genes in each list does not exceed 10%, the expected proportion in the merged list is actually higher (because true positives will tend to occur across several tumor types, while false positives will tend to be random singletons). A rigorous solution is to analyze the gene × tumor-type pairs as ~400,000 distinct hypotheses (~18,400 genes × 22 types) and apply an FDR of q ≤ 0.1. This analysis yielded 403 significant pairs, which involve 219 distinct genes. We refer to this set as the Cancer5000-S (for “stringent”) genes. (All but six of the genes are contained in the Cancer5000 set.) Of the 403 significant pairs, at most 10% (that is ~40) are expected to be false positives. Assuming conservatively that the 40 pairs affect 40 distinct genes, we expect 179 of the 219 genes to be true cancer genes. Below, we discuss genes from both the Cancer5000 and Cancer5000-S sets.
Coverage of known cancer genes
We first asked whether all cancer genes that have been discovered and validated to date can be identified by hypothesis-free genomic analysis. As a reference set, we used the Cancer Gene Census (CGC), which is a manually curated catalog of cancer genes. The current version (v65) contains 130 cancer genes driven by somatic point mutations (as well as additional genes mutated by other mechanisms), of which 82 are associated with one or more of the 21 tumor types studied here.
Of these 82 genes, 60 were identified in our Cancer5000 set. Of the remaining 22 genes, (i) 8 fell just below significance in our data set; (ii) 6 appear in the CGC based on focused studies of the gene in very large samples (typically, >1000); and (iii) 8 genes harbored few mutations and appear to lack adequate evidence to justify association with any of the tumor types we studied. The first two categories would clearly be captured with larger sample sizes.
Analysis of novel candidate cancer genes
Of the 219 genes in the Cancer5000-S set, 81 are neither listed in the CGC as affected by point mutations in these tumor types (v65) nor discussed in papers published as of this writing (Supplementary Table 4). (The list includes 3 genes that appear in tables in published papers based on mutations in a handful of samples, but were not noted or interpreted in the text.) Of the 41 additional genes in the Cancer5000 (but not Cancer5000-S) set, none are in the CGC but 3 are reported in recent publications (Supplementary Table 4).
We closely analyzed these 81 ‘novel’ genes to look for connections with cancer biology, together with a mutational pattern consistent with the biology. Where loss-of-function would be expected, we looked for an excess of disruptive changes, such as nonsense and frameshift mutations. Where gain-of-function, we examined whether the overall collection of mutations included hotspots – resulting in recurrent changes at identical or nearby amino acids (often causing precisely the same change). Conversely, where we observed distinctive mutation patterns, we examined whether they are consistent with known biology.
As discussed above, the Cancer5000-S set is expected by design to contain ~40 false positives. Assuming conservatively that these false positives fall exclusively in the novel set, we expect ~41 of the novel genes to be true positives.
In fact, we identified strong and consistent connections to cancer for at least 21 of the novel genes in the Cancer5000-S set. Among the 38 additional novel genes in the larger Cancer5000 set, we found 12 additional strong candidates. (References supporting the biological roles of the genes are provided in Supplementary Table 5.) We briefly describe these 33 genes not previously been reported as significantly mutated in cancer:
(1) Anti-proliferation
Four genes encode anti-proliferative proteins, in which loss-of-function mutations would be expected to contribute to oncogenesis. A striking example is ARHGAP35 (previously called GRLF1), which encodes a Rho GTPase activating protein, for which only a single tumor type reaches statistical significance on its own, but which gives a strong signal (q = 2 × 10−12) in the combined set of ~5000 tumors (83 missense, 38 nonsense, 16 frameshift and 2 splice site). Notably, the gene resides in a small genomic region that is focally deleted in many tumors. Other examples are MGA, whose product competes with Myc for binding to Max and which resides in small focal deletions (containing ≤4 genes) in ovarian and various epithelial cancers; the interferon regulatory factor IRF6, which is known to have tumor suppressive roles in keratinocytes and is mutated in head and neck squamous cancer; and the delta/notch-like EGF-repeat gene DNER.
(2) Proliferation
Six additional genes encode proteins that are clearly involved in cell proliferation: RHEB, RHOA, SOS1, ELF3, SGK1 and MYOCD. Notably RHEB and RHOA encode small GTPases, in which recurrent mutations affect the 9-amino-acid effector domain (ED). For RHEB, five tumors (2 endometrial and 3 kidney clear cell cancer) carry Y35N mutations, which alter the first amino acid of the ED. For RHOA, six tumors (all head and neck) carry mutations affecting the ED: these include five E40Q mutations and a single Y42I mutation, which alter the seventh and ninth amino acids, respectively, of the ED. SOS1 encodes a guanine nucleotide exchange factor that promotes activation of Ras, in which gain-of-function mutations might contribute to oncogenesis. Consistent with this notion, SOS1 carries N233Y mutations in six tumors (four endometrial and two lung adenocarcinoma) and R552 alterations in three tumors (two endometrial and one AML). Strikingly, the same R522 alterations in SOS1 have been found as germline mutations causing Noonan syndrome and been shown to cause gain-of-function resulting in Ras activation. ELF3 encodes an ETS-domain transcription factor that functions in cell differentiation; it carries many truncating mutations in bladder and colon cancer. Myocardin (MYOCD), which encodes a transcriptional regulator involved in differentiation and cell migration, has a cluster of 9 mutations at amino acids 750–770 (7 in melanoma, 1 head and neck, 1 lung adenocarcinoma) with a hotspot of four at S763. The retinoid × receptor alpha RXRA, which forms a heterodimer with retinoic acid receptors to regulate cell growth and survival, shows a clear hotspot of recurrent mutations at S427 in bladder cancer and nearby mutations in lung, head and neck, and esophageal cancers.
(3) Apoptosis
Five genes encode pro-apoptotic factors, in which loss-of-function mutations would be expected to promote oncogenesis. These genes encode alpha kinase 2 (ALPK2); Bcl2-associated factor 1 (BCLAF1); a MAP kinase (MAP4K3) reported to post-transcriptionally regulate the apoptotic proteins Puma, Bad and Bim; a zinc-finger protein (ZNF750, which harbors many early frameshift and nonsense mutations in head and neck cancer and is the only known gene residing in a small current focal deletion in head and neck and lung squamous cancers); and Tumor Necrosis Factor (TNF, which harbors mutations in five diffuse large B-cell lymphomas that are tightly clustered in the region encoding the membrane/cytoplasmic domain, rather than the soluble Tnf protein).
(4) Genome stability
Six genes encode proteins related to genome stability. These include CEP76 (encoding a centrosomal protein, whose depletion drives aberrant amplification of centrioles), which harbors early nonsense mutations in many tumor types and resides in a focal deletion peak in acute myeloid leukemia; RAD21 (encoding a protein crucial for chromosome segregation and double-strand break repair), which is mutated at significant rates in acute myeloid leukemia and also harbors mutations in other tumor types; the p53-binding protein TP53BP1 (encoding a check-point protein that binds to double-strand breaks), which does not reach significance in any single tumor type, but is significant in the combined data set due to truncating mutations in many tumor types; TPX2 (encoding a protein involved in mitotic spindle formation, whose depletion leads to aneuploidy); and ZRANB3 (encoding a translocase that helps to rescue stalled replication forks). In addition, STX2 encodes a protein required for cytokinesis, whose disruption may promote aneuploidy; STX2 harbors recurrent mutations at R107 in lung and endometrial tumors.
(5) Chromatin regulation
Five genes are associated with chromatin regulation. SETDB1 encodes a H3K9 histone methyltransferase (SETD2, which encodes a H3K36 histone methyltransferase, has been shown previously to be mutated in cancer). MBD1 encodes a protein that binds methylated-CpG and is required for SETDB1 activity; it contains 5 mutations in endometrial cancer in the N-terminal methyl binding domain. EZH1 encodes a H3K27 histone methyltransferase; it does not reach significance in any individual tumor type, but is significant in the combined set due to truncating mutations in multiple tumor types. EZH1 shows a similar pattern of mutations as seen in the well-established cancer gene EZH2, with truncating mutations along the gene and a hotspot of mutations within the SET domain. CHD8 encodes a chromatin helicase DNA binding protein (like the known cancer gene CHD4) that suppresses the beta-catenin-Wnt signaling pathway. The histone protein HIST1H4E is mutated in multiple tumor types; two other histone genes, HIST1H1E and HIST1H3B, have previously been reported as significantly mutated in CLL and DLBCL, respectively.
(6) Immune evasion
Three genes encode proteins whose loss is expected to help tumor evade immune attack; they all recurrently subject to truncating mutations across several tumor types. These include the major histocompatibility protein HLA-B (loss of the HLA-A gene has been implicated in lung cancer), TAP1 (which processes intracellular peptides for presentation to the immune system) and CD1D (which presents lipid antigens to natural killer cells), the last of which shows a cluster of truncating mutations at the internalization domain that are likely to abolish antigen presentation function.
(7) RNA processing
Three genes are associated with RNA processing and metabolism. PCBP1, whose protein product blocks translation of certain mRNAs by binding to Poly-C regions of mRNAs, carries two mutations in each of two nearby leucines (Leu100 and Leu102) that mediate dimerization of the protein’s KH domains. We speculate that disruption of PCBP1 leads to increased translation of one or more pro-oncogenic mRNAs. QKI encodes an RNA-binding protein that regulates pre-mRNA splicing, including the known cancer gene CDKN1B; the gene harbors C-terminal truncating mutations in several tumor types that likely remove the nuclear localization signal; and the gene resides in a recurrent deletion peak in glioblastoma and ovarian cancer. Finally, the ribosomal protein gene RPL5 contains early truncating mutations in glioblastoma and other tumor types and resides in a focally deleted region in glioblastoma; heterozygous loss of certain ribosomal proteins has been reported to contribute to cancer.
(8) Protein homeostasis
One gene, TRIM23, is involved in protein homeostasis. It encodes a ubiquitin E3 ligase and harbors recurrent mutations at N93 (4 tumors) and D289 (3 tumors). Mutations in this gene may promote cancer by altering the substrate specificity of the E3 ligase in a manner that leads to accumulation of an oncogenic protein.
Beyond these 33 genes, the set of 81 novel genes is likely to contain additional true cancer genes. For example, we omitted genes with connections to cancer (such as HSP90AB1, PPM1D, ITGB7) where we could not easily reconcile the function in cancer with the observed pattern of mutations. In addition, we likely overlooked additional candidate cancer genes because we did not identify clear connections with cancer – owing to gaps in the literature or in our knowledge.
Saturation analysis
We next explored whether the discovery of candidate cancer genes is approaching saturation or whether many more genes are likely to be found. An effective test is to perform “down-sampling” – that is, to study how the number of discoveries increases with sample size, by repeating the analysis on random subsets of samples of various smaller sizes.
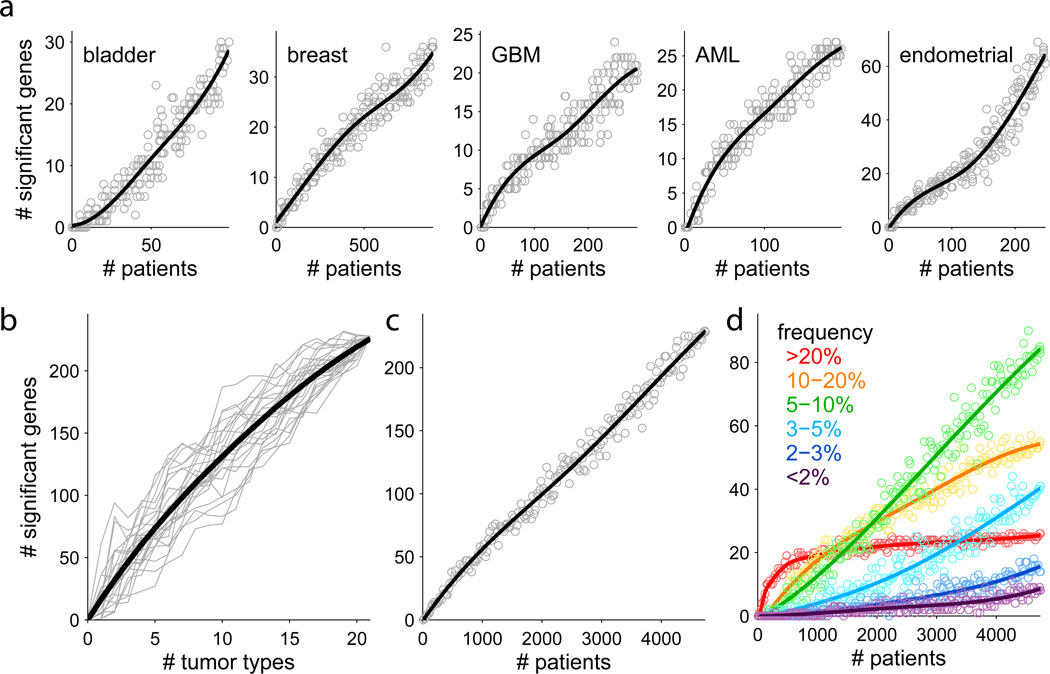
For each tumor type (omitting those with five or fewer candidate cancer genes), the number of genes increases roughly linearly with sample size (examples in Figure 4a; see also Supplementary Figure 7) – indicating that the inventory for each of the tumor types is far from complete. The number of genes also increases linearly with the number of tumor types studied (Figure 4b), suggesting that it is valuable to increase both the sample size per tumor type and the number of tumor types.
Figure 4.
Down-sampling analysis shows that gene discovery is continuing as samples and tumor types are added. a. Analysis within tumor types. Each point represents a random subset of patients. Blue line is a smoothed fit. b. Analysis by adding tumor types. Each grey line represents a random ordering of the 21 tumor types. c. Analysis by adding samples. Each point is a random subset of the 4742 patients. d. Analysis in panel c broken down by mutation frequency. Genes mutated at frequencies ≥ 20% are nearing saturation, while intermediate frequencies show steep growth. See also Supplementary Figures 7, 8.
We also studied how the total number of candidate cancer genes varies with sample size when applying the ‘stringent’ methodology used to create the Cancer5000-S set. Here too, the total number of genes increases steadily with sample size (Figure 4c). Notably, the saturation analysis varies considerably with the mutation frequency (Figure 4d). Genes mutated in >20% of tumors are approaching saturation; those mutated at frequencies of 10–20% are still rising rapidly, but at a decreasing rate; those at 5–10% are increasing linearly; and those at <5% are increasingly at an accelerating rate.
We next sought to infer the nature of the genes awaiting discovery in each tumor type. One possibility is that some of these genes are already contained in the Cancer5000 set (by virtue of their contribution to other tumor types) but have not yet reached statistical significance in the given tumor type due to insufficient sample size. To test this notion, we performed restricted hypothesis testing (RHT): For each tumor type T, we (i) omitted the tumor type, (ii) determined the set GT of genes that are significant based on the remaining tumor types, and (iii) determined which genes in GT reached significance in the omitted tumor type when correcting for multiple hypothesis testing based on only the number of genes in GT (rather than all ~18,400 genes in the genome).
The RHT analysis implicated many additional Cancer5000 genes in the individual tumor types (median 6 per tumor type, range 0–15). The number of significant gene × tumor-type pairs increased from 334 to 461 across the 21 tumor types. The RHT analysis indicates that, with somewhat larger sample size, these genes will likely reach significance in an unrestricted test (Table 1; Supplementary Table 3). For some tumor types, the number of implicated genes more than doubled: lung squamous cell carcinoma increased from 11 to 24; CLL from 7 to 15; and ovarian from 5 to 10. Notably, 3 genes now became significant in four tumor types each (ARID2, ERBB2, ARHGAP35) and 7 genes in three types each (CTNNB1, FGFR3, KRAS, PTEN, SMAD4, MLL3). While 9 of these genes are well known cancer genes, one (ARHGAP35) is absent from the current CGC list. Notably, ARHGAP35 appears in the Cancer5000 set because it is significantly mutated in endometrial cancer (although not discussed in the recent TCGA publication9), but our RHT analysis also finds it to be significant in lung adenocarcinoma, lung squamous cell carcinoma, kidney clear cell, and head and neck cancer. The genes found to be significant in additional tumor types in the RHT analysis are mutated at a median frequency of 3.4%.
However, the data also clearly show that many new candidate cancer genes remain to be discovered beyond those in the current Cancer5000 set. (1) Beyond genes becoming significant in additional tumor types, the down-sampling analysis shows that the number of novel genes is increasing sharply (using the stringent analysis used to create the Cancer5000-S set). (2) Adding additional tumor types typically adds novel ‘tumor-type-specific’ genes, which are unique to (or at vastly higher frequency in) the tumor type.
Power Analysis
Because the cancer gene catalog remains far from complete, we explored what sample sizes will be needed to approach saturation. The power to detect a gene as significantly mutated depends on the properties of the tumor type—namely, (1) the average background mutation rate for the tumor type (‘noise’), and (2) the target frequency, above the background rate, that one wishes to detect (‘signal’). It also depends on the properties of the gene – namely, its background mutation frequency relative to other genes (which depends on length and local mutation rate). We set a target of having 90% power to detect 90% of all genes. In addition, we allow for a false negative rate of 10% in detecting mutations, which increases the sample size by slightly more than 10%.
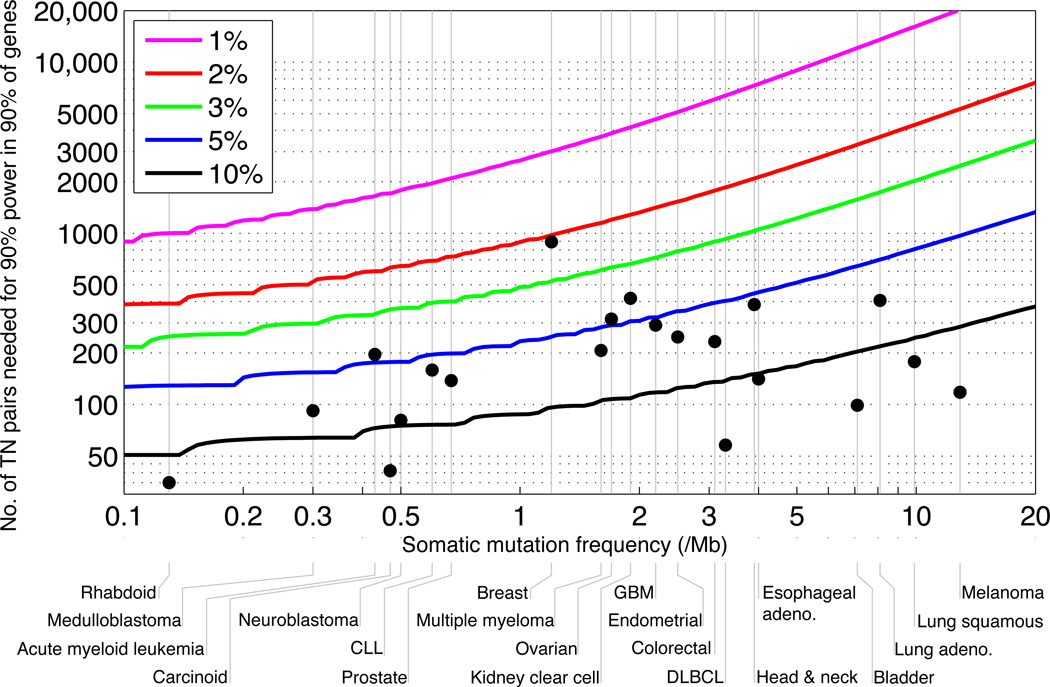
Figure 5 shows that the current collection lacks the desired power to detect genes mutated at 5% above the background rate for 17 of the 21 tumor types and even at 10% for 7 of the tumor types. These results are consistent with the down-sampling analysis showing that candidate cancer genes with frequency ≥ 20% are approaching saturation, while the number of candidate cancer genes at lower frequencies is continuing to grow rapidly with sample size.
Figure 5.
Number of samples needed to detect significantly mutated genes, as a function of a tumor type’s median background mutation frequency of (x-axis) and a cancer gene’s mutation rate above background (the various curves). Y-axis shows the number of samples needed to achieve 90% power for 90% of genes. Grey vertical lines indicate tumor type median background mutation frequencies. Black dots indicate sample sizes in the current study. For most tumor types, the current sample size is inadequate to reliably detect genes mutated at 5% or less above background. See also Supplementary Figure 9.
Creating a reasonably comprehensive catalog of candidate cancer genes mutated in ≥ 2% of patients will require between ~650 samples (for tumors with ~0.5 mutations/Mb, such as neuroblastoma) to ~5300 samples (for melanoma, with 12.9 mutations/Mb).
Discussion
Precision medicine for cancer will ultimately require having a comprehensive catalog of cancer genes to allow physicians to select the best combination therapy for each patient based on the cellular pathways disrupted in their tumor and the specific nature of the disruptions. Such a catalog will also guide therapeutic development by identifying druggable targets. In addition, the catalog and its underlying data will facilitate the interpretation of cell lines, animal models and clinical observations and will reveal patterns of co-occurrence, mutual exclusivity and lineage restriction, which may provide mechanistic insights with profound therapeutic implications.
While a handful of cancer genes are mutated at high frequency, most cancer genes mutated in most patients occur at intermediate frequencies (2–20%). To provide therapeutic options for most patients, it will thus be critical to identify and understand the pathway-level implications of all genes mutated at intermediate frequencies (2–20%).
With growing datasets across many tumor types, pan-cancer analyses are becoming of great interest.10,11 In this paper, we studied somatic point mutations in a collection of nearly 5000 tumor-normal pairs across 21 cancer types. We identified a Cancer5000 set containing 254 genes, based on merging results from each tumor type and the combined set, and a stringent Cancer5000-S set containing 219 genes, accounting for multiple hypothesis testing across the types. Nearly all previously known cancer genes in these tumor types are contained within these sets or just below statistical significance.
After eliminating genes reported in the CGC or recent papers and accounting for the expected number of false positives, the stringent Cancer5000-S set is expected to contain ~41 novel candidate cancer genes, with additional candidate cancer genes expected in the larger Cancer5000 set. Upon close inspection, we found 33 genes (21 in the stringent set and 12 more in the larger set) with strong functional connections to cancer and mutation patterns consistent with the presumed function. These genes fall within known ‘hallmarks’ of cancer3,12, including cell proliferation, apoptosis, genome stability, chromatin regulation, immune evasion, RNA processing and protein homeostasis. Follow-up studies will be required to confirm and understand the functional impact of the mutations in these genes.
Beyond identifying new candidate cancer genes, our study demonstrates that we are far from having a complete catalog of cancer genes – with many genes at clinically important frequencies within individual tumor types and across cancer as a whole still awaiting identification. The number of such genes is still increasing steeply with the number of samples and the number of tumor types studied. Importantly, these new candidate cancer genes are not rare. Substantial ongoing increases are seen in each of the 10–20%, 5–10% and 2–5% ranges (Figure 4d). Notably, of the 33 novel genes above, five are mutated at frequencies greater than 10% and fifteen at frequencies greater than 5%.
Creating a comprehensive catalog of genes in which somatic point mutations propel cancer at both high (>20%) and intermediate (2–20%) frequency will require analyzing an average of ~1700 tumors for each of at least 50 tumor types. (Currently defined tumor types may be divided, based on genomic information, into distinct subtypes, each of which should be analyzed on its own. The ultimate number of tumor types will thus be defined iteratively by molecular analysis.)
Analysis should include both point mutations (as studied here), as well as other types of functional variation7. Genomic studies of such large numbers of samples is no longer prohibitive, in light of the one-million-fold decrease in the cost of DNA sequencing over the past decade. Given the devastating human toll of cancer, with nearly 8 million deaths annually worldwide13, completing the genomic analysis of this disease should be a biomedical imperative.
Full Methods
Mutation data and preprocessing
Mutation data was obtained as follows. For TCGA tumor types, mutation data was downloaded from the Synapse website, from the following link: <https://www.synapse.org/#!Synapse:syn1729383>. For non-TCGA tumor types, sequencing data was downloaded from dbGaP (accession numbers listed at end of main text) and processed through Firehose, the Broad Institute’s analysis platform <http://www.broadinstitute.org/cancer/cga/Firehose>. For tumor types that were originally aligned to builld hg18, liftOver (<http://genome.ucsc.edu/cgi-bin/hgLiftOver>) was used to convert the coordinates of each mutation to build hg19. All mutation data was then combined into a single MAF file. Duplicate patients and duplicate mutations were removed. To standardize the definition of a “splice-site” mutation, any mutation affecting the two bases before or after a splice junction, was labeled as a splice-site mutation. Filtering was performed as follows. In order to remove common sequencing artifacts or residual germline variation, each mutation in the combined MAF file was subjected to a “Panel of Normals” filtering process using a panel of over 4000 BAM files from normal samples. For each mutation, the position of the mutation was examined in each normal BAM file. Mutations observed in the panel of normals were removed from the MAF. The final MAF is available at http://www.tumorportal.org/.
MutSig significance calculations
Three significance metrics were calculated for each gene, using the previously described methods MutSigCV, MutSigCL, and MutSigFN. These measure the significance, respectively, of mutation Burden, Clustering, and Functional Impact (Fig. S3). MutSigCV was described previously6. MutSigCV determines the p-value for observing the given quantity of nonsilent mutations in the gene, given the background model determined by silent (and noncoding) mutations in the same gene and the neighboring genes of covariate space that form its “bagel”. MutSigCL and MutSigFN were used previously8 but were not given names in that work. Here we name the methods to reflect the type of evidence of positive selection that they are designed to detect. MutSigCL and MutSigFN measure the significance of the positional clustering of the mutations observed, as well as the significance of the tendency for mutations to occur at positions that are highly evolutionarily conserved (using conservation as a proxy for probably functional impact). MutSigCL and MutSigFN are permutation-based methods and their p-values are calculated as follows: The observed nonsilent coding mutations in the gene are permuted T times (to simulate the null hypothesis, T=108 for the most significant genes), randomly reassigning their positions, but preserving their mutational “category”, as determined by local sequence context. We used the following context categories: transitions at CpG dinucleotides, transitions at other C:G basepairs, transversions at C:G basepairs, mutations at A:T basepairs, and indels. Indels are unconstrained in where they can move to in the permutations. For each of the random permutations, two scores are calculated: SCL and SFN, measuring the amount of clustering and conservation respectively. SCL is defined to be the fraction of mutations occuring in hotspots. A hotspot is defined as a three-basepair region of the gene containing many mutations: at least 2, and at least 2% of the total mutations. SFN is defined to be the mean of the basepair-level conservation values for the position of each non-silent mutation, as obtained from an alignment of 45 vertebrate genomes to the human genome, the UCSC “phyloP46way” track, which can be downloaded from <http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/README.txt>. To determine a pCL, the p-value for the observed degree of positional clustering, the observed value of SCL (computed for the mutations actually observed), was compared to the distribution of SCL obtained from the random permutations, and the p-value was defined to be the fraction of random permutations in which SCL was at least as large as the observed SCL. The p-value for the conservation of the mutated positions, pFN, was computed analogously. Finally, we noted that the gene AJUBA was referred to in some analyses by the alternative name JUB; after reconciling this naming difference, the gene was significant and added to the list of significant genes.
Combining MutSig statistics
The three MutSig tests described above (MutSigCV, MutSigCL, and MutSigFN) were combined into a single final p-value as follows. First, a joint p-value (CL+FN) for the observed clustering and conservation was calculated from the joint probability distribution of the random permutations. Next, this was combined with the MutSigCV p-value using two methods: (i) The Fisher method of combining p-values from independent tests <http://en.wikipedia.org/wiki/Fisher's_method>; (ii) The truncated product method (TPM) for combining p-values, which rewards highly significant p-values in any one of the tests The combined p-values for both methods were extremely similar. We examined the performance of each of the three metrics separately and each pairwise combination of two metrics. The results of these analyses are presented in Supplementary Table 1 (last tab) and summarized in Supplementary Table 5.
Multiple hypothesis corrections
In the analysis of each tumor type, a total of 18,388 genes were analyzed. In order to correct for these multiple hypotheses, the final MutSig p-values were converted to FDR (q-values) using the method of Benjamini and Hochberg, and genes with q≤0.1 were declared to be significantly mutated. This was also done for the analysis of the combined cohort. Genes with q≤0.1 in any tumor-type analysis or in the combined-cohort analysis were declared to be a member of the Cancer5000 list of significant cancer genes.
To correct for the 22 analyses thus combined (corresponding to 22 chances for each gene to become significant), a further level of multiple hypothesis correction was applied. A list was made of the 18,388 genes × 22 analyses = 404,536 hypotheses. The Benjamini-Hochberg method was applied to this full set, yielding new FDR q-values. Any gene involved in these gene × tumor-type pairs was declared to be a member of the stringently corrected Cancer5000-S list of genes.
Downsampling analyses
In order to analyze the dependence of the number of significantly mutated cancer-associated genes upon the size of the dataset being analyzed, down-sampling was performed. Three different down-sampling analyses are described: (1) down-sampling within each tumor type; and (2) down-sampling of the number of different tumor types; and (3) down-sampling of the full Cancer5000-S procedure.
(1) Down-sampling within each tumor type (Figure 4a, Supplementary Figure 7): For each tumor type, the MutSig analysis was repeated for a set of many smaller subsets of patients from that tumor type. The sizes of the subsets were chosen to regularly sample the interval from zero patients to the final total number of patients that were in the full analysis. For each of the random subsets thus defined, we repeated the full MutSig calculation (MutSigCV + MutSigCL + MutSigFN) and combined the results of the three tests as described above. This allowed us to determine which genes remained significant when analyzing each smaller subset. We counted how many of the genes remained significant at each smaller set size, and plotted this number as a smoothed function of set size. This allowed us to demonstrate that the number of significantly mutated genes detected is continuing to rise steeply in each tumor type. We also repeated this same analysis for the full combined dataset (4742 patients), with similar results.
(2) Down-sampling of the number of different tumor types (Figure 4b). In order to examine the effect of adding whole tumor types, we performed the following analysis. We constructed 25 random orderings of the 21 tumor types, and for each ordering, we constructed 20 subsets by sequentially adding whole tumor types according to that ordering. Then we repeated the whole MutSig analysis for each of these subsets. This yielded a set of curves showing how the number of significantly mutated genes increased as a function of the number of tumor types included in the analysis. The curve depended on the exact ordering of the tumor types as they were added, but all curves showed a steady increase in the number of genes, even at the highest numbers of tumor types. This demonstrated the importance of continuing to sample additional tumor types. We also repeated the analysis with subtraction of the expected number of false positives (Supplementary Figure 8a); the results were qualitatively unchanged.
(3) Down-sampling of the full Cancer5000-S procedure (Figure 4c): We repeated our procedure of constructing the Cancer5000-S list by applying the stringent procedure of correction for the ~400,000 hypothesis (18,388 genes × 22 analyses), and computed how many genes remained significant at each smaller set size. We plotted the number of significantly mutated genes detected as a function of set size. This yielded a similar curve to (1), with the number of significant genes continuing to rise steeply even at the largest set sizes. We also repeated the analysis with subtraction of the expected number of false positives (Supplementary Figure 8b); the results were qualitatively unchanged. Furthermore, we stratified the genes according to their frequency (calculated as the maximal frequency across tumor types), and plotted separate curves for each of the following frequency categories: 20% and above, 10–20%, 5–10%, 3–5%, 2–3%, and below 2%. This clearly demonstrated that the 20%+ genes have largely all been discovered. In contrast, genes at lower frequencies are continuing to be discovered (Figure 4d). Note that rerunning the analysis produces slightly different results in every run since the calculation of p-values has a stochastic component. The genes at the edge of significance (i.e. ones with q-value close to 0.1) may be declared as significant or insignificant with respect to the cutoff of q=0.1 in different analyses. This slight fluctuation is standard for permutation-based methods.
Power calculations
Power analysis was performed using a binomial power model. We first calculated the probability, p0, that a patient will have at least one non-silent mutation in a particular gene from the background model. The calculation is based on the length of the gene, L (in coding bases), the background mutation rate, μ, (in mutations / base), the gene-specific mutation rate factor, fg, (calculated by MutSigCV), the 3:1 typical ratio of non-silent to silent mutations; p0=1−(1− μ*fg)(L*3/4). We used L=1500, and fg = 3.9 (representing the 90th percentile of fg*Lg/1500 across the ~18,000 genes and fg=1 for the 50th percentile gene). We then calculated the signal we want to detect, p1=p0 + r * (1−m); where r is the frequency of non-silent mutations in the population (above background) that a gene is mutated and m is the mis-detection rate of the mutation (we took m=0.1). The power was then calculated as the probability to obtain a p-value ≤ 0.1/20,000, calculated as the tail of the binomial of N trials (i.e. N patients) using p0, when in fact the binomial p=p1. To obtain Figure 5 and Supplementary Figure 9 we found the values of N that yielded 90% power as a function of μ and r.
Supplementary Material
Acknowledgements
This work was conducted as part of TCGA, a project of the National Cancer Institute and the National Human Genome Research Institute. We are grateful to Travis I. Zack, Steven E. Schumacher, and Rameen Beroukhim for sharing their copy-number analyses before publication.
Footnotes
Author Contributions
G.G., E.S.L., T.R.G., M.M., L.A.G., and S.B.G. conceived the project and provided leadership. M.S.L., G.G., P.S., and C.H.M. analysed the data and contributed to scientific discussions. M.S.L., E.S.L., and G.G. wrote the paper.
Accession numbers
The data analyzed in this manuscript has been deposited in Synapse (http://www.synapse.org), accession number syn1729383, and in dbGaP (http://www.ncbi.nlm.nih.gov/gap), accession numbers phs000330.v1.p1, phs000348.v1.p1, phs000369.v1.p1, phs000370.v1.p1, phs000374.v1.p1, phs000435.v2.p1, phs000447.v1.p1, phs000450.v1.p1, phs000452.v1.p1, phs000467.v6.p1, phs000488.v1.p1, phs000504.v1.p1, phs000508.v1.p1, phs000579.v1.p1, phs000598.v1.p1.
The authors declare the following competing financial interest: a patent application has been filed related to this work.
References
- 1.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research, N et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamborero D, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.