Abstract
Mast cells orchestrate the allergic response through the release of pro-inflammatory mediators, which is driven by the fusion of cytoplasmic secretory granules with the plasma membrane. During this process, SNARE proteins including Syntaxin4, SNAP23 and VAMP8 play a key role. Following stimulation, the kinase IKKβ interacts with and phosphorylates the t-SNARE SNAP23. Phosphorylated SNAP23 then associates with Syntaxin4 and the v-SNARE VAMP8 to form a ternary SNARE complex, which drives membrane fusion and mediator release. Interestingly, mast cell degranulation is impaired following exposure to bacteria such as Escherichia coli (E. coli). However, the molecular mechanism(s) by which this occurs is unknown. Here, we show that E. coli exposure rapidly and additively inhibits degranulation in the RBL-2H3 rat mast cell line. Following co-culture with E. coli, the interaction between IKKβ and SNAP23 is disrupted, resulting in the hypophosphorylation of SNAP23. Subsequent formation of the ternary SNARE complex between SNAP23, Syntaxin4 and VAMP8 is strongly reduced. Collectively, these results demonstrate that E. coli exposure inhibits the formation of VAMP8-containing exocytic SNARE complexes and thus the release of VAMP8-dependent granules by interfering with SNAP23 phosphorylation.
Keywords: Mast cell, SNAREs, degranulation, bacterial infection, Escherichia coli, SNAP23, Syntaxin4, VAMP8, intracellular trafficking, membrane fusion
Introduction
Microbiota play a major role in regulating immune responses (1, 2). Interestingly, changes in microbiota have been observed in patients with allergic diseases (3, 4). Moreover, allergic inflammation is exacerbated in germ-free mice suggesting that presence of bacteria may influence the allergic response, during which mast cells play a central role. (5).
Mast cells are localized at the host-environment interface in connective and mucosal tissues, in particular the gut where the prevalence of bacteria is high (6, 7). During the first exposure to an allergen, IgE is produced and binds its high affinity receptor, FcεRI, on the surface of mast cells. As the allergen is reintroduced, it binds directly to IgE on the surface of mast cells, thus cross-linking FcεRI. FcεRI-mediated activation results in the rapid release of proinflammatory mediators including histamine and proteases as well as the production of cytokines (6, 8, 9). Based on their localization in microbe-rich tissues, mast cells may be a crucial link to understanding how bacterial exposure or stimulation influences the development of allergic inflammation.
Several studies have established that bacteria differentially regulate the secretion of mast cell-derived mediators (10). While some bacteria such as Mycobacterium tuberculosis (11), Mycoplasma pneumonia (12) and Streptococcus pneumonia (13) activate mast cells to secrete cytokines and induce degranulation, other bacteria such as probiotics (14-17) and E. coli inhibit degranulation in human and mouse mast cells (18, 19). Although E. coli exposure reduces serotonin and β-hexosaminidase secretion, it induces the release of histamine in mouse models (20, 21). Thus, bacterial exposure may play a regulatory role by which certain bacteria selectively modulate the hyper-reactivity of mast cells to circulating allergen. However, the molecular mechanisms involved in this phenomenon are unclear.
Mast cell degranulation is largely mediated by the exocytic SNARE proteins. In addition to the t-SNAREs SNAP23 and Syntaxin4, several v-SNAREs have been implicated in this process including VAMP2, VAMP8 and VAMP7. However, their function is dependent on mast cell subsets and types of granules (22-25). Data suggests that VAMP8 regulates the release of a subset of secretory granules in rodent mast cells, where VAMP8 significantly colocalizes with serotonin and cathepsin D, but is absent from histamine-containing granules (26, 27). Additionally, while bone marrow-derived mast cells generated from VAMP8-deficient mice have profound defects in β-hexosaminidase, serotonin, and cathepsin D release, they exhibit no defect in histamine or TNFα secretion (26). Although VAMP2 interacts with Syntaxin4 and SNAP23 in a stimulus-dependent manner, a functional role for this particular v-SNARE in mediator release has yet to be determined (23, 28, 29). In contrast, both VAMP8 and VAMP7 are required for degranulation in cord blood-derived human mast cells (25).
Here, we demonstrate that co-culturing mast cells with E. coli induces a profound decrease in SNAP23 phosphorylation and ternary SNARE complex assembly, both of which are required for exocytosis, resulting in the inhibition of FcεRI-dependent degranulation.
Results
Translationally active E. coli rapidly and additively inhibit RBL-2H3 mast cell degranulation
We investigated the impact of E. coli exposure in the RBL-2H3 (RBL) rat mast cell line, a commonly used model to study the mechanisms of mast cell function (27, 28, 30).
First, we determined whether E. coli interfered with RBL degranulation and which multiplicity of infection (MOI) would induce the optimal effect. RBLs were co-cultured with increasing MOIs of E. coli for 2h. Then the kinetics of β-hexosaminidase secretion was assessed for anti-DNP IgE sensitized RBLs stimulated with DNP-BSA. For comparative purposes, the amount of β-hexosaminidase released at 60min in the control population was considered 100%. As shown in Figure S1A, E. coli inhibits FcεRI-mediated β-hexosaminidase release in a dose-dependent manner. A significant effect is observed at an MOI of 1,000 and becomes maximal at an MOI of 10,000. This result is consistent with the effect of a single co-culture with E. coli observed in mouse mast cell lines and in primary peritoneal mast cells (18). To further test E. coli-induced inhibition, we bypassed FcεRI and directly stimulated RBLs using phorbol myristate acetate (PMA) and ionomycin. PMA activates Protein Kinase C (PKC), while ionomycin releases the intracellular calcium pool, which are two critical signals for mast cell degranulation (31). In these conditions, RBL degranulation is still inhibited following a single exposure, with significant inhibition only achieved at an MOI of 10,000 (Fig S1B). This could be due to the fact that PMA/Ionomycin is a potent stimulant and small differences may be undetectable. Consistent with this possibility, FcεRI-mediated stimulation results in ~35% secretion at 60min in control cells, while PMA/Ionomycin stimulation induces ~78% total β-hexosaminidase release (data not shown). Alternatively, an FcεRI-dependent signaling component may be impacted by E. coli exposure as PMA/Ionomycin bypasses these proximal signaling events. The reduction in β-hexosaminidase secretion was not due to differences in the total intracellular pool of β-hexosaminidase between control cells and those incubated with E. coli (Fig S1A, inset), suggesting that E. coli did not induce mast cell degranulation during the exposure.
Interestingly, we observed that only translationally active E. coli is able to inhibit secretion (Fig S1C, left and center panels). However, E. coli-conditioned medium alone does not inhibit degranulation (Fig S1C, right panel). These data suggest at least two possibilities: 1) the inhibition of mast cell degranulation requires E. coli to be translationally active and any bacterial factor(s) responsible for the effect are locally secreted, probably in close contact with mast cells, and/or 2) a membrane factor is responsible for the inhibition, but is denatured during the heat-inactivation process. Further investigation will be required to distinguish between these possibilities.
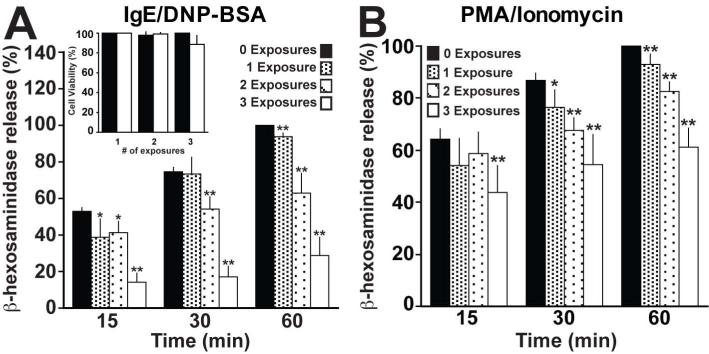
Next, we examined the extent to which multiple E. coli exposures would affect degranulation. RBLs were incubated with E. coli at an MOI of 10,000 for 2h either once, twice or three times, with each incubation occurring 24h apart. The MOI of 10,000 was chosen because it induced the maximal effect (Fig S1A & B). The cells were then stimulated with either anti-DNP IgE/DNP-BSA or PMA/ionomycin. In these conditions, we observed an exposure-dependent inhibition of β-hexosaminidase release (Fig 1A & B). After 60min of stimulation with IgE/DNP-BSA, one exposure results in ~7% inhibition, while two exposures lead to a decrease of ~37% and three exposures to a decrease of ~71%. A similar decrease is observed in cells stimulated with PMA/ionomycin. This inhibition is not due to reduced cell viability as shown in Figure 1A (inset).
Figure 1. E. coli exposures additively inhibits mast cell degranulation.
(A) 5×105 RBL-2H3 cells were exposed to E. coli multiple times for 2h at an MOI of 10,000. Each exposure occurred 24h apart. The cells were then transferred to 96 well plates at a density of 5×104 viable cells per well. Following sensitization with 100ng/ml anti-DNP IgE for 2h, the cells were stimulated with 100ng/ml DNP-BSA for the indicated times. Cell viability prior to stimulation was assessed by trypan blue exclusion (inset). (B) RBLs were treated as in A, but stimulated with 1μM PMA/1μM Ionomycin. The amount of β-hexosaminidase released at 60min in the control population was considered 100%. Graphs depict the mean ± the standard deviation of 5 independent experiments conducted in duplicate. In all graphs, asterisks denote statistically significant p values, where * ≤ 0.05 and ** ≤ 0.01.
Collectively, these results demonstrate that not only does co-culture with E. coli rapidly inhibit mast cell degranulation, but also that repetitive exposures enhance the inhibitory effect. This is consistent with the impact of E. coli on peritoneal mast cells, in that it takes at least 5 days to regain normal levels of degranulation following a single co-culture with E. coli (18).
FcεRI surface expression is not affected by E. coli exposure
One possible explanation for the reduction in FcεRI-mediated degranulation in mast cells co-cultured with E. coli is changes in the surface levels of FcεRI. In fact, using human mast cells, Kulka et al observed a significant reduction in FcεRI surface levels following overnight incubation with E. coli (19). To test this possibility, FcεRI surface levels in RBLs cultured in the presence or absence of E. coli for 2h were examined using immunofluorescence microscopy. As shown in Figure S2A, the surface expression of FcεRI is similar in RBLs cultured with or without E. coli. This observation was confirmed using flow cytometry. Irrespective of being exposed once (Fig S2B, left panel) or thrice (Fig S2B, right panel) to E. coli, FcεRI expression remains constant. These data suggest that the inhibition of FcεRI-mediated degranulation following incubation with E. coli is not due to changes in the surface expression of FcεRI. These observations are consistent with the fact that RBLs exposed to E. coli still secrete less when stimulated by PMA/ionomycin (Fig S1B). Under certain conditions however, FcεRI surface expression has been shown to be reduced (19, 32, 33). This discrepancy may be due to differences in the exposure conditions as reductions in FcεRI expression are reported when mast cells are co-cultured for 4h or longer with bacteria (19, 32, 33).
E. coli exposure impacts exocytic membrane fusion in mast cells
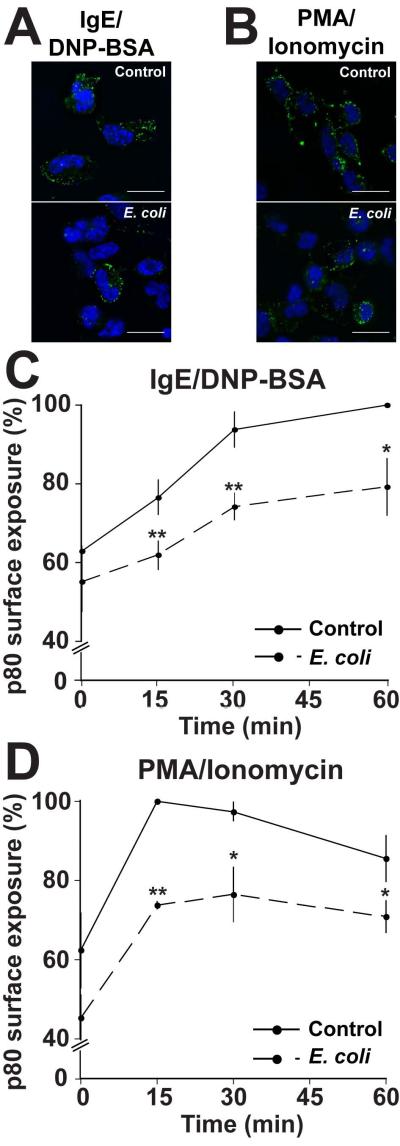
Given that equivalent pools of β-hexosaminidase are available for release, yet secretion is reduced following incubation with E. coli (Fig S1A, inset), granule-plasma membrane fusion is likely compromised. This event constitutes the final step in the degranulation process, and blocking it will result in reduced secretion. To test this hypothesis, we cultured RBLs with or without E. coli for 2h, prior to stimulation with anti-DNP IgE/DNP-BSA or PMA/Ionomycin for 60min. Next, the presence of the protein p80 on the cell surface was measured by immunofluorescence microscopy. p80 is an integral membrane protein found in the lumen of mast cell secretory granules (34), which becomes available on the cell surface upon stimulation. The presence of p80 on the cell surface has been shown to correlate with degranulation in RBL-2H3 mast cells (34, 35). In control cells stimulated by either IgE/DNP-BSA (Fig 2A, top panel) or PMA/Ionomycin (Fig 2B, top panel), p80 was observed as uniform spots on the cell surface. In contrast, when RBLs were co-cultured with E. coli, a noticeable reduction in p80 surface levels was observed (Fig 2A and B, lower panels). Not only is the amount of p80 per cell reduced, but also fewer cells were labeled with p80. To quantify the levels of p80, we analyzed p80 surface expression by flow cytometry. In unstimulated cells cultured in the absence (control) or presence of E. coli, low levels of p80 surface expression were recorded, likely due to constitutive exocytosis (Fig 2C and D, t=0) (34). Note that at t=0, lower levels of spontaneous release are observed in E. coli-treated cells compared with the control suggesting that E. coli also interferes with constitutive secretion of β-hexosaminidase in mast cells. Upon stimulation, we observed a significant increase in p80 surface expression in control cells using both anti-DNP IgE/DNP-BSA or PMA/Ionomycin stimuli (Fig 2C and D, solid lines). In cells co-cultured with E. coli, although stimulation induces the appearance of p80 on the surface, we observed a rapid and significant reduction in p80 levels compared with the non-exposed control cells. At t=15min in E. coli-exposed cells stimulated with IgE/DNP-BSA and PMA/Ionomycin, p80 surface levels were reduced by 15% and 26%, respectively (Fig 2C and D, dashed lines). These data suggest that membrane fusion is inhibited in mast cells co-cultured with E. coli.
Figure 2. E. coli exposure impairs membrane fusion during degranulation.
(A) RBLs were plated on coverslips at a density of 5×104 cells/coverslip 24hr prior to being incubated with (lower panel) or without (top panel-control) E. coli at an MOI of 10,000 for 2h. The cells were then sensitized with 100ng/ml anti-DNP IgE and stimulated with 100ng/ml DNP-BSA for 1h. Cells were fixed and stained with anti-p80 and anti-mouse IgG1 AlexaFluor488-conjugated antibodies. Nuclei were stained with Hoechst. Coverslips were analyzed by immunofluorescence microscopy. Scale bars = 20μm. Representative of 3 independent experiments. (B) RBLs were treated as described in A, but stimulated with 1μM PMA/1μM Ionomycin. Scale bars = 20μm. Representative of 3 independent experiments. (C) 4×106 RBLs were plated in 10cm tissue culture plates 24h prior to being incubated with or without E. coli at an MOI of 10,000 for 2h. The cells were harvested, sensitized with 800ng/ml anti-DNP IgE at a concentration of 2×106 cells/ml and stimulated with 800ng/ml DNP-BSA for 0, 15, 30 or 60min. The cells were fixed and labeled as described in A then analyzed by flow cytometry. (D) RBLs were treated as described in C, but stimulated with 1μM PMA/1μM Ionomycin. Graphs depict the mean fluorescence intensity of p80 staining ± the standard deviation of 3 independent experiments. The maximal p80 fluorescence intensity in the control population was considered 100% (t=60 for IgE/DNP-BSA and t=15min for PMA/Ionomycin). In all graphs, asterisks denote statistically significant p values, where * ≤ 0.05 and ** ≤ 0.01.
E. coli exposure specifically influences the function of the exocytic SNARE SNAP23
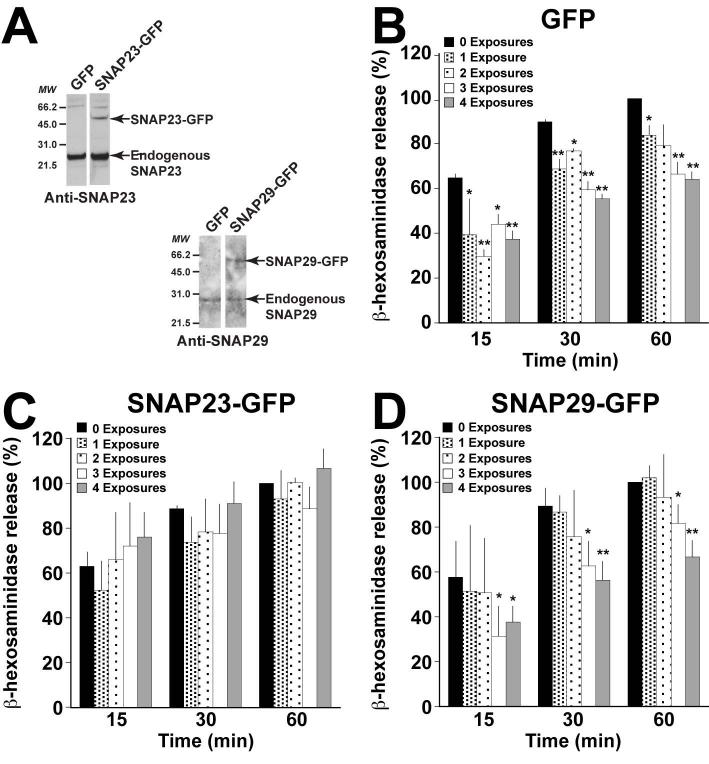
Since incubating mast cells with E. coli results in a reduction of membrane fusion, we anticipate that the exocytic SNARE machinery is affected in this process. The mast cell secretion machinery is composed of Syntaxin4, SNAP23 and VAMP8 in which SNAP23 is a critical component (23, 29, 36, 37). One way to inhibit membrane fusion is by reducing the amount of functional exocytic SNAREs, in particular the amount of functional SNAP23 (28, 29). To assess this possibility, we generated stable RBL clones overexpressing SNAP23-GFP and tested whether this overexpression was able to reverse the secretion phenotype caused by E. coli exposure. As a control, we overexpressed GFP alone. The overexpression of SNAP23-GFP was confirmed by Western blot (Fig 3A). The impact of E. coli on β-hexosaminidase release in these stable RBL populations was assessed following multiple exposures and stimulation with PMA/Ionomycin, as described above. For comparative purposes, the amount of β-hexosaminidase released after 60min of stimulation for each unexposed population was considered as 100%. Similar to non-transfected RBLs (Fig 1B), E. coli co-culture additively inhibits β-hexosaminidase secretion in the GFP control population (Fig 3B). At 60min, one exposure leads to ~17% inhibition, while three and four exposures inhibit ~34% and 37%, respectively. This amount of inhibition is comparable to non-transfected RBLs exposed three times with E. coli (~39% inhibition at 60min). Interestingly, degranulation in cells overexpressing SNAP23-GFP is not significantly impaired by incubation with E. coli (Fig 3C). Even with four exposures, we were unable to detect a difference in β-hexosaminidase release between SNAP23-GFP expressing cells cultured with or without E. coli (107% versus 100%). To confirm that this phenotype was specific to the exocytic SNARE SNAP23, we measured the impact of E. coli co-culture on degranulation in SNAP29-GFP expressing cells (Fig 3A). Previously, we have shown that SNAP29, a homolog of SNAP23, is not involved in mast cell exocytosis (38), and therefore is an appropriate control for SNAP23. When SNAP29-GFP expressing cells are incubated with E. coli, degranulation is inhibited in an exposure-dependent manner (Fig 3D). After 60min of stimulation, four exposures to E. coli reduces β-hexosaminidase secretion by ~35%, similar to the GFP control. These data suggest that E. coli co-culture with mast cells specifically impacts the exocytic SNARE SNAP23.
Figure 3. Overexpression of SNAP23 compensates for the inhibitory effect of E. coli exposure.
(A) Stable populations of RBL-2H3 cells overexpressing GFP, SNAP23-GFP or SNAP29-GFP were generated. The expression of SNAP23-GFP and SNAP29-GFP were analyzed by Western blot using anti-SNAP23 and anti-SNAP29 antibodies. In each population, two bands were detected for SNAP23 and SNAP29, corresponding to the endogenous protein (lower band) and the GFP-fusion protein (upper band). Stable populations expressing GFP (B), SNAP23-GFP (C) and SNAP29-GFP (D) were cultured with or without E. coli, as described in Fig 1A, either 1, 2, 3, or 4 times with each exposure occurring 24h apart. After each exposure, 5×104 viable cells were plated in 96 well plates in a volume of 100μl per well and allowed to recover for 2h at 37°C. The cells were then stimulated with 1μM PMA/1μM Ionomycin for the indicated times and β-hexosaminidase release was quantified. The amount of β-hexosaminidase released at 60min in each unexposed population was considered 100%. Graphs represent the mean ± the standard deviation of 5 independent experiments conducted in duplicate. In all graphs, asterisks denote statistically significant p values, where * ≤ 0.05 and ** ≤ 0.01.
E. coli exposure impairs SNAP23 phosphorylation during stimulation without affecting its localization in lipid rafts
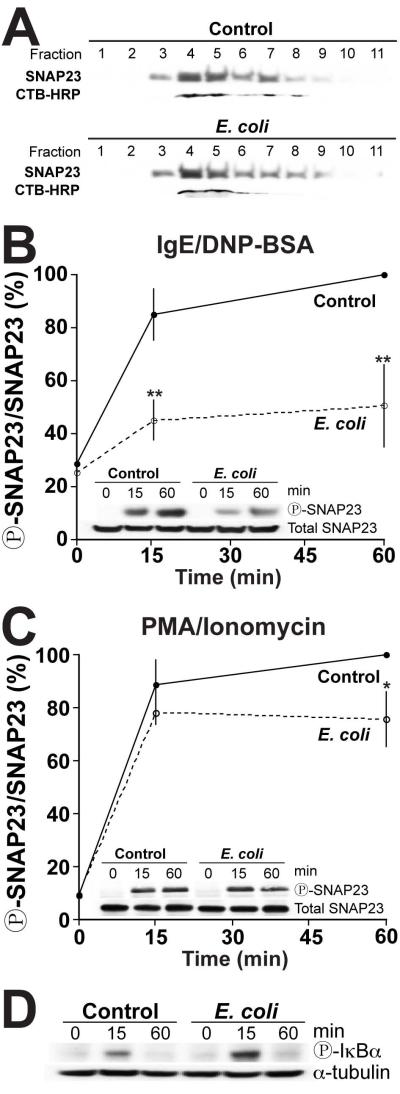
SNAP23 is significantly localized in lipid rafts within the plasma membrane, where it triggers the association with Syntaxin 4 to mediate membrane fusion (23, 29). Thus, we assessed whether the reduced degranulation was caused by changes in the association of SNAP23 with these particular membrane sub-domains. To do so, lipid rafts were isolated by density centrifugation. Lipid rafts were identified by CTB-HRP labeling which specifically localizes to these domains (23, 29). As seen in Figure 4A, we did not observe any changes in the association of SNAP23 with lipid rafts. SNAP23 is enriched in fractions 4 and 5 with CTB-HRP in both control and E. coli-treated cells, suggesting that the inhibition of mast cell degranulation is not due to the mis-localization of SNAP23.
Figure 4. SNAP23 is hypo-phosphorylated in E. coli exposed mast cells, while IKKβ remains functional.
(A) RBL-2H3 cells cultured with or without (control) E. coli for 2h, followed by stimulation in suspension with 1μM PMA/1μM Ionomycin for 30min. Lipid rafts were isolated by density centrifugation as described in the Methods. 20μl of each fraction was analyzed by Western blot using anti-SNAP23 antibody. Lipid rafts were identified using cholera toxin subunit B-HRP (CTB). Representative of 3 independent experiments. (B and C) 2×106 RBLs/ml cultured with or without (control) E. coli at an MOI of 10,000 for 2h. The cells were harvested and stimulated with 800ng/ml anti-DNP IgE and 800ng/ml DNP-BSA (B) or with 1μM PMA/1μM Ionomycin (C) for 0, 15 or 60min. The cells were then lysed and SNAP23 was immunoprecipitated from 1mg of total protein for each condition. Samples were analyzed by Western blot using anti-SNAP23 and anti-SNAP23120 antibodies (insets). The band intensities were determined by densitometry. Graphs represent the mean band intensity ratio of SNAP23120 to total SNAP23 ± the standard deviation of 3 independent experiments. The SNAP23120 to total SNAP23 ratio of the control population at 60min was considered 100%. In all graphs, asterisks denote statistically significant p values, where * ≤ 0.05 and * ≤ 0.01. (D) RBLs were cultured with or without (control) E. coli (MOI 10,000) for 2h. The cells were then harvested and stimulated in suspension with 1μM PMA/1μM Ionomycin for 0, 15 or 60min before being lysed. 50μg of total protein from exposed and control whole cell lysates were analyzed by Western blot using anti-phospho-IκBα and anti-alpha tubulin antibodies. Representative of 3 independent experiments.
Upon stimulation, SNAP23 is phosphorylated at serine residues 95 and 120 by IKKβ (28, 29). In vivo and in vitro, this phosphorylation event is essential for degranulation to occur (29). If SNAP23 phosphorylation is compromised, mast cell degranulation will be reduced (28). Therefore, we investigated whether co-culturing mast cells with E. coli interfered with this event. To do so, SNAP23 was immunoprecipitated from stimulated RBLs cultured with or without E. coli. SNAP23 phosphorylation at serine residue 120 was determined using Western blot. In control cells, SNAP23 is rapidly phosphorylated within 15min of stimulation with IgE/DNP-BSA or PMA/Ionomycin (Fig 4B and C, control), consistent with previous observations (28, 29). Interestingly, when cells are incubated with E. coli, SNAP23 phosphorylation is significantly reduced by ~50% at both 15 and 60min post-stimulation with IgE/DNP-BSA (Fig 4B) and by ~25% at 60min with PMA/Ionomycin (Fig 4C). The differences in SNAP23 phosphorylation using IgE/DNP-BSA and PMA/Ionomycin stimulation correlates with the level of β-hexosaminidase secretion observed previously where E. coli exposure inhibited secretion about twice as much with IgE/DNP-BSA stimulation (87%, Fig S1A) compared with PMA/Ionomycin stimulation (47%, Fig S1B). Although we previously observed a significant inhibition of degranulation at 15min with PMA/Ionomycin stimulation (Fig S1B), we did not observe a significant reduction in SNAP23 phosphorylation at the same time point. This discrepancy is likely due to the fact that SNAP23 phosphorylation was analyzed in cells that were stimulated in suspension (Fig 4C), whereas degranulation was analyzed with adherent cells (Fig S1B). It has been previously shown that the kinetics of degranulation is delayed in suspension cells compared to adherent cells (39).
Although PMA directly activates PKC, which has been shown to phosphorylate SNAP23 in vitro (28), PKC does not appear to be directly phosphorylating SNAP23 in vivo. In fact, in mast cells derived from IKKβ−/− mice there is little to no stimulus-induced phosphorylation of SNAP23 suggesting that IKKβ is the major kinase responsible for SNAP23 phosphorylation and that PKC acts upstream of IKKβ (29).
We then determined whether SNAP23 hypophosphorylation was due to changes in IKKβ activity. In addition to phosphorylating SNAP23, IKKβ is also the major kinase that phosphorylates IκBα during NFκB signaling in response to FcεRI-mediated activation as well as Toll-like receptor engagement (40-42). We then tested IKKβ activity by monitoring IκBα phosphorylation following co-culture with or without E. coli using Western blot. In cells cultured with E. coli, we observed an increase in IκBα phosphorylation following stimulation compared with the control cells (Fig 4D), suggesting that IKKβ is still enzymatically active following co-culture with E. coli. The enhanced phosphorylation of IκBα may be due to Toll-like receptor (TLR) engagement during the culture period, which would activate the NFκB pathway resulting in IκBα phosphorylation.
IKKβ binding to SNAP23 is decreased in mast cells exposed to E. coli
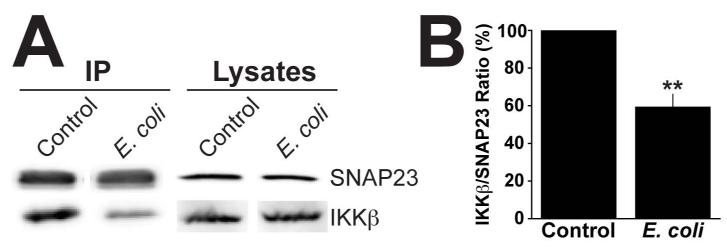
Upon stimulation, IKKβ interacts with and phosphorylates SNAP23. Therefore, we investigated whether the reduction in SNAP23 phosphorylation was due to impairment in the interaction between SNAP23 and IKKβ. Initially, we tested whether IKKβ recruitment into lipid rafts containing SNAP23 was impaired. However, we were unable to visualize IKKβ in the rafts, likely due to the lack of sensitivity of the antibody (data not shown). We then assessed the direct interaction between both of these proteins. To do so, we immunoprecipitated SNAP23 from stimulated RBLs cultured with or without E. coli, and analyzed the binding of IKKβ by Western blot. As shown in Fig 5A (control), IKKβ co-immunoprecipitates with SNAP23, in agreement with Suzuki et al (29). However, in cells cultured with E. coli, we observed a 40% reduction in the amount of IKKβ that co-immunoprecipitates with SNAP23 (Fig 5A and B). Together, these data suggest that E. coli exposure impairs the interaction between IKKβ and SNAP23 resulting in the hypophosphorylation of SNAP23.
Figure 5. Binding of IKKβ to SNAP23 is reduced following E. coli exposure.
(A) RBLs were incubated with or without (control) E. coli at an MOI of 10,000 for 2h. The cells were harvested and stimulated for 30min with 1μM PMA/1μM Ionomycin at a concentration of 2×106 cells/ml, lysed and SNAP23 was immunoprecipitated from 1mg of total protein. Immunoprecipitated samples and 40μg of whole cell lysates were analyzed by Western blot with anti-SNAP23 and -IKKβ antibodies. (B) Graph represents the mean band intensity ratio of IKKβ to total SNAP23 ± the standard deviation of 3 independent experiments. The IKKβ/SNAP23 ratio of the control population was considered 100%. Asterisks denote statistically significant p values, where ** ≤ 0.01.
Ternary exocytic SNARE complex formation is inhibited in E. coli exposed mast cells
The exocytic SNARE machinery plays a critical role in degranulation since it catalyzes membrane fusion. SNAP23, which is phosphorylated upon mast cell activation (23, 28, 29), binds its cognate t-SNARE Syntaxin4 to form the exocytic t-SNARE complex (23, 27). The v-SNAREs VAMP2 and VAMP8 in turn interact with the t-SNARE complex to form ternary SNARE complexes (23, 29, 43). Since SNAP23 phosphorylation in mast cells is significantly reduced following incubation with E. coli (Fig 4B & C), ternary SNARE complex formation should also be impaired. To test this hypothesis, ternary SNARE complex formation was examined by co-immunoprecipitation and Western blot analysis of stimulated RBLs cultured with or without E. coli. To stabilize SNARE complexes, we treated the cells with N-Ethylmaleimide (NEM) prior to cell lysis. NEM blocks NEM-sensitive factor (NSF), which is responsible for SNARE complex disassembly. As a result, NEM enhances the accumulation of SNARE complexes (27, 43-45). As shown in Fig 6A, Syntaxin4 co-immunoprecipitates with SNAP23 in the absence of E. coli exposure. However, there is a 23% reduction in the co-immunoprecipitation of Syntaxin4 when the cells are cultured with E. coli prior to stimulation (Fig 6A and B, Syntaxin4), indicating that t-SNARE complex formation is impaired.
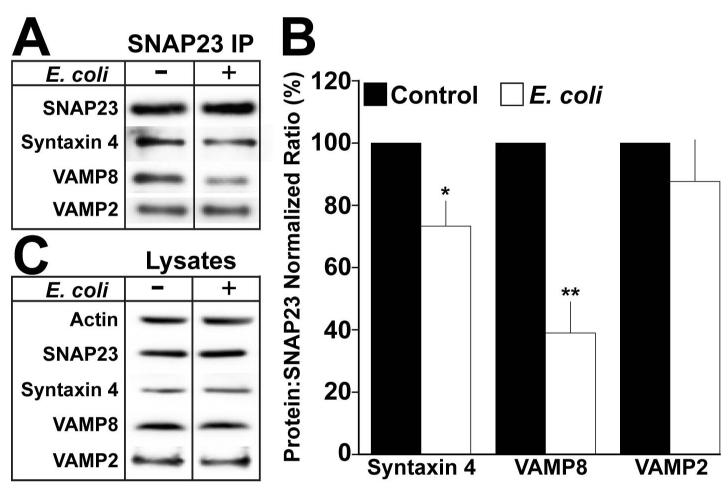
Figure 6. Exocytic SNARE complex formation is impaired in E. coli exposed mast cells.
(A) RBLs were cultured with or without (control) E. coli for 2h at an MOI of 10,000. The cells were harvested and stimulated for 30min with 1μM PMA/1μM Ionomycin at a concentration of 2×106 cells/ml. Cells were treated with 1mM NEM for 15min. Next, NEM was inactivated with 2mM DTT for 15min on ice. NEM-treated cells were washed and lysed. 1mg of total protein was immunoprecipitated using anti-SNAP23 antibody. Samples were analyzed by Western blot using anti-SNAP23, -Syntaxin4, -VAMP8, and –VAMP2 antibodies. Each blot is representative of at least 3 independent experiments. (B) Quantification of the mean band intensity ratio’s of the indicated protein to SNAP23 ± the standard deviation of at least 3 independent experiments. The protein:SNAP23 ratio of the control population was considered 100%. Asterisks denote statistically significant p values, where * ≤ 0.05 and ** ≤ 0.01. (C) 40μg of total protein from exposed or control whole cell lysates was analyzed by Western blot as described in A.
The assembly of a competent t-SNARE complex is essential for the formation of SNARE complexes with cognate v-SNAREs (46, 47). Thus, we examined the interactions between SNAP23 and its cognate exocytic v-SNAREs VAMP2 and VAMP8. E. coli exposure profoundly impairs the interaction between SNAP23 and VAMP8 by ~59% (Fig 6A and B, VAMP8). However, similar amounts of VAMP2 co-immunoprecipitate with SNAP23 whether the cells are cultured with or without E. coli (Fig 6A and B, VAMP2). The reduction in SNARE complex formation is not due to differences in the expression of SNAP23, Syntaxin4, or VAMP8 as shown in the lysates (Fig 6C). Altogether, these results suggest that E. coli exposure specifically impairs the formation of VAMP8-dependent exocytic SNARE complexes.
Discussion
Mast cells play an integral role in the development of allergic inflammation through the rapid release of prestored pro-inflammatory mediators (48). Importantly, different bacteria have been shown to interfere with mast cell degranulation (14-19, 32). E. coli for instance, has been reported to inhibit FcεRI-mediated secretion in primary mast cells (18, 19). In this report, we demonstrate that, similar to primary mast cells and mouse mast cell lines, E. coli exposure inhibits FcεRI-mediated and PMA/Ionomycin-induced β-hexosaminidase secretion in RBL-2H3 rat mast cells (18). Increasing MOIs as well as repeated E. coli exposures inhibit mast cell degranulation in a cumulative manner, suggesting that both the magnitude (i.e. MOI) and the chronicity of the infection are important factors in inhibiting mast cell degranulation. Chronic exposure to bacteria is important for regulating immune responses and inflammation, especially in the gut where exposure to the bacterial microflora is extensive (1). Collectively, these results suggest that the microenvironment within which mast cells reside modulates their activation (10-19).
The mechanism by which bacteria inhibit FcεRI-mediated degranulation in mast cells is unknown. Here, we dissected the cascade of events leading to the inhibition of mast cell degranulation and the role of the exocytic SNARE machinery in this process. We discovered that culturing mast cells with E. coli reduces the interaction between SNAP23 and its kinase IKKβ, which in turn leads to SNAP23 hypo-phosphorylation and a decrease in exocytic SNARE complex formation following activation.
SNARE protein phosphorylation is an important signal in regulated exocytosis. In particular, SNAP23 phosphorylation plays a pivotal role in regulated secretion, and has been reported in a number of cell types (28, 49-51). In mast cells, SNAP23 phosphorylation on Ser95 and Ser120 represents an important step for exocytic SNARE protein function and complex assembly (28). Several kinases are able to phosphorylate SNAP23 in vitro, including PKC (28, 49, 50) and IKKβ (29), although only IKKβ has been shown to be involved in this process in vivo (29). In fact, little to no phosphorylation of SNAP23 is detected in the absence of IKKβ (29). The phosphorylation of SNAP23 by IKKβ has also been observed during platelet secretion thus further supporting a role for the SNAP23:IKKβ axis in regulated secretion (51). Here, we have shown that E. coli exposure impairs this critical checkpoint in SNARE complex formation, which has significant consequences for membrane fusion and mast cell degranulation.
In mast cells cultured with E. coli, SNAP23 phosphorylation is reduced following FcεRI- or PMA/Ionomycin-mediated stimulation. Less stringent inhibition was observed with PMA/Ionomycin-induced than with FcεRI-mediated stimulation following E. coli exposure suggesting a possible compensatory mechanism controlled by PKC activation. However, the association of IKKβ with SNAP23 is still significantly reduced in mast cells co-cultured with E. coli and stimulated with PMA/Ionomycin suggesting that even if PKC activation partially compensates for SNAP23 phosphorylation, the impairment in IKKβ recruitment plays a significant role in the inhibition of SNAP23 phosphorylation. Further experiments will be necessary to investigate the extent to which PKC is impacted during bacterial exposure.
Previous studies have shown that the phosphorylation of SNAP23 is essential for mast cell degranulation and SNARE complex assembly (28, 29). In the present study, we showed that SNAP23 hypo-phosphorylation induced by E. coli results in a decrease in SNARE complex formation. In particular, incubating mast cells with E. coli leads to the inhibition of VAMP8-containing SNARE complexes, while VAMP2-containing complexes are not affected. Since VAMP2 and VAMP8 are localized to different secretory granules (27), these data demonstrate that E. coli exposure interferes with the release of a subset of secretory granules, and does not block exocytosis as a whole. In fact, only VAMP8 has been shown to be specifically involved in serotonin and β-hexosaminidase secretion (26). While VAMP8−/− mice are defective in β-hexosaminidase and serotonin release, VAMP2−/− mice show normal levels of degranulation. Interestingly, VAMP8−/− mice exhibited no defect in histamine (26) or cytokine secretion (43) suggesting that the fusion of subsets of secretory granules in mast cells are regulated by different SNARE proteins. Supporting this hypothesis, E. coli exposure appears to inhibit β-hexosaminidase (this study) and serotonin release (18), but induces histamine (20) and cytokine secretion (52). Since not all exocytic events are similarly affected by SNAP23 hypo-phosphorylation, our results also suggest that each VAMP-containing vesicle might fuse with different exocytic t-SNARE complexes that contain SNAP23. Alternatively, the requirement for SNAP23 phosphorylation might be different for each v-SNARE. Collectively, these data suggest that culturing mast cells with E. coli specifically blocks the fusion of VAMP8-containing β-hexosaminidase granules. In view of this evidence, one can hypothesize that E. coli exposure redirects the reactivity of mast cells towards an immune response against the invading pathogen (53, 54).
Although the bacterial component(s) that initiates the inhibitory cascade and impairs the IKKβ SNAP23 interaction and SNAP23 phosphorylation remains to be identified, the enhanced phosphorylation of IκBα that we observed in E. coli exposed cells (Fig 4D) may provide some insight. In addition to inducing degranulation, FcεRI crosslinking also activates the NFκB pathway, which involves the IKKβ-dependent phosphorylation of IκBα (40). Furthermore, bacterial pattern recognition receptors also activate the NFκB pathway (42). Both degranulation and the NFκB pathway require IKKβ activity (29). Our data suggests that the reduced interaction between IKKβ and SNAP23 may be due to the fact that IKKβ is titrated away from the “degranulation pathway” and towards the NFκB pathway. Thus, less IKKβ would be available to phosphorylate SNAP23. Given that the increased IκBα phosphorylation is stimulus-dependent, it is possible that bacterial exposure may “prime” the NFκB pathway for antigen-mediated activation. The signaling pathway induced by Toll-like receptor (TLR) activation may explain this phenomenon. Mast cells express a number of TLRs, which play an important role in immune responses to bacterial infection (55). Surface expressed TLR4 and TLR2 recognize bacterial lipopolysaccharide and peptidoglycan, respectively (55). Treatment of mast cells with TLR2/4 agonists prior to FcεRI-mediated stimulation inhibits mast cell degranulation (56) and dampens allergic inflammation (57-59) while inducing cytokine secretion (42). Of particular interest is the involvement of the kinase IKKβ in TLR signaling pathways and SNAP23 phosphorylation (29, 55), which suggests that TLRs are strong candidates for initiating the inhibition of mast cell degranulation. Although TLR4 agonists alone inhibit degranulation, heat-inactivated E. coli do not inhibit mast cell secretion (Fig S1C, left panel) suggesting that heat stable antigens like LPS are not involved in this process. Whether other TLRs or surface receptors play a role in this inhibitory cascade requires further investigation.
In summary, these data present a novel mechanism by which E. coli exposures influence mast cell degranulation through specific changes in membrane fusion events. This effect most likely preserves the secretion of protective cytokines while reducing the release of mediators that drive allergic inflammation. It will be important to determine which events upstream SNARE complex formation in mast cells are impaired as a result of bacterial exposure. In addition, identifying the bacterial component(s) that contributes to the inhibition of mast cell degranulation will be essential for developing ways to artificially regulate mast cell activation and ultimately the development of allergy.
Materials and Methods
Cell line and bacterial strain
The rat basophilic leukemia (RBL-2H3) cell line was obtained from the ATCC (CRL-2256) and cultured as previously described (27, 38). Briefly, RBL-2H3 cells were maintained in complete DMEM (DMEM supplemented with 15mM HEPES, 10% fetal bovine serum, 2mM L-glutamine, 100 units/ml penicillin and 100μg/ml streptomycin). The E. coli strain BL21 (DE3) [Invitrogen] was grown to the stationary phase overnight in Luria broth for all assays.
RBL-2H3 transfection and generation of stable cell lines
RBL-2H3 cells were transfected with SNAP23-EGFP-N3, SNAP29-EGFP-N3 or empty EGFP-N3 vectors as previously described (38). The expression of GFP fusion proteins was assessed by Western blot.
Antibodies
Rabbit anti-SNAP23, -Syntaxin4, -VAMP8, –Actin and mouse anti-alpha tubulin were purchased from Sigma. Rabbit anti-VAMP2 was obtained from Synaptic Systems. Anti-p80 (clone 5G10, mouse) and –phospho-SNAP23 (Serine 120, rabbit) were kind gifts from Drs. J. Bonifacino and P. Roche, respectively. Anti-IKKβ (rabbit) was purchased from Cell Signaling and anti-FcεRIα (mouse) was obtained from Abcam. Mouse anti-SNAP23 and -phospho IκBα were purchased from Santa Cruz Biotechnology. HRP-conjugated Cholera toxin subunit B and Goat anti-mouse IgG1-AlexaFluor488 conjugated antibody was obtained from Invitrogen. Sheep anti-mouse and donkey anti-rabbit HRP-conjugated antibodies were purchased from GE Healthcare.
β-hexosaminidase assay
RBL-2H3 cells were plated at 5×104 cells/well in 100μl of complete DMEM in 96 well flat bottom plates 24h prior to the assay. For single exposure experiments, E. coli was grown to the stationary phase, washed and diluted in DMEM without antibiotics to multiplicities of infection (MOI) of 100, 1,000 and 10,000 per 200μl of medium. E. coli-conditioned medium was generated by culturing E. coli at MOIs 10,000 (1×) and 20,000 (2×) in DMEM without antibiotics for 2h, followed by centrifugation at 100,000×g for 1h and filtration (0.2μm). E. coli was heat-killed for 45min at 70°C. The cells were washed with DMEM without antibiotics then 200μl of the appropriate MOI was added to the cells followed by incubation at 37°C for 2h. Control cells received 200μl of DMEM without antibiotics. To inhibit protein translation, 100μg/ml chloramphenicol was added during the 2h incubation. The wells were then thoroughly washed to remove extracellular bacteria. For FcεRI-mediated stimulation, the cells were sensitized with 200μl of 100ng/ml anti-dinitrophenol (DNP) IgE (Sigma) in DMEM supplemented with 100μg/ml gentamicin for 1h at 37°C. Cells to be stimulated with phorbol myristate acetate (PMA) and Ionomycin received 200μl of DMEM supplemented with 100μg/ml gentamicin and were incubated for 1h at 37°C. The cells were then washed three times with β-hex medium (DMEM without phenol red containing 500μg/ml bovine serum albumin (BSA) and 2mM L-glutamine). For IgE-mediated stimulation, FcεRI was cross-linked with 200μl of 100ng/ml DNP-BSA (Sigma) in β-hex medium and the cells were incubated for 15, 30 or 60min at 37°C. For PMA/Ionomycin stimulation, the cells were stimulated with 200μl of 1μM PMA (Fisher) and 1μM Ionomycin (Fisher) in β-hex medium and incubated for 15, 30 or 60min at 37°C. At each time point a sample of the supernatant was removed for analysis. β-hexosaminidase was quantified as described (38).
To determine the effect of multiple E. coli exposures on β-hexosaminidase release, 5×105 RBL-2H3 cells were plated in 10cm tissue culture plates 24h prior to the first exposure. For each exposure, the cells were co-cultured with E. coli as described above for 2h at an MOI of 10,000 in 10ml of DMEM without antibiotics.
The cells were then washed thoroughly to remove extracellular bacteria and incubated overnight with DMEM supplemented with 100μg/ml gentamicin at 37°C. Each exposure occurred 24h apart. After the final exposure, the cells were incubated overnight at 37°C in DMEM containing 100μg/ml gentamicin. The cells were then harvested and 5×104 viable cells (as determined by trypan blue exclusion) were plated in 96 well plates in DMEM and incubated at 37°C for 30min until they adhered to the plate. The cells were then sensitized with 200μl of 100ng/ml anti-DNP IgE or media alone (for PMA/Ionomcyin stimulation) for 2h at 37°C followed by stimulation as described above.
Flow cytometry
4×106 RBL-2H3 cells were cultured with or without E. coli at an MOI of 10,000 in 10cm culture plates, harvested and stimulated in suspension with either 800ng/ml anti-DNP IgE and 800ng/ml DNP-BSA or 1μM PMA/1μM Ionomycin at a concentration of 2×106 cells/ml. Following stimulation, cells were fixed with 2% paraformaldehyde (PFA) on ice for 30min. The cells were washed with PBS, blocked for 30min with FACs buffer (1% BSA, 1mM EDTA in PBS, pH 7.4) containing 20% goat serum and resuspended in primary antibodies indicated in the Figure legends for 1h on ice. Cell pellets were washed with FACs buffer by centrifugation and then stained with goat anti-mouse IgG1 AlexaFluor488-conjugated antibody (Invitrogen) for 30min on ice. Samples were washed, resuspended in FACs buffer and analyzed using the FACS Calibur Flow Cytometer (BD). Data was analyzed using FlowJo software.
Immunofluorescence microscopy
5×104 RBL-2H3 cells were seeded onto coverslips 24h prior to the experiment in complete DMEM. Cells were cultured with or without E. coli at an MOI of 10,000 for 2h. Extracellular bacteria were removed by thorough washes with PBS. To assess surface levels of FcεRI, cells were fixed in 2% PFA for 30min, quenched with 50mM ammonium chloride for 15min, and stained as described below. For FcεRI-mediated stimulation the cells were sensitized with 100ng/ml anti-DNP IgE for 1h at 37°C following exposure. The cells were stimulated with 100ng/ml DNP-BSA for 60min at 37°C, then fixed and quenched. For PMA/Ionomycin stimulation, following E. coli exposure the cells were incubated at 37°C for 1h, stimulated with 1μM PMA/1μM Ionomycin for 60min then fixed and quenched. To label surface proteins, the coverslips were blocked with blocking buffer (20% goat serum, 0.1% BSA in PBS, pH 7.4) for 1h and incubated with primary antibodies diluted in blocking buffer as indicated in the Figure legends for 1h. The coverslips were washed and incubated with goat anti-mouse IgG1 AlexaFluor488-conjugated antibody diluted in blocking buffer for 1h. The cells were washed and nuclei were labeled with 1μg/ml Hoechst (Invitrogen). Coverslips were mounted with Prolong Gold Anti-fade reagent (Invitrogen). Images were acquired using a Nikon TiE inverted immunofluorescence microscope with a 60× oil immersion lens and NIS Elements AR 3.2 software (Nikon). Image analysis was conducted using ImageJ (NIH).
Immunoprecipitation
1.6×107 RBL-2H3 cells were cultured with or without E. coli at an MOI of 10,000 for 2h in 150mm plates. Following the exposure, the cells were harvested using Cellstripper (Invitrogen) and resuspended to 2×106 cells/ml in DMEM containing 100μg/ml gentamicin at 37°C for 1h. For FcεRI-mediated stimulation, the cells were incubated with 800ng/ml of anti-DNP IgE during the hour recovery. The cells were washed three times by centrifugation with β-hex medium then resuspended to 2×106 cells/ml with either 1μM PMA/1μM Ionomycin or 800ng/ml DNP-BSA in β-hex medium and incubated at 37°C for the indicated time. Ice-cold PBS was added to the cell suspension to stop degranulation and the cells were pelleted by centrifugation at 4°C. Stimulants were removed by washing the cell pellets with ice-cold PBS followed by centrifugation. To stabilize SNARE complexes, the cell pellets were treated with N-Ethylmaleimide (NEM) as previously described (27) before cell lysis. Cells were lysed in lysis buffer (1% NP-40, 1mM EDTA, 1mM EGTA in TBS, pH 7.4) at a concentration of 3×107 cells/ml for 1h at 4°C followed by centrifugation at 14,000rpm for 20min. 3×107 cells were used for immunoprecipitation. 1mg of total protein was incubated with 30μl of BSA-blocked ProteinG Plus Agarose beads (Millipore) bound with 10μg of anti-SNAP23 (mouse) antibody overnight at 4°C. The beads were then washed with lysis buffer, eluted with sodium dodecyl sulfate-containing sample buffer and boiled for 5min at 95°C. Samples were analyzed by SDS-PAGE and Western blot.
Gel electrophoresis and Western blotting
Samples were run on 10% Bis-Tris gels (Invitrogen) and then transferred to PVDF membrane (Bioexpress). The membranes were blocked with wash buffer (25mM Tris, 250mM Sodium chloride, 0.2% Tween-20, pH 7.6) containing 5% milk for 1h and immunoblotted with primary antibodies diluted in wash buffer containing 5% BSA as indicated in the Figure legends for 1h. The membranes were then washed with wash buffer and incubated with HRP-conjugated secondary antibodies diluted in wash buffer containing 5% milk for 1h. Following several washes, the blots were revealed with WesternBright Quantum ECL (Bioexpress). Images were captured using a FluorChem M imager with CCD camera (ProteinSimple) and band intensities were analyzed using Alphaview Software (ProteinSimple).
Lipid raft preparation
RBL-2H3 cells were co-cultured with or without (control) E. coli at an MOI of 10,000 for 2h, then harvested and stimulated in suspension with 1μM PMA/1μM Ionomycin for 30min at 7×106 cells/ml. The cells were washed with cold PBS and resuspended to 2.5×108 cells/ml in cold MBS (25mM MES, pH 6.5 and 150mM NaCl) containing 1% Triton-X100, 1mM Na3VO4, 10mM NaF, 2mM PMSF, 5μg/ml Leupeptin, 2μg/ml Pepstatin A for 20min on ice followed by 20 passages in a dounce homogenizer. 400μl of lysate was mixed with 400μl of 80% sucrose and overlaid with 2.2ml of 30% and 1.4ml of 5% sucrose in a 4.2ml centrifuge tube. All sucrose solutions were prepared in MBS. The gradient was then centrifuged for 18h at 200,000×g at 4°C. 400μl fractions were collected from the top of the gradient and analyzed by Western blot.
Statistical analysis
The Students t-test (two-tailed) was used to compare the means of different populations. Data are considered statistically significant at p values ≤0.05.
Supplementary Material
Figure S1: E. coli, which must be translationally active, rapidly inhibits RBL-2H3 mast cell degranulation. (A) RBLs were plated in 96 well plates at a density of 5×104 viable cells per well in 100μl of DMEM 24h prior to being cultured with increasing MOI’s of E. coli for 2h. Following sensitization with 100ng/ml anti-DNP IgE for 1h and stimulation with 100ng/ml DNP-BSA for the indicated times, β-hexosaminidase release was quantified. Total intracellular β-hexosaminidase content was determined after E. coli co-culture for each MOI (inset). (B) RBLs were cultured with or without E. coli as described in A, but stimulated with 1μM PMA/1μM Ionomycin and β-hexosaminidase release was quantified. Graphs depict the mean ± the standard deviation of 5 independent experiments conducted in duplicate. The amount of β-hexosaminidase released at 60min in the control population was considered 100%. (C) RBLs were plated in 96 well plates at a density of 5×104 viable cells per well in 100μl of DMEM 24h prior to being cultured with or without (control) heat-killed E. coli (left), chloramphenicol-treated E. coli (center) or E. coli-conditioned medium (right) for 2h. The cells were then sensitized with 100ng/ml anti-DNP IgE for 1h, stimulated with 100ng/ml DNP-BSA for 1h and β-hexosaminidase release was quantified. The amount of β-hexosaminidase released at 60min in the control population was considered 100%. Graphs depict the mean ± the standard deviation of 3 independent experiments conducted in triplicate. In all graphs, asterisks denote statistically significant p values, where * ≤ 0.05 and * ≤ 0.01.
Figure S2: FcεRI surface expression is not affected by E. coli exposure. (A) RBLs were seeded on coverslips at a density of 5×104 cells/coverslip 24hr prior to being incubated with (right panel) or without (left panel-control) E. coli at an MOI of 10,000 for 2h. The cells were then washed, fixed and stained with anti-FcεRI and anti-mouse IgG1 AlexaFluor488-conjugated antibodies. Nuclei were stained with Hoechst. Surface levels of FcεRI were determined using immunofluorescence microscopy. Scale bars = 20μm. Representative of 3 independent experiments (B) RBLs were cultured with or without E. coli at an MOI of 10,000 either once (left panel) or three times (right panel), labeled as described in A and surface levels were quantified using flow cytometry. Graphs represent the mean fluorescence intensity of FcεRI staining from 3 independent experiments ± the standard deviation.
Synopsis.
Mast cell degranulation plays a major role in allergic diseases. Here, we report that co-culturing mast cells with E. coli inhibits FcεRI-mediated degranulation by impacting the function of the exocytic SNARE fusion machinery. In particular, E. coli co-culture reduces the IKKβ-dependent phosphorylation of the t-SNARE SNAP23, thus impairing the formation of ternary exocytic SNARE complexes and ultimately mediator release.
Acknowledgements
This work was indirectly supported by the National Institutes of Health grant AI073486 to F.P. We thank V. Caldwell for technical assistance and the members of the Paumet lab for their helpful discussions during the course of this study. We are grateful to Drs. K. Suzuki and P. Roche for providing DNA constructs and technical advice. We also thank Dr. C. Tkaczyk for critical reading of the manuscript, and Drs. J. Bonifacino and P. Roche for respectively providing anti-p80 (clone 5G10) and anti-phospho-SNAP23 (Serine 120) antibodies. We are grateful to Dr. M. Mahoney for technical assistance in isolating lipid rafts. The authors declare no conflict of interest.
References
- 1.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol. 2012;24(1):58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt M, Cookson W. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardenas PA, Cooper PJ, Cox MJ, Chico M, Arias C, Moffatt MF, Cookson WO. Upper airways microbiota in antibiotic-naive wheezing and healthy infants from the tropics of rural Ecuador. PLoS One. 2012;7(10):e46803. doi: 10.1371/journal.pone.0046803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbst T, Sichelstiel A, Schar C, Yadava K, Burki K, Cahenzli J, McCoy K, Marsland BJ, Harris NL. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184(2):198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 6.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CM, Tsai M. Mast cells as “tunable” effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 7.Beil WJ, Schulz M, Wefelmeyer U. Mast cell granule composition and tissue location--a close correlation. Histol Histopathol. 2000;15(3):937–946. doi: 10.14670/HH-15.937. [DOI] [PubMed] [Google Scholar]
- 8.Blank U, Rivera J. The ins and outs of IgE-dependent mast-cell exocytosis. Trends Immunol. 2004;25(5):266–273. doi: 10.1016/j.it.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997;77(4):1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 10.Wesolowski J, Paumet F. The impact of bacterial infection on mast cell degranulation. Immunol Res. 2011;51(2-3):215–226. doi: 10.1007/s12026-011-8250-x. [DOI] [PubMed] [Google Scholar]
- 11.Munoz S, Hernandez-Pando R, Abraham S, Enciso J. Mast cell activation by Mycobacterium tuberculosis: mediator release and role of CD48. J Immunol. 2003;170:5590–5596. doi: 10.4049/jimmunol.170.11.5590. [DOI] [PubMed] [Google Scholar]
- 12.Hoek KL, Cassell GH, Duffy LB, Atkinson TP. Mycoplasma pneumoniae-induced activation and cytokine production in rodent mast cells. J Allergy Clin Immunol. 2002;109(3):470–476. doi: 10.1067/mai.2002.121951. [DOI] [PubMed] [Google Scholar]
- 13.Barbuti G, Moschioni M, Censini S, Covacci A, Montecucco C, Montemurro P. Streptococcus pneumoniae induces mast cell degranulation. Int J Med Microbiol. 2006;296(4-5):325–329. doi: 10.1016/j.ijmm.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Kawahara T. Inhibitory effect of heat-killed Lactobacillus strain on immunoglobulin E-mediated degranulation and late-phase immune reactions of mouse bone marrow-derived mast cells. Animal Sc J. 2010;81:714–721. doi: 10.1111/j.1740-0929.2010.00788.x. [DOI] [PubMed] [Google Scholar]
- 15.Schiffer C, Lalanne AI, Cassard L, Mancardi DA, Malbec O, Bruhns P, Dif F, Daeron M. A strain of Lactobacillus casei inhibits the effector phase of immune inflammation. J Immunol. 2011;187(5):2646–2655. doi: 10.4049/jimmunol.1002415. [DOI] [PubMed] [Google Scholar]
- 16.Forsythe P, Wang B, Khambati I, Kunze WA. Systemic effects of ingested Lactobacillus rhamnosus: inhibition of mast cell membrane potassium (IKCa) current and degranulation. PLoS One. 2012;7(7):e41234. doi: 10.1371/journal.pone.0041234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harata G, He F, Takahashi K, Hosono A, Kawase M, Kubota A, Hiramatsu M, Kaminogawa S. Bifidobacterium suppresses IgE-mediated degranulation of rat basophilic leukemia (RBL-2H3) cells. Microbiol Immunol. 2010;54:54–57. doi: 10.1111/j.1348-0421.2009.00185.x. [DOI] [PubMed] [Google Scholar]
- 18.Magerl M, Lammel V, Siebenhaar F, Zuberbier T, Metz M, Maurer M. Non-pathogenic commensal Escherichia coli bacteria can inhibit degranulation of mast cells. Exp Dermatol. 2008;17:427–435. doi: 10.1111/j.1600-0625.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 19.Kulka M, Fukuishi N, Rottem M, Mekori Y, Metcalfe D. Mast cells, which interact with Escherichia coli, up-regulate genes associated with innate immunity and become less responsive to FceRI-mediated activation. J Leukoc Biol. 2006;79:339–350. doi: 10.1189/jlb.1004600. [DOI] [PubMed] [Google Scholar]
- 20.Malaviya R, Ross E, Jakschik B, Abraham S. Mast cell degranulation induced by type I fimbriated Escherichia coli in mice. J Clin Invest. 1994;93:1645–1653. doi: 10.1172/JCI117146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malaviya R, Ikeda T, Abraham S, Malaviya R. Contribution of mast cells to bacterial clearance and their proliferation during experimental cystitis induced by type 1 fimbriated E. coli. Immunol Lett. 2004;91:103–111. doi: 10.1016/j.imlet.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Lippert U, Ferrari DM, Jahn R. Endobrevin/VAMP8 mediates exocytotic release of hexosaminidase from rat basophilic leukaemia cells. FEBS Lett. 2007;581(18):3479–3484. doi: 10.1016/j.febslet.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 23.Puri N, Roche PA. Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis. Traffic. 2006;7(11):1482–1494. doi: 10.1111/j.1600-0854.2006.00490.x. [DOI] [PubMed] [Google Scholar]
- 24.Hibi T, Hirashima N, Nakanishi M. Rat basophilic leukemia cells express syntaxin-3 and VAMP-7 in granule membranes. Biochem Biophys Res Commun. 2000;271(1):36–41. doi: 10.1006/bbrc.2000.2591. [DOI] [PubMed] [Google Scholar]
- 25.Sander L, Frank S, Bolat S, Blank U, Galli T, Bigalke H, Bischoff S, Lrentz A. Vesicle associated membrane protein (VAMP)-7 and VAMP-8, but not VAMP-2 or VAMP-3, are required for activation-induced degranulation of mature human mast cells. Eur J Immunol. 2008;38:855–863. doi: 10.1002/eji.200737634. [DOI] [PubMed] [Google Scholar]
- 26.Puri N, Roche P. Mast cells possess distinct secretory granule subsets whose exocytosis is regulated by different SNARE isoforms. PNAS. 2008;105:2580–2585. doi: 10.1073/pnas.0707854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paumet F, Le Mao J, Martin S, Galli T, David B, Blank U, Roa M. Soluble NSF attachment protein receptors (SNAREs) in RBL-2H3 mast cells: functional role of syntaxin 4 in exocytosis and identification of a vesicle-associated membrane protein 8-containing secretory compartment. J Immunol. 2000;164(11):5850–5857. doi: 10.4049/jimmunol.164.11.5850. [DOI] [PubMed] [Google Scholar]
- 28.Hepp R, Puri N, Hohenstein A, Crawford G, Whiteheart S, Roche P. Phosphorylation of SNAP-23 regulates exocytosis from mast cells. J Biol Chem. 2005;280:6610–6620. doi: 10.1074/jbc.M412126200. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki K, Verma I. Phosphorylation of SNAP-23 by IkB kinase 2 regulates mast cell degranulation. Cell. 2008;134:485–495. doi: 10.1016/j.cell.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roa M, Paumet F, Le Mao J, David B, Blank U. Involvement of the ras-like GTPase rab3d in RBL-2H3 mast cell exocytosis following stimulation via high affinity IgE receptors (Fc epsilonRI) J Immunol. 1997;159(6):2815–2823. [PubMed] [Google Scholar]
- 31.Ludowyke R, Kawasugi K, French P. PMA and calcium ionophore induce myosin and F-actin rearrangement during histamine secretion from RBL-2H3 cells. Cell Motility and the Cytoskeleton. 1994;29:354–365. doi: 10.1002/cm.970290408. [DOI] [PubMed] [Google Scholar]
- 32.Oksaharju A, Kankainen M, Kekkonen RA, Lindstedt KA, Kovanen PT, Korpela R, Miettinen M. Probiotic Lactobacillus rhamnosus downregulates FCER1 and HRH4 expression in human mast cells. World J Gastroenterol. 2011;17(6):750–759. doi: 10.3748/wjg.v17.i6.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshioka M, Fukuishi N, Iriguchi S, Ohsaki K, Yamanobe H, Inukai A, Kurihara D, Imajo N, Yasui Y, Matsui N, Tsujita T, Ishii A, Seya T, Takahama M, Akagi M. Lipoteichoic acid downregulates FcepsilonRI expression on human mast cells through Toll-like receptor 2. J Allergy Clin Immunol. 2007;120(2):452–461. doi: 10.1016/j.jaci.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 34.Bonifacino J, Perez P, Klausner R, Sandoval I. Study of the transit of an integral membrane protein from secretory granules through the plasma membrane of secreting Rat Basophilic Leukemia cells using a specific monoclonal antibody. J Cell Biol. 1986;105:516–522. doi: 10.1083/jcb.102.2.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonifacino JS, Yuan L, Sandoval IV. Internalization and recycling to serotonin-containing granules of the 80K integral membrane protein exposed on the surface of secreting rat basophilic leukaemia cells. J Cell Sci. 1989;92(Pt 4):701–712. doi: 10.1242/jcs.92.4.701. [DOI] [PubMed] [Google Scholar]
- 36.Guo Z, Turner C, Castle D. Relocation of the t-SNARE SNAP23 from lamellipodia-like cell surface projections regulates compound exocytosis in mast cells. Cell. 1998;94:537–548. doi: 10.1016/s0092-8674(00)81594-9. [DOI] [PubMed] [Google Scholar]
- 37.Salinas E, Quintanar-Stephano A, Cordova LE, Ouintanar JL. Allergen-sensitization increases mast-cell expression of the exocytotic proteins SNAP-23 and syntaxin 4, which are involved in histamine secretion. J Investig Allergol Clin Immunol. 2008;18(5):366–371. [PubMed] [Google Scholar]
- 38.Wesolowski J, Caldwell V, Paumet F. A Novel Function for SNAP29 (Synaptosomal-Associated Protein of 29 kDa) in Mast Cell Phagocytosis. PLoS One. 2012;7(11):e49886. doi: 10.1371/journal.pone.0049886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen R, Corwith K, Holowka D, Baird B. Spatiotemporal resolution of mast cell granule exocytosis reveals correlation with Ca2+ wave initiation. J Cell Sci. 2012;125(Pt 12):2986–2994. doi: 10.1242/jcs.102632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Y, Power MR, Li B, Lin TJ. Inhibition of IKK down-regulates antigen + IgE-induced TNF production by mast cells: a role for the IKK-IkappaB-NF-kappaB pathway in IgE-dependent mast cell activation. J Leukoc Biol. 2005;77(6):975–983. doi: 10.1189/jlb.0204115. [DOI] [PubMed] [Google Scholar]
- 41.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science. 1999;284(5412):309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 42.Supajatura V, Ushio H, Nakao A, Akira S, Okumura K, Ra C, Ogawa H. Differential responses of mast cell Toll-like receptors 2 and 4 in allergy and innate immunity. J Clin Invest. 2002;109(10):1351–1359. doi: 10.1172/JCI14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiwari N, Wang C-C, Brochetta C, Ke G, Vita F, Qi Z, Rivera J, Soranzo M, Zabucchi G, Hong W, Blank U. VAMP-8 segregates mast cell preformed mediator exocytosis from cytokine trafficking pathways. Blood. 2008;111:3665–3674. doi: 10.1182/blood-2007-07-103309. [DOI] [PubMed] [Google Scholar]
- 44.Beckers C, Block M, Glick B, Rothman J, Balch W. Vesicular transport between the endoplasmic reticulum and the Golgi stack requires the NEM-sensitive fusion protein. Nature. 1989;339:397–398. doi: 10.1038/339397a0. [DOI] [PubMed] [Google Scholar]
- 45.Banerjee A, Barry V, DasGupta B, Martin T. N-Ethylmaleimide-sensitive factor acts at a prefusion ATP-dependent step in Ca2+-activated exocytosis. J Biol Chem. 1996;271:20223–20226. doi: 10.1074/jbc.271.34.20223. [DOI] [PubMed] [Google Scholar]
- 46.Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger AT. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272(44):28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- 47.Fasshauer D, Bruns D, Shen B, Jahn R, Brunger AT. A structural change occurs upon binding of syntaxin to SNAP-25. J Biol Chem. 1997;272(7):4582–4590. doi: 10.1074/jbc.272.7.4582. [DOI] [PubMed] [Google Scholar]
- 48.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18(5):693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polgar J, Lane WS, Chung SH, Houng AK, Reed GL. Phosphorylation of SNAP-23 in activated human platelets. J Biol Chem. 2003;278(45):44369–44376. doi: 10.1074/jbc.M307864200. [DOI] [PubMed] [Google Scholar]
- 50.Yasuda K, Itakura M, Aoyagi K, Sugaya T, Nagata E, Ihara H, Takahashi M. PKC-dependent inhibition of CA2+-dependent exocytosis from astrocytes. Glia. 2011;59(1):143–151. doi: 10.1002/glia.21083. [DOI] [PubMed] [Google Scholar]
- 51.Karim ZA, Zhang J, Banerjee M, Chicka MC, Al Hawas R, Hamilton TR, Roche PA, Whiteheart SW. IkappaB kinase phosphorylation of SNAP-23 controls platelet secretion. Blood. 2013;121(22):4567–4574. doi: 10.1182/blood-2012-11-470468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malaviya R, Gao Z, Thankavel K, van der Merwe PA, Abraham SN. The mast cell tumor necrosis factor alpha response to FimH-expressing Escherichia coli is mediated by the glycosylphosphatidylinositol-anchored molecule CD48. Proc Natl Acad Sci U S A. 1999;96(14):8110–8115. doi: 10.1073/pnas.96.14.8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shelburne CP, Nakano H, St John AL, Chan C, McLachlan JB, Gunn MD, Staats HF, Abraham SN. Mast cells augment adaptive immunity by orchestrating dendritic cell trafficking through infected tissues. Cell Host Microbe. 2009;6(4):331–342. doi: 10.1016/j.chom.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abraham S, St John A. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10:440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandig H, Bulfone-Paus S. TLR signaling in mast cells: common and unique features. Front Immunol. 2012;3:185. doi: 10.3389/fimmu.2012.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasakura K, Takahashi K, Aizawa T, Hosono A, Kaminogawa S. A TLR2 ligand suppresses allergic inflammatory reactions by acting directly on mast cells. Int Arch Allergy Immunol. 2009;150(4):359–369. doi: 10.1159/000226237. [DOI] [PubMed] [Google Scholar]
- 57.Nigo YI, Yamashita M, Hirahara K, Shinnakasu R, Inami M, Kimura M, Hasegawa A, Kohno Y, Nakayama T. Regulation of allergic airway inflammation through Toll-like receptor 4-mediated modification of mast cell function. Proc Natl Acad Sci U S A. 2006;103(7):2286–2291. doi: 10.1073/pnas.0510685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchs B, Knothe S, Rochlitzer S, Nassimi M, Greweling M, Lauenstein HD, Nassenstein C, Muller M, Ebensen T, Dittrich AM, Krug N, Guzman CA, Braun A. A Toll-like receptor 2/6 agonist reduces allergic airway inflammation in chronic respiratory sensitisation to Timothy grass pollen antigens. Int Arch Allergy Immunol. 2010;152(2):131–139. doi: 10.1159/000265534. [DOI] [PubMed] [Google Scholar]
- 59.Velasco G, Campo M, Manrique OJ, Bellou A, He H, Arestides RS, Schaub B, Perkins DL, Finn PW. Toll-like receptor 4 or 2 agonists decrease allergic inflammation. Am J Respir Cell Mol Biol. 2005;32(3):218–224. doi: 10.1165/rcmb.2003-0435OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: E. coli, which must be translationally active, rapidly inhibits RBL-2H3 mast cell degranulation. (A) RBLs were plated in 96 well plates at a density of 5×104 viable cells per well in 100μl of DMEM 24h prior to being cultured with increasing MOI’s of E. coli for 2h. Following sensitization with 100ng/ml anti-DNP IgE for 1h and stimulation with 100ng/ml DNP-BSA for the indicated times, β-hexosaminidase release was quantified. Total intracellular β-hexosaminidase content was determined after E. coli co-culture for each MOI (inset). (B) RBLs were cultured with or without E. coli as described in A, but stimulated with 1μM PMA/1μM Ionomycin and β-hexosaminidase release was quantified. Graphs depict the mean ± the standard deviation of 5 independent experiments conducted in duplicate. The amount of β-hexosaminidase released at 60min in the control population was considered 100%. (C) RBLs were plated in 96 well plates at a density of 5×104 viable cells per well in 100μl of DMEM 24h prior to being cultured with or without (control) heat-killed E. coli (left), chloramphenicol-treated E. coli (center) or E. coli-conditioned medium (right) for 2h. The cells were then sensitized with 100ng/ml anti-DNP IgE for 1h, stimulated with 100ng/ml DNP-BSA for 1h and β-hexosaminidase release was quantified. The amount of β-hexosaminidase released at 60min in the control population was considered 100%. Graphs depict the mean ± the standard deviation of 3 independent experiments conducted in triplicate. In all graphs, asterisks denote statistically significant p values, where * ≤ 0.05 and * ≤ 0.01.
Figure S2: FcεRI surface expression is not affected by E. coli exposure. (A) RBLs were seeded on coverslips at a density of 5×104 cells/coverslip 24hr prior to being incubated with (right panel) or without (left panel-control) E. coli at an MOI of 10,000 for 2h. The cells were then washed, fixed and stained with anti-FcεRI and anti-mouse IgG1 AlexaFluor488-conjugated antibodies. Nuclei were stained with Hoechst. Surface levels of FcεRI were determined using immunofluorescence microscopy. Scale bars = 20μm. Representative of 3 independent experiments (B) RBLs were cultured with or without E. coli at an MOI of 10,000 either once (left panel) or three times (right panel), labeled as described in A and surface levels were quantified using flow cytometry. Graphs represent the mean fluorescence intensity of FcεRI staining from 3 independent experiments ± the standard deviation.