Abstract
Embryonic stem cells (ESCs) can give rise to all the differentiated cell types of the organism, including neurons. However, the efficiency and specificity of neural differentiation protocols still needs to be improved in order to plan their use in cell replacement therapies. In this study, we modified a monolayer differentiation protocol by selecting green fluorescent protein (GFP) positive neural precursors with fluorescence-activated cell sorting (FACS). The enhancement of neural differentiation was obtained by positively selecting for neural precursors, while specific neuronal subtypes spontaneously differentiated without additional cues; a comparable but delayed behavior was also observed in the GFP negative population, indicating that sorting settings per se eliminated nonneural and undifferentiated ESCs. This highly reproducible approach could be applied as a strategy to enhance neuronal differentiation and could be the first step toward the selection of pure populations of neurons, to be generated by the administration of specific factors in high throughput screening assays.
Key words: : mESC, FACS, neural differentiation, neurons
Introduction
Embryonic stem cells (ESCs) are well-established tools to recapitulate embryonic development in vitro and have been extensively studied for their potential to give rise to cell types deriving from the three germlayers.1 The acquisition of a neuroectodermal fate is an evolutionary conserved mechanism involving few signaling pathways, whose expression is spatiotemporally regulated and modulated.2 Despite the high complexity of the in vivo development, neural lineages can be easily derived with proper differentiation protocols and with minimal media3 via formation of embryoid bodies4–6 or by using adherent cultures.7,8 One of the major concerns about the differentiation of pure neuronal populations from ESCs is the need to administer growth factors or other compounds to the culture medium, like inhibitors of signaling pathways,9–11 as well as specific supplements.8,12,13 Despite the use of expensive compounds, pure populations of neurons are still difficult to obtain; for this reason, fluorescent knock-in cell lines have been generated starting from ESCs8,14 in order to allow for physical separation of the desired progenitors by fluorescence-activated cell sorting (FACS). In this study, the Sox1-green fluorescent protein (GFP) knock-in cell line 46C8 was differentiated with a simple one-step neural protocol, without supplements and growth factors, modified from existing literature.7 Taking advantage of GFP, fluorescent Sox1–expressing cells were efficiently purified as neural precursors; both positive and negative sorted cells were replated and neuronal differentiation was monitored throughout the protocol, before and after sorting, and compared with the unsorted population. Quantitative reverse-transcription polymerase chain reaction (RT-qPCR) as well as immunocytochemistry (ICC) and Western blotting analyses confirmed neuronal differentiation of GFP positive (GFP+) cells and allowed us to verify the positioning potential along rostrocaudal and dorsoventral axes as well as the ability of purified cells to give rise to several neuronal subtypes. Interestingly, GFP negative (GFP−) cells also showed the acquisition of neural fate, similarly to what we observed with their GFP positive counterpart, but in a delayed manner. Moreover, our data show that differentiating sorted GFP positive cells do not show any specific regionalization and that different neuronal subtypes can be obtained without additional factors. This protocol could thus be considered an initial step toward the generation of highly pure neuronal populations, to be obtained by the addition of specific signals. It could also be used as an efficient cell assay for the high throughput screening of new compounds able to drive differentiation toward specific neuronal subtypes.
Materials and Methods
Embryonic stem cell culture and differentiation
The mouse feeder-independent embryonic stem cells lines 46C (Sox1-GFP knock-in8, kindly provided by A. Smith, University of Cambridge, United Kingdom) and E14Tg2a.415 were grown on gelatin-coated dishes in ESC medium: Glasgow Minimum Essential Medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% ESC-certified fetal calf serum (Millipore, Billerica, MA), 2 mM L-glutamine and 100 U/mL penicillin/streptomycin (Lonza, Basel, Switzerland), 1 mM sodium pyruvate and 0.1 mM nonessential amino acids (Gibco, Life Technologies, Carlsbad, CA), 0.05 mM beta-mercaptoethanol (Sigma-Aldrich) and 103 U/mL LIF (Millipore). Cells were differentiated as previously described,7 with minor adjustments. Briefly, 1000 cells/cm2 were plated on gelatin-coated culture dishes. At day 0, ESC medium was replaced by knockout serum replacement (KSR) medium: knockout Dulbecco's modified Eagle's medium (Gibco) supplemented with 15% (pre-sorting) or 5% (post-sorting) KSR (Invitrogen, Life Technologies, Carlsbad, CA), 2 mM L-glutamine, 100 U/mL penicillin/streptomycin (Lonza), and 0.1 mM beta-mercaptoethanol (Sigma-Aldrich). Medium was replaced every two days until day 13. Two later time points were analyzed for ICC, day 15 and day 20, for GFP− and GFP+ populations respectively. Control unsorted cells were grown in 15% KSR medium throughout the protocol and then collected at day 13 for comparison with sorted samples.
Fluorescence-activated cell sorting
For GFP time course analyses, 46C and negative control E14Tg2a.4 cells were trypsinized, counted, and resuspended in phosphate-buffered saline (PBS)/0.5% bovine serum albumin (BSA)/2mM EDTA at the concentration of 106 cells/mL. GFP positive cells were counted daily from day 0 to day 7 of the differentiation protocol using the FACSCanto BD cytometer (Beckton Dickinson, Franklin Lakes, NJ) and FACSDiva software, and 10,000 events were recorded for each time point from three independent experiments. For sorting analyses, at day 5 cells were trypsinized, counted, and resuspended in PBS/0.02% EDTA (Sigma-Aldrich) at the concentration of 3–5×106 cells/mL. The cell suspension was then passed through a 70 μm preseparation filter (Miltenyi Biotec, Bergisch Gladbach, Germany) and transferred in sterile polypropylene tubes. FACS Aria BD II (Beckton Dickinson) and FACSDiva software were used. A 100-μm nozzle was chosen and proper electronic gates of side scatter and forward scatter parameters were set in order to exclude cell debris and dead cells. The GFP negative control cells E14Tg2a.4 at the same day of differentiation were analyzed in order to assess the minimal fluorescein isothiocyanate (FITC) baseline, which was set at 103 fluorescence units. Sorting was performed in sterile conditions throughout the experiment, with a maximum flow rate of 2000 events/second in order to preserve precision of the purification and cell viability. Sorted cells were recovered in sterile polypropylene tubes containing differentiation medium, counted, transferred to sterile 15-mL Falcon centrifuge tubes, and spun down. Fifty thousand cells per cm2 were plated on gelatin-coated 24-well plates (Corning, Corning, NY) in 5% KSR medium and cultured until day 13.
RNA extraction and RT-qPCR
Cells were collected at different time points, RNA and proteins were extracted from the same samples with a Nucleospin RNA/Protein kit (Macherey-Nagel, Düren, Germany), according to the manufacturer's instructions. Samples were quantified and 1 μg of total RNA was retrotranscribed using Superscript III VILO RT (Invitrogen). RT-qPCR was performed on 5 ng of cDNA, provided in technical triplicates, with KAPA SYBR Green (Resnova, Genzano di Roma, Italy) using a Biorad (Hercules, CA) 384 machine. Samples were amplified for 40 cycles using the following parameters: 95°C for 30 sec, 60°C for 30 sec, and 72°C for 40 sec. Primer sequences are listed in Supplementary Table S1. Results were analyzed with Biorad CFX manager software and cycle threshold (Ct) data from three replicas were normalized on β-actin amplification, used as reference gene. Finally, ΔCt were normalized on control undifferentiated cells at day 0. The resulting ΔΔCt data were merged from three independent differentiation and sorting experiments, and the fold change values were displayed in histograms.
Protein extraction and Western blot analyses
Proteins were extracted using the Nucleospin RNA/Protein kit (Macherey-Nagel) and quantified using the Bicinchoninic acid method (Pierce, Rockford, IL). Thirty micrograms of protein were loaded onto a 10% or 6% denaturing polyacrylamide gel. Gels were run for 1 h at 150 V and proteins were transferred onto a nitrocellulose membrane using the iBlot device (Life Technologies). Membranes were blocked with 5% nonfat dry milk (Santa Cruz Biotechnologies, Dallas, Texas). Primary antibodies were diluted in 2% milk and membranes washed with Tris buffered saline–Tween solution. The following antibodies and dilutions were used: Nestin 1:200 (Millipore); βIII-Tubulin 1:1,000 (Covance, Princeton, NJ); Synaptophysin1 1:1000 (Synaptic Systems, Goettingen, Germany); Post Synaptic Density (PSD)-95 1:2000 (Cell Signaling, Danvers, MA); horseradish peroxidase–conjugated anti-rabbit or anti-mouse 1:10,000 (Jackson ImmunoLab, West Grove, PA). Membranes were incubated with enhanced chemiluminescence (GE Healthcare, Fairfield, CT) and signals were acquired with Chemidoc (Biorad).
Immunocytochemistry
Cells were fixed with paraformaldehyde 4% for 15 min at room temperature. After a wash in PBS, blocking was performed with BSA 5% and 0.01% Triton X-100. The following primary antibodies dilutions were used: anti-Nestin 1:200 (Millipore-Merck); anti-βIII Tubulin 1:1000 (Covance); anti-MAP2 1:200 (Santa Cruz Biotechnologies); anti-glial fibrillary acidic protein 1:800 (Sigma); anti-Synaptophysin 1 (anti-Syn1) 1:200 (Synaptic Systems); and anti-glutamatergic vesicular transporter 2 (anti-VGlut2) 1:500 (Synaptic Systems). Goat anti-mouse or anti-rabbit Alexa Fluor 488 or 594 (Life Technologies) were diluted 1:1000 in blocking solution and used as secondary antibodies. Hoechst 1:10,000 was used to counterstain nuclei, and images were acquired at Zeiss (Oberkochen, Germany) ApoTome.2 microscope with AxioVision 4.8 software.
Statistical analyses
Data in RT-qPCR experiments displaying Sox1-GFP+, Sox1-GFP−, and unsorted cells are represented as the mean of three biological replicas±standard error of the mean (SEM). Statistical significance was assessed with unpaired Student's t-test between Sox1-GFP+ and bulk unsorted populations at day 13; p-value of <0.05 was considered statistically significant.
Results
Sorting of Sox1-GFP cells avoids non neuroectodermal cell fate commitment
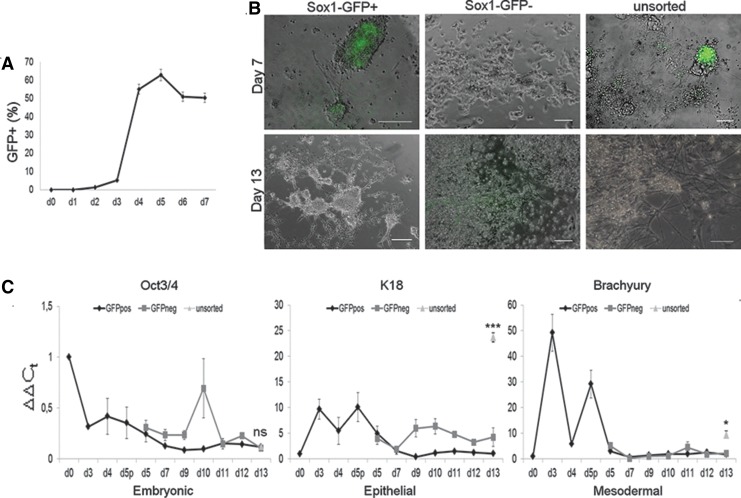
The mouse knock-in embryonic stem cell line 46C Sox1-GFP8 was cultured in monolayer on gelatin-coated dishes with minimal medium containing KSR. As previously shown,7 this medium is able to support neural differentiation of mouse ESCs (mESCs) without additional growth factors or embryoid bodies formation. Transferrin, insulin, and albumin are the only proteins contained in KSR16 and are sufficient to sustain neural survival and proliferation while giving little or no bias toward specific cell identities.17,18 In fact, the absence of other specific growth factors leads to the differentiation of a very heterogeneous population, which expresses mesodermal markers along with neuroectodermal genes.7 In this study, we improved this very simple protocol in order to select neuroectodermal precursors and obtain a purer neural population. We cultured both the Sox1-GFP cells and the E14Tg2a.4 parental ESC line, and we assessed the reproducibility of the protocol between the two cell lines (data not shown). We first performed a cytofluorimetric analysis in order to quantify the number of GFP-expressing cells at different time points (Fig. 1A). In 46C cells cultured with this protocol, the GFP, corresponding to Sox1 expression, started to be expressed around day 3. The analyses showed a highly reproducible increase in GFP positive cells from day 3, reaching a peak at day 5 and then slightly decreasing. We thus decided to perform FACS analysis at day 5. E14 cells at day 5 of differentiation were used as GFP negative control to set the proper FITC baseline (Supplementary Fig. S1A, left panel). The Sox1–GFP positive (GFP+) population was easily recognized and separated: we decided to choose the brighter subpopulation for further analysis, as well as the GFP negative (GFP−) population. Cells with an intermediate amount of GFP expression were discarded in order to avoid the presence of the earliest precursors19 and also to avoid cross-contamination between the two groups (Supplementary Fig. S1A, left panel and black box). GFP+ cells represented about 65% of the total population (Fig. 1A), a slightly lower percentage with respect to other published results, in which the differentiation media were supplemented with signaling pathway antagonists like Dkk1 and Lefty A11 or with neural-specific supplements like N2 and B278; this evidence clearly indicates that the absence of specific growth factors and supplements can impair the more efficient acquisition of a neuroectodermal fate. Finally, we checked for the purity of the sorting procedures by performing a further FACS analysis on the two sorted populations (GFP− and GFP+; Supplementary Fig. S1A, right panel). Different cell densities were tested for replating after sorting, ranging from 2×104 to 25×104 cells/cm2: due to the best output in terms of viability and/or neurogenic potential (Supplementary Fig. S1B), the 5×104 cells/cm2 density was chosen. Along with replating, we decided to make a further improvement to the original protocol by lowering KSR percentage from 15% to 5% in order to increase differentiation.20 GFP was slowly turned off starting from day 7 in both GFP+ and unsorted cells (Fig. 1B, top left and top right respectively). Cells were collected for molecular analyses at day 7, day 9, and then every other day starting from day 10. At day 13, cells showed good viability and a neuronal-like morphology (Fig. 1B, bottom left), with dense neurite outgrowth comparable with that observed in the unsorted population (Fig. 1B, bottom right). For this reason, the protocol was stopped and cells collected or fixed for molecular and immunocytochemical analyses. Interestingly, GFP− cells, which were completely GFP negative at day 7 (Fig. 1B, top middle), started expressing GFP around day 10 and fluorescence peaked at day 13 (Fig. 1B, bottom middle). Total RNA was extracted from all the samples and RT-qPCR analyses were performed in order to assess the differentiation potential of the cells. Unsorted cells collected at day 13 were analyzed in order to provide a direct comparison with the published differentiation protocol. First of all, we assessed whether pluripotent cells and nonneural ectodermal precursors such as epithelial cells were present in our cultures. As shown in Fig. 1C, the expression of Oct3/4 decreases from the third day of the culture; it is maintained at levels lower than undifferentiated cells (day 0) until the end, in both GFP+ (black line) and GFP− (dark gray line) samples, as well as in the unsorted population (light gray indicator), indicating that most of the cells were already committed to differentiate at the time of sorting, regardless of the purification procedure. GFP− cells only showed an increase of Oct3/4 expression at day 10 (d10): this could suggest the presence of undifferentiated ESCs in the GFP− population that, after recovering from the stress of the sorting procedure, are able to undergo intense proliferation, as observed between day 7 and 13 (see Fig.1B, middle panels). Early keratinocyte marker keratin (K)1821 also decreased at day 5 (d5) immediately after sorting. Interestingly, the unsorted population showed a significantly higher expression level of K18, suggesting that the purification step was able to discard nonneural ectoderm. The expression of the mesodermal marker Brachyury,22 which increased at day 5 of differentiation after a significant down-regulation at day 4, was again down-regulated after sorting in both GFP+ and GFP− populations; once again, the unsorted population showed a significant higher expression of this mesodermal marker. This finding indicates that FACS analysis efficiently discarded nonneuroectodermal precursors and also suggests that the mesodermal progenitors could have been contained in the middle population that was excluded by the sorting, since in unsorted cells we also find cells with nonneural morphology (data not shown). We also checked for endodermal markers such as Sox17, but no expression was observed throughout the protocol in both GFP+ and GFP− cells, as well as in the unsorted population, in line with what has been previously described.7 Altogether, this data indicates that sorting was able to discard the non neuroectodermal lineages from both GFP+ and GFP− subpopulations.
FIG. 1.
Differentiation potential of sorted Sox1-green fluorescent protein (Sox1-GFP) mouse embryonic stem cells (mESCs). (A) Time course analysis showing the percentage of Sox1-GFP positive cells during neural differentiation from day 0 (d0) to day 7 (d7). Error bars represent±SEM with n=3 independent experiments. (B) Brightfield pictures of sorted Sox1-GFP+ (left panels), Sox1-GFP− cells (middle panels) and unsorted cells (right panels) at day 7 (top) and at day 13 (bottom). Scale bars, 100 μm. (C) Quantitative reverse-transcription polymerase chain reaction (RT-qPCR) showing the expression of the pluripotency marker Oct3/4, the epithelial marker Keratin (K)18 and the mesodermal marker Brachyury, expressed as ΔΔCt values, in GFP+ (black lines) and GFP− (dark gray lines) from day 0 (d0) to day 13 (d13), and in unsorted cells (light gray indicator) at day 13. Error bars represent±SEM with n=3 independent experiments. d5p, day 5 pre-sorting; ns, not significant. *p<0.05, ***p<0.001. Ct, cycle threshold.
Sorted Sox1-GFP neural precursors undergo neuronal differentiation
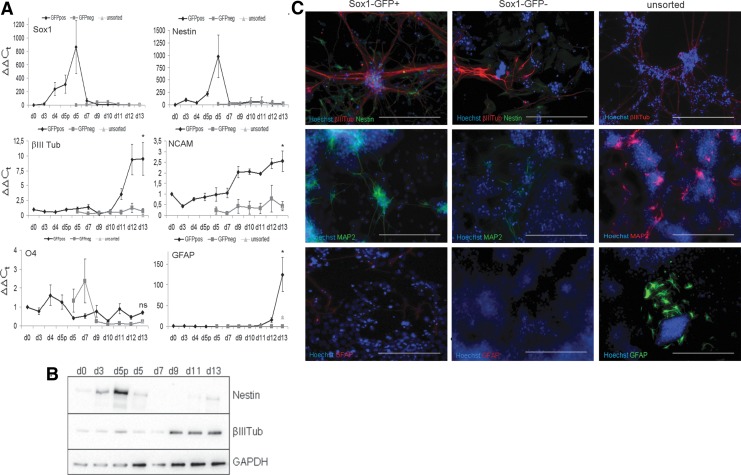
We subsequently assessed the expression of neuroectodermal precursors' markers. Figure 2A shows a RT-qPCR panel in which the extent of neural differentiation was investigated: we analyzed the neural precursors marker Sox1 and Nestin, the neuronal markers βIII Tubulin and neural cell adhesion molecule (NCAM), the glial marker GFAP and the oligodendrocyte marker O4.23 Sox1 expression perfectly paralleled GFP behavior, with a robust increase between days 3 and 4 and a peak at day 5 (d5p); sorting consistently separated Sox1-expressing cells from the Sox1-GFP negative subpopulation (d5, black line and dark gray line respectively). Despite the GFP appearance observed in culture (Fig. 1B), GFP− cells (dark gray line) only showed a very slight increase in Sox1 expression around day 9–10, suggesting that the total amount of Sox1 transcript in the GFP− population is too low with respect to the day 5 sorted GFP+ cells and cannot be properly appreciated on the histograms. However, Sox1 expression completely turned off at the end of the protocol in all three of the populations analyzed (GFP+, GFP−, and unsorted), as expected from the acquisition of a more mature neuronal fate. Nestin expression rapidly increased during the first 5 days of differentiation and was highly upregulated in GFP+ sorted cells (Fig. 2A d5, black line; Supplementary Fig. S2A), in a way comparable to Sox1 behavior. This finding is in line with what previously known about temporal expression of early neural markers24,26 and was also confirmed by Western blot and ICC analyses (Fig. 2B, C and Supplementary Fig. S2A respectively). Interestingly, Nestin expression also slightly increased in the GFP− samples (dark gray lines), further suggesting that GFP− cells are acquiring a neural fate in a delayed fashion (Supplementary Fig. S2A, d7 GFP−). At the same time, the expression of the later marker βIII Tubulin23 and NCAM increased at the end of the protocol in GFP+ cells, and their levels were significantly higher than those of the unsorted cells. ICC analyses performed at day 13 (Fig. 2C) confirmed the expression and localization of βIII Tubulin in all three populations (Sox1-GFP+, Sox1-GFP−, and unsorted). Neuronal terminal differentiation was assessed by MAP2 staining,27 confirming that mature postmitotic neurons were present in Sox1-GFP+ as well as in unsorted cells, to a lower extent. In contrast, GFP− cells did not show a robust expression of these later markers at day 13, as was also confirmed by ICC analysis (Fig. 2C). However, at day 15, only two days later, the GFP− population showed a widespread localization of Nestin and βIII Tubulin proteins, as well as MAP2 expression, thus confirming the delayed acquisition of a neural fate (Supplementary Fig. S2B). Interestingly, βIII Tubulin positive neurites in GFP negative cells show a less regular localization with respect to the GFP+ and unsorted cells (Fig. 2C), where neurites usually appear as radial outgrowths from a group of cell bodies. Glial differentiation was instead verified by GFAP expression. Even though expressed at the end of the protocol only in GFP+ cells (Fig. 2A), which show a significantly higher amount of GFAP transcript with respect to the unsorted population, this glia-specific protein was poorly expressed in culture, as assessed by ICC (Fig. 2C). This observation indicates that day 13 might be too early to assess for glial fate, in line with what is known from literature.23 Moreover, O4 expression also did not show any difference among undifferentiated and differentiating cells, or among GFP+, GFP−, and unsorted populations, suggesting that the oligodendrocyte lineage is not arising at the time points within this protocol. These data demonstrate that sorting followed by an optimized cell density for replating and a simple culture medium modification was able not only to improve the acquisition of neuroectodermal fate with respect to the unsorted population, but also to provide a more robust neuronal differentiation.
FIG. 2.
Neural induction of sorted Sox1-GFP cells. (A) RT-qPCR analyses showing the expression of Sox1, Nestin, βIII Tubulin, NCAM, O4, and GFAP markers during differentiation, expressed as ΔΔCt values, in GFP+ (black lines) and GFP− (dark gray lines) from day 0 (d0) to day 13 (d13) and in unsorted cells (light gray indicator) at day 13. Error bars represent±SEM with n=3 independent experiments. d5p, day 5 pre-sorting; ns, not significant. *p<0.05. (B) Western blot analysis showing the expression of Nestin and βIII-Tubulin during differentiation. GAPDH was used for normalization. (C) Immunocytochemistry showing expression of Nestin, βIII Tubulin, MAP2, and GFAP in Sox1-GFP+ (left), Sox1-GFP− (middle), and unsorted (right) cells at day 13 of differentiation. Hoechst 33342 (Life Technologies) was used to counterstain nuclei. Scale bars, 100 μm.
Differentiated neurons acquire a heterogeneous anteroposterior and dorsoventral positional identity
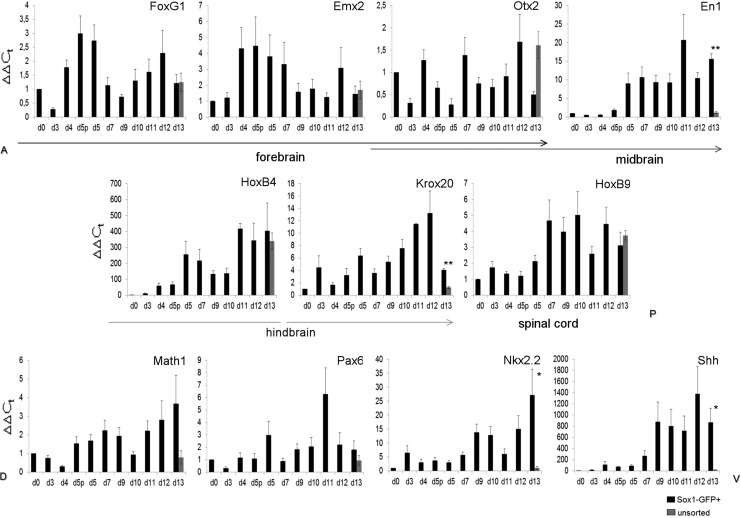
We further investigated the positional identity of the terminally differentiated neurons. Due to the delayed behavior of GFP− cells, we focused our attention only on GFP+ cells. Figure 3 shows the results of RT-qPCR analyses before sorting and on sorted Sox1-GFP+ cells. The expression of forebrain (FoxG1, Emx2), fore-midbrain (Otx2), midbrain (En1), hindbrain (HoxB4, Krox20), and spinal cord (HoxB9) markers along the anteroposterior (A-P) axis of the developing neural tube was assessed. All of the analyzed markers appeared to be expressed with no significant differences with respect to the unsorted population at day 13 (d13, gray bar); the only exceptions were midbrain marker En1 and hindbrain marker Krox20, which were significantly more expressed at the end of the protocol in Sox1-GFP+ cells (d13, black bars). This evidence suggests that despite the positional heterogeneity, a trend toward mid- and hindbrain can be recognized in the sorted population, consistent with recent findings showing that the absence of exogenous signals allow the generation of midbrain neurons.28 Analyzing dorsoventral (D-V) patterning, we observed that sorted cells, upon differentiation, express Math1, Pax6, Nkx2.2, and Shh transcripts.6,7 Interestingly, at the end of the differentiation protocol the expression of dorsal markers Math1 and Pax6 was comparable between the Sox1-GFP+ (black bar) and unsorted (gray bar) populations, while ventral markers Nkx2.2 and Shh were significantly upregulated in sorted cells. These data suggest that the purified population could be committed toward a more ventral fate differentiation, but this hypothesis needs to be further validated. In conclusion, these results confirmed that sorted cells can give rise to a wide range of neuronal types found in vivo along the A-P and D-V axes, including the most anterior part of the developing neural tube. These data also suggest that sorted cells are not biased toward a specific region; interestingly, they can potentially give rise also to the more rostral neurons along the rostrocaudal axis, thus making this improved simple protocol suitable for the differentiation of telencephalic neurons. In summary, sorting purification and medium modifications added to this differentiation protocol did not impair the possibility of obtaining a variety of neuronal cells widely distributed along the neural tube.
FIG. 3.
Positional identity of sorted Sox1-GFP cells. RT-qPCR showing the expression of anteroposterior (A-P) and dorsoventral (D-V) markers during differentiation, expressed as ΔΔCt values, in GFP+ (black lines) and GFP− (dark gray lines) from day 0 (d0) to day 13 (d13), and in unsorted cells (light gray indicator) at day 13. d5p, day 5 pre-sorting. Error bars represent±SEM with n=3 independent experiments. *p<0.05, **p value<0.01.
Differentiated neurons widely express neuronal subtype markers
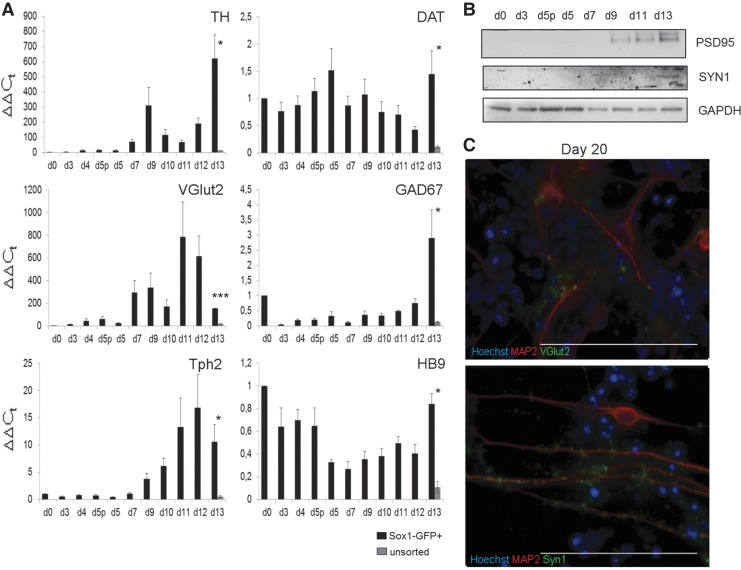
We finally checked for the presence of specific neuronal populations among the differentiated cells before sorting and in the sorted Sox1-GFP+ population. The early tyrosine hydroxylase (TH) and later dopamine transporter (DAT) dopaminergic markers,29 the VGlut2),30 the GABAergic biosynthetic enzyme GAD67,31 the serotonergic tryptophan hydroxylase 2 (Tph2)32 and the motor neuron marker HB933 were analyzed by RT-qPCR (Fig. 4A). First of all we compared their expression between the Sox1-GFP+ (black bar) and unsorted (gray bar) populations at the end of the protocol. All markers were expressed and significantly upregulated in the purified cells with respect to the unsorted population. In particular, TH, VGlut2, and Tph2 showed the most robust up-regulation with respect to undifferentiated cells and after sorting. Specifically, most dopaminergic and serotonergic neurons are born in the midbrain and hindbrain respectively,34 so that these data could be consistent with the observed trend toward mid- and hindbrain (see Fig. 3). They also suggest that the corresponding neuronal subtypes could be enriched in our cultures; moreover, the absence of upregulation of DAT might signify that cells are still in an early phase of terminal differentiation, as DAT is considered to be a late marker of mature dopaminergic neurons.35 Finally, to analyze the extent of differentiation, we checked for the presence of the synaptic proteins (Syn136 and PSD,9537 which are a pre- and post-synaptic marker respectively. Western blot analyses confirmed their expression (Fig. 4B) at the latest time points of the differentiation protocol. However, it was difficult to observe their localization by ICC at day 13, suggesting that the formation of functional synapses is undergoing but not yet terminated. For this reason, we decided to perform ICC analysis at day 20. At this later time point, we observed the localization of Syn1 in a spotted fashion along neurites (Fig. 4C, upper panel). A similar pattern of expression was also observed for the vesicular protein VGlut2 (Fig. 4C, lower panel), confirming that glutamatergic neurons were indeed present. From these observations, we can thus conclude that our modifications to the differentiation protocol can potentially allow a more efficient differentiation toward specific neuronal subtypes whose markers were otherwise very poorly expressed in the unsorted cells.
FIG. 4.
Neuronal subtype specification of sorted cells. (A) RT-qPCR showing the expression of markers for dopaminergic (TH and DAT), glutamatergic (VGlut2), GABAergic (GAD67), serotoninergic (Tph2), and motor (HB9) neuronal subtypes, expressed as ΔΔCt values, in GFP+ (black bars) from day 0 to day 13, and in unsorted cells (gray bar) at day 13. Error bars represent±SEM with n=3 independent experiments. d5p, day 5 pre-sorting. *p<0.05, ***p<0.001. (B) Western blot analysis showing the expression of the presynaptic marker Synaptophysin 1 (SYN1) and the post-synaptic marker Post Synaptic Density (PSD) 95. GAPDH was used to normalize signals. (C) Immunocytochemistry showing expression of MAP2 and VGlut2 (upper panel) and MAP2 and SYN1 (lower panel) in Sox1-GFP+ cells at day 20. Hoechst 33342 (Life Technologies) was used to counterstain nuclei. Scale bar, 50 μm.
Discussion
Sorting of Sox1-GFP is an efficient yet established method used to improve the purification of neural precursors;8,11,19 however, it is usually associated to the administration of growth factors and supplements, expensive and possibly difficult to modulate. Thus, obtaining pure neuronal precursors without the expensive support of exogenous factors appears to be an appealing and efficient system, which can subsequently lead to the differentiation of pure neuronal populations, to be used for cell replacement therapeutic approaches. In the present study we show the setup of cell sorting by FACS to improve a very simple one-step neural differentiation protocol, suitable for high throughput screenings. In fact, we were able to select neural cells, while nonneuroectodermal lineages were efficiently discarded by the sorting procedure. We also demonstrated that the combination between the optimal cell density for replating and a simple culture medium modification, obtained by lowering KSR concentration, was able not only to improve the acquisition of a neuroectodermal fate with respect to the unsorted population, but also to provide a more robust neuronal differentiation. The replated cells showed the upregulation of several neuronal subtypes markers, widely positioned along the rostro-caudal and dorso-ventral axes of the developing neural tube. In particular, the neurons we obtained did not display any robust specific positional identity, showing only a trend toward mid-hindbrain, which is not unexpected due to the lack of specific growth factors in our culture.28 In particular, glutamatergic, serotoninergic, and GABAergic neurons markers were upregulated and, at the latest time point analyzed, synaptic markers were also shown to be expressed, indicating that the purified cells were able to undergo terminal differentiation. Interestingly, the Sox1-GFP negative population that was chosen in order to avoid the cells displaying low to intermediate amounts of GFP was also able to undergo neuronal differentiation. This finding can be discussed in light of some published results demonstrating the presence of lateral induction during in vitro monolayer neural differentiation of mESCs.38 In this paper, the authors show that Notch signaling, while promoting neural induction, is also able to inhibit nonneural differentiation. Although the analysis of Notch signaling was beyond the scope of this work, we speculated that in this culture system, the earliest Sox1-GFP positive population was able to activate the neuroectodermal commitment in part of the neighboring GFP negative cells while probably inhibiting the differentiation towards other lineages. In addition to this, our FACS settings efficiently discarded the nonneural cells, thus improving the less efficient original protocol. In conclusion, we designed a culture system able to generate an almost pure population of neural precursors. We demonstrated that sorting purification and modifications added to this differentiation protocol did not impair the possibility to obtain a variety of neuronal cells, widely distributed along the neural tube. This system displays a strong applicative potential, as the introduction in the culture of growth factors or small molecules could allow for the generation of highly pure specific neuronal populations. Moreover, it could be used in high throughput screening to select for new compounds able to drive differentiation toward specific neuronal subtypes.
Supplementary Material
Abbreviations Used
- A-P
Anteroposterior
- BSA
Bovine serum albumin
- Ct
cycle threshold
- DAT
Dopamine transporter
- D-V
Dorsoventral
- ESC
Embryonic stem cell
- FACS
Fluorescence-activated cell sorting
- FITC
Fluorescein isothiocyanate
- GAD67
GABAergic neuronal subtype
- GFAP
glial fibrillary acidic protein
- GFP
Sox1-green fluorescent protein
- ICC
Immunocytochemistry
- KSR
Knockout serum replacement
- NCAM
neural cell adhesion molecule
- PBS
Phosphate-buffered saline
- PSD
Post synaptic density
- RT-qPCR
Quantitative reverse-transcription polymerase chain reaction
- Syn1
Synaptophysin 1
- TH
Tyrosine hydroxylase
- Tph2
Tryptophan hydroxylase 2
- VGlut2
Glutamatergic vesicular transporter 2
Author Disclosure Statement
No competing financial interests exist.
Acknowledgments
The authors acknowledge National Heart Lung and Blood Institute-BayGenomics and National Center for Research Resources-Mutant Mouse Regional Resource Center (University of California, Davis) for the E14Tg2A cell line and Austin Smith for the Sox1-GFP 46C cells. The authors would like to thank Isabella Pesce for help with FACS analyses and Patrizia Paoli and Giorgia Moser for administrative support. The authors are grateful to Angela Bozza for helpful discussions and Luciano Conti for helpful discussions and critical reading of the manuscript. This work was supported by a University of Trento Startup Grant (S.C. and Y.B.) and by Cassa di Risparmio di Trento e Rovereto Grant n. 2011.0251 (S.C.).
References
- 1.Keller G.Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155 [DOI] [PubMed] [Google Scholar]
- 2.Petros TJ, Tyson JA, Anderson SA. Pluripotent stem cells for the study of CNS development. Front Mol Neurosci. 2011;4:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tropepe V, Hitoshi S, Sirard C, et al. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001; 30:65–78 [DOI] [PubMed] [Google Scholar]
- 4.Bibel M, Richter J, Schrenk K, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nature Neurosci. 2004;7:1003–1009 [DOI] [PubMed] [Google Scholar]
- 5.Gaspard N, Bouschet T, Hourez R, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357 [DOI] [PubMed] [Google Scholar]
- 6.Okabe S, Forsberg-Nilsson K, Spiro C, et al. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev 2000; 59: 124–142 [DOI] [PubMed] [Google Scholar]
- 7.Fico A, Manganelli G, Simeone M, et al. High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cells Dev. 2008;17:573–584 [DOI] [PubMed] [Google Scholar]
- 8.Ying Q, Stavridis M, Griffiths D, et al. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nature Biotechnol. 2003;2:183–186 [DOI] [PubMed] [Google Scholar]
- 9.Chambers SM, Fasano C, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnol. 2009;27:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan K, Chang H, Rolletschek A, et al. Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell Tissue Res. 2001;305:171–176 [DOI] [PubMed] [Google Scholar]
- 11.Watanabe K, Kamiya D, Nishiyama A, et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nature Neurosci. 2005;8:288–296 [DOI] [PubMed] [Google Scholar]
- 12.Lee SH, Lumelsky N, Studer L, et al. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nature Biotechnol 2000;18:675–679 [DOI] [PubMed] [Google Scholar]
- 13.Yamazoe H, Kobori M, Murakami Y, et al. One-step induction of neurons from mouse embryonic stem cells in serum-free media containing vitamin B12 and heparin. Cell Transplant. 2006;15:135–145 [DOI] [PubMed] [Google Scholar]
- 14.Wataya T, Ando S, Muguruma K, et al. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc Natl Acad Sci USA. 2008;105:11796–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper M, Hardy K, Handyside A, et al. HPRT-deficient (Lesch–Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295 [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Gonzalo F, Izpisùa-Belmonte JC. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PloS One. 2008;1:e1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dìaz B, Pimentel B, de Pablo F, et al. Apoptotic cell death of proliferating neuroepithelial cells in the embryonic retina is prevented by insulin. European J Neurosci. 1999;11:1624–1632 [DOI] [PubMed] [Google Scholar]
- 18.Erickson RI, Paucar AA, Jackson RL, et al. Role of insulin and transferrin in neural progenitor survival and proliferation. J Neurosci Res. 2008;86:1884–1894 [DOI] [PubMed] [Google Scholar]
- 19.Abranches E, Silva M, Pradier L, et al. Neural differentiation of embryonic stem cells in vitro: a roadmap to neurogenesis in the embryo. PLoS One. 2009;4:e6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhry M, Vitalis TZ, Bowen BD, et al. Basal medium composition and serum or serum replacement concentration influences on the maintenance of murine embryonic stem cells. Cytotechnology. 2008;58:173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalom-Feuerstein R, Lena AM, Zhou H, et al. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2010;18:887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada Y, Shimazaki T, Sobue G, et al. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev Biol. 2004;275:124–142 [DOI] [PubMed] [Google Scholar]
- 23.Qian X, Shen Q, Goderie SK, et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 2000;28:69–80 [DOI] [PubMed] [Google Scholar]
- 24.Aubert J, Stavridis MP, Tweedie S, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1–gfp knock-in mice. Proc Natl Acad Sci USA. 2003;100:11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pevny LH, Sockanathan S, Placzek M.et al. A role for SOX1 in neural determination. Development. 2000;125:1967–1978 [DOI] [PubMed] [Google Scholar]
- 26.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201 [DOI] [PubMed] [Google Scholar]
- 27.De Camilli P, Miller PE, Navone F, et al. Distribution of microtubule-associated protein 2 in the nervous system of the rat studied by immunofluorescence. Neuroscience. 1984;4:817–846 [PubMed] [Google Scholar]
- 28.Bertacchi M, Pandolfini L, Murenu E, et al. The positional identity of mouse ES cell-generated neurons is affected by BMP signaling. Cell Mol Life Sci. 2013;70:1095–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallén A, Perlman T. Transcriptional control of dopamine neuron development. Ann N Y Acad Sci. 2003;991:48–60 [DOI] [PubMed] [Google Scholar]
- 30.Takamori S, Rhee JS, Rosenmund C, et al. Identification of differentiation-associated brain-specific phosphate transporter as a second vesicular glutamate transporter (VGLUT2). J Neurosci. 2001;21:RC182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin DL, Rimvall K. Regulation of gamma-aminobutyric acid synthesis in the brain. J Neurochem. 1993;60:395–407 [DOI] [PubMed] [Google Scholar]
- 32.Håkanson R, Lombard des Gouttes MN, Owman C. Activities of tryptophan hydroxylase, dopa decarboxylase, and monoamine oxidase as correlated with the appearance of monoamines in developing rat pineal gland. Life Sci. 1967;6:2577–2585 [DOI] [PubMed] [Google Scholar]
- 33.Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell 1998;95:67–80 [DOI] [PubMed] [Google Scholar]
- 34.Hynes M, Rosenthal A. Specification of dopaminergic and serotonergic neurons in the vertebrate CNS. Curr Opin Neurobiol. 1999;9:26–36 [DOI] [PubMed] [Google Scholar]
- 35.Tan XF, Jin GH, Tian ML, et al. The co-transduction of Nurr1 and Brn4 genes induces the differentiation of neural stem cells into dopaminergic neurons. Cell Biol Int. 1998;351217–351223 [DOI] [PubMed] [Google Scholar]
- 36.Südhof TC, Lottspeich F, Greengard P, et al. A synaptic vesicle protein with a novel cytoplasmic domain and four transmembrane regions. Science 1987;238:1142–1144 [DOI] [PubMed] [Google Scholar]
- 37.Cho KM, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 1992;9:929–942 [DOI] [PubMed] [Google Scholar]
- 38.Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.