Summary
The short interspersed elements (SINEs) Alu and B2 are retrotransposons that litter the human and mouse genomes, respectively. Given their abundance, the manner in which these elements impact the host genome and what their biological functions might be is of significant interest. Finding that Alu and B2 SINEs are transcribed, both as distinct RNA polymerase III transcripts and as part of RNA polymerase II transcripts, and that these SINE encoded RNAs indeed have biological functions has refuted the historical notion that SINEs are merely “junk DNA.” This article reviews currently known cellular functions of both RNA polymerase II and RNA polymerase III transcribed Alu and B2 RNAs. These RNAs, in different forms, control gene expression by participating in processes as diverse as mRNA transcriptional control, A-to-I editing, nuclear retention, and alternative splicing. Future studies will likely reveal additional contributions of Alu and B2 RNAs as regulators of gene expression.
Keywords: Alu, B2, SINE, ncRNA
Introduction
With recent estimates of approximately one insertion every 20 births and having reached over 1 million copies, Alu is the most successful retrotransposon in the human genome (1, 2). B1 and B2 occupy approximately 5% of the mouse genome with about 550,000 and 350,000 copies, respectively (3). Alu, B1 and B2 belong to a class of retrotransposons known as short interspersed elements (SINEs) that amplify throughout their respective genomes via a RNA intermediate (4). While these SINEs contain an internal RNA polymerase III (Pol III) promoter they do not encode the machinery necessary to propagate throughout the genome (4). To accomplish this, SINEs utilize the reverse transcriptase and nuclease encoded by long interspersed elements (LINEs) to integrate into the host genome (5, 6).
Given the abundance of Alu, B1 and B2 SINEs there is a great deal of interest in how they impact the host genome and what, if any, biological functions led to their maintenance. Curiously, retrotransposition of SINEs can have a detrimental effect on the host genome, being implicated in various disease states including cancer, yet these elements have flourished for millions of years (7). Although long regarded as being “junk DNA”, the discovery that the Pol III transcripts encoded by these SINEs are greatly increased upon various cell stresses led to the hypothesis that SINE RNAs may serve biological functions, thereby leading to the selection and maintenance of SINEs in the host cell genome (8). Moreover, recent work has revealed several interesting implications and potential functions of Alu and B2 RNAs embedded within Pol II mRNA transcripts. Therefore, Alu and B2 RNA have the potential to function in enhancing transcriptome diversity, as well as in controlling gene expression in trans as Pol III transcripts and in cis as part of Pol II transcribed mRNAs. This review will focus on the currently known functions and contributions to the host cell of both Pol II and Pol III transcribed Alu and B2 RNAs. The impact of SINEs on the host genome has been extensively reviewed elsewhere (4, 7, 9-12).
Biological Functions of Pol III Transcribed Alu and B2 RNAs
Alu RNA, the Pol III transcript, is ~300 nucleotides in length and consists of two 7SL-like arms joined by an A-rich linker and an A-rich tail on the 3’ end (13). B1 SINEs, also thought to have derived from 7SL RNA, are transcribed by Pol III into B1 RNA, which is ~135 nucleotides long and approximates the left arm of Alu (8, 14). B2 SINEs are derived from tRNA and encode the ~200 nucleotide B2 RNA (15). B1, B2, and Alu SINEs contain A and B boxes required for their transcription by the Pol III machinery. Transcription of Alu and B2 SINEs by Pol III is highly regulated by both cisand trans-regulatory factors (16, 17).
In human cells, full length, Pol III transcribed Alu RNA is present at low levels, however, upon physiological stresses Alu RNA levels significantly and transiently increase (18). The increase in Alu RNA transcripts upon heat-shock correlates with genome-wide chromatin remodeling, potentially increasing the accessibility of the internal Alu promoter elements (19). Correspondingly, B1 and B2 RNA transcripts increase in abundance upon heat shock in mouse cells (18, 20). Viral infection, ethanol treatment, and UV exposure have also been shown to increase Alu, B1 and B2 RNA levels. For example, infection of human cell lines with herpes simplex virus (HSV) resulted in increased transcription of Alu SINEs (17, 21, 22). The stimulation of transcription of Alu elements by HSV was presumably due to increased activity of transcription factor TFIIIC, which binds the B box in internal Alu promoters (17). Similarly, in mouse cells, minute virus of mice, a parvovirus, was shown to increase Pol III derived B1 and B2 RNAs: 24 hours post infection B1 and B2 RNA levels were significantly elevated and showed continued increase 72 hours post transfection (23). Transformation of mouse cells with simian virus 40 (SV40) has also been shown to increase the activity of TFIIIC resulting in heightened levels of B2 RNA (24). Transcriptional activation of SINEs by DNA damaging agents has also been reported. Specifically, prolonged exposure of up to 96 hours to the chemotherapeutics Cisplatin and Etoposide activated transcription of B1, B2 and Alu SINEs (25). B2 RNA also increased with UV exposure, and Alu and B2 RNAs increased with exposure to gamma radiation (25).
Because it is possible that the observed increases in SINE transcription in response to heat-shock or other stresses was an artifact of tissue culture and not biologically relevant, stress-induced SINE expression was analyzed in vivo (26). Analysis of heat-stressed mice revealed maximal increases in B1 and B2 RNAs between 1 and 3 hours of recovery, with a similar response by HSP70 mRNA. SINE RNA expression patterns were similar in tissue from liver, kidney, and spleen. Additionally, a marked increase in B1 and B2 RNAs was observed in response to ethanol intake while HSP27 was not induced (26). As the percent ethanol decreased during recovery, the expression of B1 and B2 RNAs also decreased.
The observations that the abundance of Alu and B2 RNAs increased in response to various stresses raised the possibility that these RNAs had biological functions. An initial indication that Alu RNA could serve a biological role came from the observation that Alu RNA can negatively regulate the activity of the dsRNA-activated protein kinase (PKR) (27). PKR plays a significant role in antiviral defense by phosphorylating eukaryotic initiation factor-2 alpha (eIF-2α), which results in translational inhibition (28). Overexpression of Alu RNA in cell-based assays resulted in increased expression levels of a reporter construct. Moreover, Alu RNA was shown to bind PKR and repress its activity (27).
A biological role for developmentally induced transcription of B2 SINEs has been determined. Tissue-specific transcription of a B2 SINE facilitates the temporal activation of genes within the mouse growth hormone locus by changing the local chromatin structure (29). The B2 SINE is bidirectionally transcribed by both Pol II and Pol III, the former of which occurs in a developmentally regulated fashion. Transcription by both polymerases is necessary and sufficient to change the growth hormone locus from a repressive heterochromatic environment to a more permissive euchromatic one such that developmentally pertinent genes can be expressed.
Transacting Repression of RNA Polymerase II Transcription by B2 and Alu RNAs
In eukaryotes, Pol II is the enzyme responsible for the synthesis of mRNA, working in concert with general transcription factors to drive promoter-specific transcription (30). Gene-specific Pol II transcription is tightly regulated by a multiplicity of protein activators, repressors, and co-activators, as well as DNA elements and chromatin structure (31, 32). More recently, non-coding RNAs (ncRNAs) have been identified as having regulatory roles at many different steps in the transcription process (33). Transcriptional repression by B2 and Alu RNAs were among the first examples of ncRNAs having a transacting regulatory function in the process of mRNA transcription. These discoveries provided strong support for the notion that Alu and B2 SINEs are more than simply parasites of the genome, by showing that they indeed have a function that likely led to their selection and maintenance.
Along with the increase in SINE RNA expression upon heat-shock, general transcription of Pol II transcribed genes decreases. For example, housekeeping genes such as beta-actin and hexokinase II show similar levels of reduction upon heat-shock (34, 35). Not all protein-encoding genes are repressed upon heat-shock; heat-shock protein transcripts, such as HSP70, show a robust increase. Alu and B2 RNAs were shown to be responsible for the general transcriptional repression of housekeeping genes in response to heat-shock. Knockdown of Alu RNA or B2 RNA in human or mouse cells, respectively, attenuated the decrease in mRNA levels of housekeeping genes seen upon heat-shock, while the HSP70 transcript was still robustly induced (34, 35).
A series of in vitro studies showed that Alu and B2 RNAs directly bind Pol II, are part of complexes assembled on promoter DNA, and potently inhibit RNA synthesis (34, 36). This led to the hypothesis that upon heat-shock, B2 and Alu RNAs assemble into complexes at the promoters of repressed genes, via their association with Pol II, where they block transcription. To test this hypothesis in cells, the occupancy of Pol II at heat-shock repressed genes was determined using Chromatin Immuno-precipitation (ChIP) assays. In response to heat-shock in both mouse and human cells, the occupancy of Pol II increased in the promoter regions of repressed genes and simultaneously decreased in the downstream regions of the genes (34). In order to detect the occupancy of Alu and B2 RNAs, a variation of the ChIP assay was utilized in which a biotinylated antisense oligonucleotide against either Alu or B2 RNA was used to pull down chromatin, and genomic regions of interest were then amplified by PCR. These experiments showed that the promoters of heat shock repressed genes contained Alu and B2 RNAs, whereas the promoter of the HSP70 gene did not (34). These data support a model in which Alu and B2 RNAs, upon heat-shock, repress mRNA transcription in trans by assembling with Pol II at the promoters of repressed genes (Figure 1). The transacting transcriptional repression by Alu and B2 RNAs is likely a common response to a variety of physiological stresses, but this idea remains to be explored experimentally. Additionally, future work will need to address how heat-shock induced genes are protected from repression by B2 and Alu RNAs, and how repression of housekeeping genes is alleviated as cells recover from stress.
Figure 1.
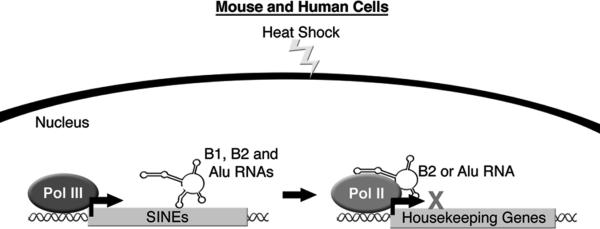
Upon heat shock, Pol III transcription of B1 and B2 RNAs in mouse cells and Alu RNA in human cells increases. B2 or Alu RNA can inhibit Pol II mRNA synthesis by entering complexes at the promoters of repressed genes.
Alu and B2 RNAs do not share any sequence similarity or obvious conserved secondary structures within their functional regions, but appear to have the same function in cells. Alu SINEs are thought to have the same ancestral origin as B1 SINEs; indeed the secondary structure of Alu RNA consists of two B1 RNA-like arms (13, 14, 37). In vitro, B1 RNA can tightly bind Pol II with a KD of 2 nM, however, unlike B2 RNA and Alu RNA does not repress transcription at low nM concentrations (34, 36). When Alu RNA is separated into its left and right B1-like arms, it was observed that much like B1 RNA, the left arm of Alu RNA binds Pol II but cannot potently repress transcription (34). By contrast, the right arm of Alu RNA functions much like B2 RNA in that it can both bind Pol II and repress transcription. These data suggest that Alu RNA has taken on the transcriptional functions of both B1 and B2 RNAs.
A Connection Between Alu RNAs and microRNAs
MicroRNAs (miRNAs) are ~22 nucleotide long noncoding RNAs that function as negative regulators of gene expression by targeting mRNAs for cleavage or translational repression (38). Some of the miRNAs encoded in human chromosome 19 miRNA cluster (C19MC) are thought to be transcribed by Pol III, essentially utilizing Alu as an upstream promoter element (39). Stimuli resulting in active transcription of the upstream Alu element may also result in active transcription of the downstream miRNA. The discovery that a subset of miRNAs from C19MC appear to target the most conserved regions of sense Alu RNA raises the intriguing possibility that these miRNAs could function as regulators of Alu RNA upon cell stress (40).
The Biological Functions of Alu RNAs Embedded in mRNA
An analysis of Alu SINE distribution in the genome revealed that nearly 75% of identified protein-encoding genes contained an Alu insertion with a strong preference for intronic regions (41). Alu insertions in gene rich regions result in a high level of Alu sequence in mRNA transcripts, referred to as embedded Alu RNA. Recently, embedded Alu RNAs have been shown to have an increasingly important role in RNA editing, gene silencing, and alternative splicing. It is not yet clear if embedded B2 RNA has the same biological impact as embedded Alu RNA.
Alu RNA as a Substrate for A-to-I Editing
By changing the RNA sequence relative to that of the genomic DNA sequence, RNA editing machinery has the capability to greatly enhance proteome diversity. A specific type of RNA editing, which has been shown to be crucial in proper development of vertebrates, is deamination of adenosine to produce inosine within dsRNA (42). A-to-I editing is catalyzed by a family of three identified enzymes known as Adenosine Deaminases that act on RNA (ADAR1-3). ADAR1 and ADAR2 are ubiquitously expressed while expression of ADAR3, whose editing activity has not yet been observed, is restricted to brain tissue (43). When adenosine is changed to inosine it is recognized by the cellular translation and splicing machinery as guanine, which can change the meaning of a codon or create an alternative splice site. Beyond this, A-to-I editing has the capacity to effect RNA stability through the creation of destabilizing I-U wobble pairs or stabilizing I-C pairs (42, 44). Changes in A-to-I editing have been correlated with various disease states including depression, cancer, and amytrophic lateral sclerosis (ALS) (45-47).
The lack of well-characterized targets for A-to-I editing led three groups to identify ADAR substrates in the human genome (44, 48, 49). Since inosine is recognized by reverse transcriptase as guanine in the RNA sequence, identification of edited sites can be made by comparing cDNA to genomic DNA and locating A-to-G substitutions. The three research groups independently identified Alu as the primary target of A-to-I editing with approximately 90% of editing occurring within Alu repeats and at multiple positions within a single repeat. 13,000-14,000 sites were identified as being edited in more than 1,400 genes (44, 48).
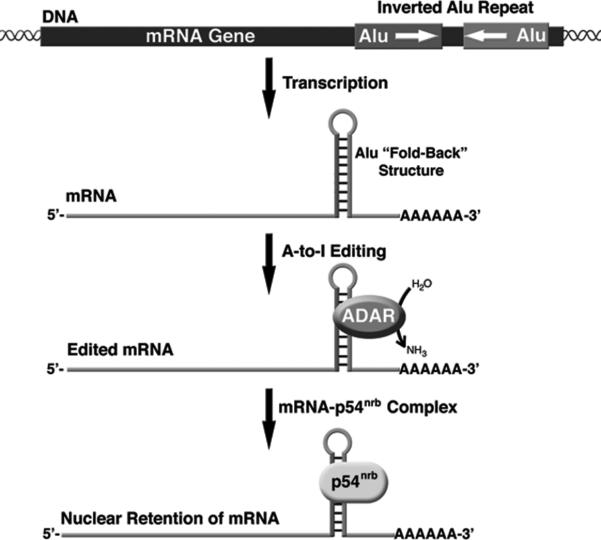
ADAR enzymes target base-paired adenosines within double stranded regions of the RNA (42). The propensity for embedded Alu RNAs to be a substrate for the ADAR enzymes is thought to be due to the ability of two nearby Alu SINEs that are in opposite orientations to anneal and give rise to an extended region of dsRNA (44, 48, 49). The vast number of Alu elements, particularly in gene rich regions, leads to a high number of inverted repeats and potential ADAR targets. The inverted repeats, generally within 2 kb, can form so called “fold-back” structures and are largely present in the introns and untranslated regions of the mRNA (Figure 2). There are hotspots for editing within Alu repeats, as well as specific adenosines that show a low degree of editing (49).
Figure 2.
Two nearby Alu sequences in an inverted orientation can form a “fold-back” structure in the 3’-UTR of a mRNA. This long stretch of dsRNA is an ideal substrate for adenosine deamination by the ADAR enzymes. The highly edited Alu RNA duplex is then bound by the RNA binding protein p54nrb and the mRNA is retained in the nucleus, thereby preventing translation.
Although ADARs target dsRNA without any apparent sequence specificity, analysis of the nucleotides neighboring edited adenosines revealed a preference for certain trinucleotide motifs within the Alu sequence. Specifically, TAG and AAG motifs are over-represented in edited trinucleotide sequences while GAN is significantly under represented (49). The editing patterns in different embedded Alu RNAs are rather diverse, which can be attributed in part to sequence divergence among repeats. Furthermore, when comparing tissue from different organs there is a wide range in the amount of editing detected in cDNA transcripts. In fact, the abundance of editing can vary from less than 1% of edited cDNA in blood, muscle, and pancreatic tissues to 8.2% and 12.8% in prostate and thymus tissues, respectively (49). Intriguingly, different regions of the brain show up to 3-fold differences in editing levels. The significant deviations in editing levels among various tissue types, indicates that the regulation of A-to-I editing is likely a complex process. The precise role of Alu RNA editing, given that almost all occurrences reside within non-coding regions of mRNAs, has yet to be fully expounded.
The mouse genome, although absent of Alu SINEs, is comprised of approximately the same portion of genomic repeats (3). In mice, the primary genomic repeats targeted for editing are embedded B1 and B2 RNAs (50). B1 SINEs were derived from the same ancestor as Alu, but A-to-I editing is at least an order of magnitude lower in mice than in humans (51). It has been proposed that embedded B1 and B2 RNAs are edited at lower levels largely due to their greater sequence divergence relative to that of Alu, thereby lowering their propensity to form long dsRNA regions (50).
Nuclear Retention of mRNA by Edited Alu Repeats
A function for A-to-I editing in gene silencing was revealed from studies of the mouse cationic amino-acid transporter 2 (CAT2) gene. CAT2 Transcribed Nuclear RNA (CTN-RNA) has an extended 3’-UTR, which contains inverted repeats of SINE origin that are extensively A-to-I edited (52). The extensive A-to-I editing of the inverted repeat strongly correlated with nuclear retention of the CTN-RNA. Although these repeats were not Alu sequences, this finding raised the possibility that highly edited inverted Alu repeats in a 3’-UTR may also result in nuclear retention of an mRNA and the consequential lack of translation. To address this, inverted Alu repeats were inserted into the 3’-UTR of an enhanced green fluorescent protein (EGFP) transgene (53). The inverted Alu repeats, from the Nicn1 and Lin28 genes, which were reported to be highly edited, dramatically repressed EGFP expression when present in the 3’-UTR. The mechanism for this repression was shown to be nuclear retention, which strongly correlated with A-to-I editing and association with the protein p54nrb (Figure 2). p54nrb was previously found to function in retaining dsRNAs in the nucleus and by binding to inosine-containing RNAs with high specificity (54). In total, 333 genes with inverted Alu repeats in the 3’-UTR were identified, indicating that nuclear retention by association with p54nrb could be an important mechanism of gene regulation (53).
Is There a Role for A-to-I editing of Pol III Transcribed B2 and Alu RNAs?
Although the identified targets of the ADAR enzymes have almost exclusively been repeats in Pol II transcribed mRNAs, it is worth speculating about the possibility that Pol III transcribed Alu and B2 RNAs are targets for A-to-I editing. We could foresee editing as a mechanism of regulating the transcriptional activities of Alu and B2 RNAs during cellular stresses. A recent study showed an increase in transcription of reporter plasmids when ADAR1 was expressed in HeLa and HEK293 cells (55). The increase in transcription appeared to be due to the editing activity of ADAR1 on an unknown RNA substrate. Presumably, this could either occur by an edited RNA activating transcription of the reporter plasmids, or ADAR1 down-regulating the activity of an RNA capable of repressing Pol II transcription. For the latter model, Pol III transcribed Alu and B2 RNAs are possible targets for regulation by A-to-I editing.
Embedded Alu RNA Can Cause Alternative Splicing
By creating multiple mRNA products from one gene, alternative splicing drastically increases transcriptome and proteome diversity. Potentially, 40-60% of human genes are alternatively spliced, with a significant portion, up to 30%, being tissue specific (56-58). Embedded Alu RNAs have been shown to contribute to alternative splicing (59, 60). Moreover, the role of embedded Alu RNA in alternative splicing suggests that Alu SINEs could have been a major driving force in human evolution by allowing different spliced forms of proteins to be sampled and the advantageous ones retained.
The consensus Alu RNA sequence, both sense and antisense, contains multiple putative donor and acceptor splice sites (Figure 3A) (61, 62). Therefore, an intronic Alu RNA has the potential to be exonized, incorporating a portion of the Alu sequence into the mature mRNA (60). Importantly, genes that contain an exonized Alu are predominately alternatively spliced, yielding isoforms with and without the Alu-derived exon (Figure 3B) (62). Given the vast abundance of Alu elements in gene rich regions, it is not surprising that an estimated 400 genes contain a portion of Alu sequence within their protein coding regions (63).
Figure 3.
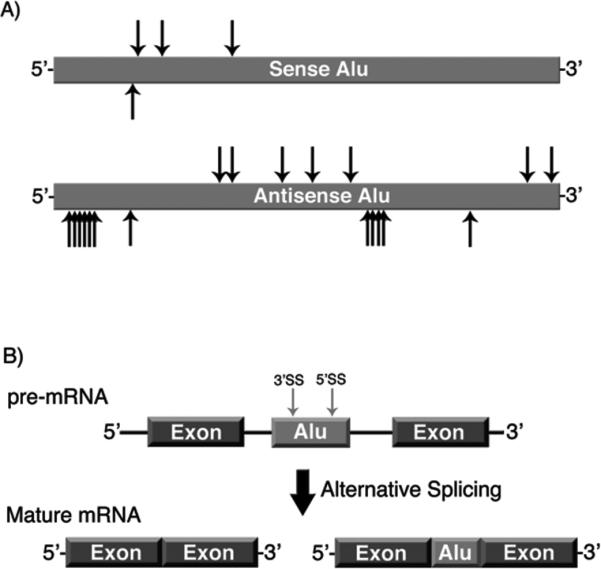
Embedded Alu RNAs can be alternatively spliced. A) Schematic of the potential splice sites in the sense and antisense Alu sequence (62). Arrows above the Alu elements indicate potential 5’ splice sites, arrows below the Alu elements indicate potential 3’ splice sites. B) Schematic illustrating exonization of an embedded antisense Alu RNA. Sequences within Alu RNA are recognized as splice sites, resulting in a mature mRNA isoform containing a portion of the Alu sequence.
Alu exonization in the ADAR2 gene is a widely studied example in which an inverted Alu sequence adds 40 amino acids to a protein isoform (60, 64). Using a minigene construct of ADAR2, it was revealed that a single nucleotide substitution within the Alu sequence transformed the alternatively spliced minigene into a nearly constitutively spliced isoform that included the Alu sequence (60). In a minigene of putative glucosyltransferase (PGT), the corresponding nucleotide substitution in a nonexonized Alu resulted in near constitutive Alu inclusion (60). This finding revealed that numerous intronic Alu RNAs might only be a single point mutation away from being constitutively exonized. These studies highlight both the potential of Alu-exonization to drive proteome diversity as well as the delicate balance between alternative and constitutive splicing of Alu-containing genes. Interestingly, the shift from alternative to constitutive inclusion of Alu sequence in the spliced ADAR2 minigene can also be achieved by deleting the left arm of Alu (65). Hence, it appears that the left arm of Alu in Pol II transcribed RNAs aids in maintaining alternative splicing, while it is the right arm of Alu in Pol III transcribed RNAs that is functional as a mRNA transcriptional repressor (34). Together, this provides support for the maintenance of the dimeric structure of Alu throughout human evolution.
Concluding Remarks
Collectively, the studies described in this review provide insight as to why Alu and B2 SINEs have been maintained throughout millions of years of evolution and highlight the intricate relationship between their encoded RNAs and the cell. Functioning as transacting regulators of gene expression, Pol III transcribed Alu and B2 RNAs can interact with Pol II and repress mRNA transcription. Inverted Alu repeats embedded in mRNA transcripts are targets for A-to-I editing, and when present in the 3’-UTR have the potential to silence gene expression. Moreover, embedded Alu RNAs exhibit a delicate balance in the regulation of splicing, having the capacity to increase proteome diversity along with the potential to cause deleterious mutations. Future studies will continue to elucidate the full contribution of SINEs and their transcripts to controlling cellular function, development, and evolution.
Acknowledgements
This work was funded by a Public Health Science grant from the National Institute of General Medical Sciences (R01 GM068414). R.D.W. was supported in part by NIH Predoctoral Training Grant T32 GM08759.
References
- 1.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 5.Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J. Mol. Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 6.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 7.Deininger PL, Batzer MA. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 8.Schmid CW. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 10.Kazazian HHJ. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 11.Beauregard A, Curcio MJ, Belfort M. The Take and Give Between Retrotransposable Elements and Their Hosts. Annu. Rev. Genet. 2008 doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- 13.Sinnett D, Richer C, Deragon JM, Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J. Biol. Chem. 1991;266:8675–8678. [PubMed] [Google Scholar]
- 14.Labuda D, Sinnett D, Richer C, Deragon JM, Striker G. Evolution of mouse B1 repeats: 7SL RNA folding pattern conserved. J. Mol. Evol. 1991;32:405–414. doi: 10.1007/BF02101280. [DOI] [PubMed] [Google Scholar]
- 15.Daniels GR, Deininger PL. Repeat sequence families derived from mammalian tRNA genes. Nature. 1985;317:819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- 16.Aleman C, Roy-Engel AM, Shaikh TH, Deininger PL. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 2000;28:4755–4761. doi: 10.1093/nar/28.23.4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang KL, Latchman DS. The herpes simplex virus immediate-early protein ICP27 stimulates the transcription of cellular Alu repeated sequences by increasing the activity of transcription factor TFIIIC. Biochem. J. 1992;284:667–673. doi: 10.1042/bj2840667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene. 2001;276:127–133. doi: 10.1016/s0378-1119(01)00639-4. [DOI] [PubMed] [Google Scholar]
- 20.Fornace AJJ, Alamo IJ, Hollander MC, Lamoreaux E. Induction of heat shock protein transcripts and B2 transcripts by various stresses in Chinese hamster cells. Exp. Cell Res. 1989;182:61–74. doi: 10.1016/0014-4827(89)90279-6. [DOI] [PubMed] [Google Scholar]
- 21.Panning B, Smiley JR. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J. Mol. Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 22.Panning B, Smiley JR. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology. 1994;202:408–417. doi: 10.1006/viro.1994.1357. [DOI] [PubMed] [Google Scholar]
- 23.Williams WP, Tamburic L, Astell CR. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology. 2004;327:233–241. doi: 10.1016/j.virol.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 24.White RJ, Stott D, Rigby PW. Regulation of RNA polymerase III transcription in response to Simian virus 40 transformation. EMBO J. 1990;9:3713–3721. doi: 10.1002/j.1460-2075.1990.tb07584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- 26.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 27.Chu WM, Ballard R, Carpick BW, Williams BR, Schmid CW. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 1998;18:58–68. doi: 10.1128/mcb.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 30.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 31.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 32.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 33.Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit. Rev. Biochem. Mol. Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 36.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 37.Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- 38.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 40.Lehnert S, Van Loo P, Thilakarathne PJ, Marynen P, Verbeke G, Schuit FC. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS ONE. 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 42.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 45.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol. Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 48.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 53.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 55.Gommans WM, Maas S. Characterization of ADAR1-mediated modulation of gene expression. Biochem. Biophys. Res. Commun. 2008;377:170–175. doi: 10.1016/j.bbrc.2008.09.109. [DOI] [PubMed] [Google Scholar]
- 56.Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- 57.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G. Intronic Alus influence alternative splicing. PLoS Genet. 2008;4:e1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 61.Makalowski W, Mitchell GA, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 62.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12(7):1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 64.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gal-Mark N, Schwartz S, Ast G. Alternative splicing of Alu exons--two arms are better than one. Nucleic Acids Res. 2008;36:2012–2023. doi: 10.1093/nar/gkn024. [DOI] [PMC free article] [PubMed] [Google Scholar]