Abstract
Effectors are essential virulence proteins produced by a broad range of parasites, including viruses, bacteria, fungi, oomycetes, protozoa, insects and nematodes. Upon entry into host cells, pathogen effectors manipulate specific physiological processes or signaling pathways to subvert host immunity. Most effectors, especially those of eukaryotic pathogens, remain functionally uncharacterized. Here, we show that two effectors from the oomycete plant pathogen Phytophthora sojae suppress RNA silencing in plants by inhibiting the biogenesis of small RNAs. Ectopic expression of these Phytophthora suppressors of RNA silencing enhances plant susceptibility to both a virus and Phytophthora, showing that some eukaryotic pathogens have evolved virulence proteins that target host RNA silencing processes to promote infection. These findings identify RNA silencing suppression as a common strategy used by pathogens across kingdoms to cause disease and are consistent with RNA silencing having key roles in host defense.
Oomycetes are a group of microbial eukaryotes that include important pathogens of plants and animals. The genus Phytophthora contains many notorious pathogens of crops. For example, the potato pathogen Phytophthora infestans triggered the Irish Famine in the nineteenth century and remains a serious problem worldwide, and P. sojae causes millions of dollars of losses annually in soybean. Until now, battling oomycete-related diseases has been challenging owing to a lack of understanding of pathogenesis.
Compared to infection by viral or bacterial pathogens, oomycete infection entails more complex defense-counterdefense crosstalk. This is reflected by the hundreds of effectors predicted from oomycete genomes1–3. The majority of these effectors have a conserved N-terminal RXLR motif, where X represents any amino acid, which mediates their intake into host cells after being secreted from the pathogens4,5. Although several effectors have been shown to suppress plant defense6, the functions of the vast majority of eukaryotic effectors remain unknown.
RNA silencing is a universal gene regulation mechanism in eukaryotes that affects many processes. Central players in RNA silencing are small RNAs of 20–30 nucleotides in length that guide the sequence-specific repression of target genes. Plants produce two major types of small RNAs, microRNAs (miRNAs) and small interfering RNAs (siRNAs)7. miRNAs are encoded by endogenous MIR family genes8, whereas siRNAs are derived from invading nucleic acids, such as viruses and transgenes, and from endogenous loci, such as repeats, transposable elements and genes7.
RNA silencing serves as a major defense mechanism against RNA viruses in plants and invertebrates9,10. Viral infection of a host induces siRNAs, which guide cleavage of viral RNAs. As a counterdefense, viral suppressors of RNA silencing (VSRs) enable efficient infection by interfering with host silencing. Small RNA–mediated post-transcriptional regulation has also been implicated in antibacterial plant defense11,12. Furthermore, three bacterial effectors can suppress the miRNA pathway in Arabidopsis thaliana13. However, whether RNA silencing regulates defense against eukaryotic pathogens remains unknown. If so, these pathogens might have evolved virulence strategies to disrupt host RNA silencing machinery.
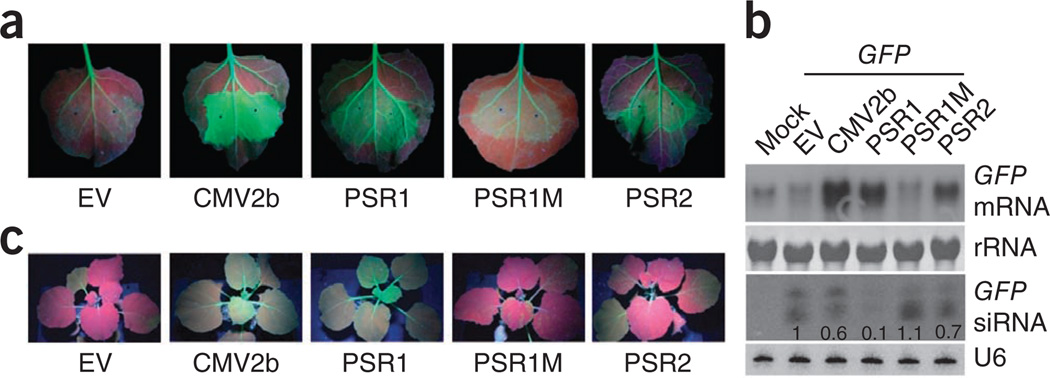
We searched for Phytophthora effectors that suppress RNA silencing in plants. Individual effector and green fluorescent protein (GFP) genes were coexpressed by Agrobacterium tumefaciens infiltration in the leaves of Nicotiana benthamiana 16c, which constitutively expresses GFP under the control of the cauliflower mosaic virus 35S promoter14. This system provides a convenient assay for the suppression of siRNA-mediated transgene silencing, as both endogenous and exogenous GFP genes are silenced by siRNAs induced by the infiltrated GFP, resulting in no or very low green fluorescence in the infiltrated zone (Fig. 1a). However, strong fluorescence can be observed if a protein capable of suppressing siRNA-mediated silencing, such as cucumber mosaic virus protein 2b (CMV2b)15, is coexpressed with GFP. Using this assay, we screened 59 P. sojae RXLR effectors (out of the approximately 400 predicted effectors with the RXLR domain) and found that PsAvh18 and PsAvh146 suppress GFP silencing (Fig. 1a). Both effectors are expressed during P. sojae infection of soybean (Supplementary Fig. 1) and are therefore designated Phytophthora suppressors of RNA silencing 1 and 2 (PSR1 and PSR2, respectively).
Figure 1.
P. sojae RXLR effectors PSR1 and PSR2 suppress transgene-mediated GFP silencing in GFP-transgenic N. benthamiana 16c plants. (a) Local RNA silencing suppression in N. benthamiana 16c transiently expressing GFP and PSR1, PSR2, PSR1M or CMV2b. Empty vector pEG100 (EV) was used as a negative control. Pictures were taken 3 d after Agrobacterium infiltration. (b) Accumulation of GFP mRNA and siRNA in infiltrated N. benthamiana 16c leaves. Samples from leaves without Agrobacterium infiltration (mock) were used as negative controls. Numbers below the siRNA blot represent the relative abundance of GFP siRNA, with the level in the leaves expressing only GFP set to 1. U6 and rRNA were used as loading controls. (c) PSR1 but not PSR2 or PSR1M suppresses systemic RNA silencing. Pictures were taken 15 d after the basal leaves of N. benthamiana 16c were infiltrated by Agrobacterium harboring 35S::GFP and either empty vector (EV) or vector expressing PSR1, PSR1M, PSR2 or CMV2b from the 35S promoter. This experiment was repeated at least three times with similar results.
We next examined the abundance of GFP siRNA and mRNA in N. benthamiana 16c leaves coexpressing 35S::GFP and the PSRs. RNA blots (Fig. 1b and Supplementary Fig. 2) showed that PSR1 expression strongly reduced the abundance of GFP siRNA, leading to increased accumulation of GFP transcript. PSR2 expression also resulted in lower levels of GFP siRNA, although the reduction was moderate compared to that caused by PSR1. Small RNA–mediated gene silencing can be established in systemic tissues, with small RNAs likely being the signals that move systemically16. Because PSR1 strongly inhibits siRNA biogenesis, it might also suppress systemic silencing. Indeed, like the viral effector CMV2b, PSR1 suppressed the systemic silencing of GFP in newly emerged leaves when it was coexpressed with GFP in a basal leaf of N. benthamiana 16c (Fig. 1c and Supplementary Fig. 3). PSR2 did not suppress systemic silencing, probably owing to its weaker effect on siRNA accumulation.
Sequence analysis showed the presence of a putative bipartite nuclear localization signal (NLS)17 in PSR1 (Supplementary Fig. 4a). Expression of PSR1 fused with yellow fluorescent protein (PSR1-YFP) in N. benthamiana confirmed that PSR1 protein localizes predominantly to plant nuclei, and substitution of the NLS residues with alanines abolished nuclear localization (Supplementary Fig. 4b). Notably, the mutant lacking the NLS (PSR1M) also largely lost its ability to suppress RNA silencing (Fig. 1 and Supplementary Fig. 3), suggesting that nuclear localization is required for PSR1 activity.
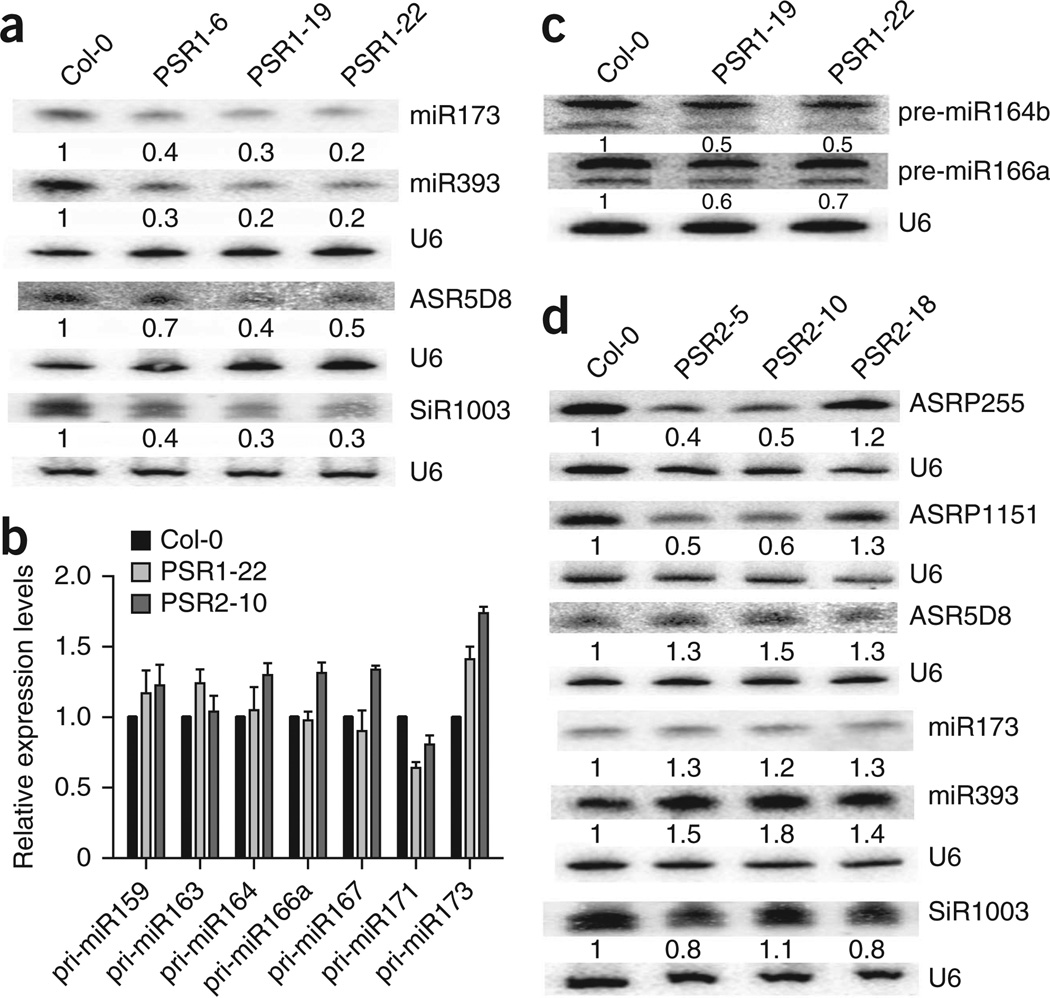
Many VSRs suppress RNA silencing by directly binding to small RNAs18. In vitro assays showed that PSR1 and PSR2 do not bind single-stranded or double-stranded 21-nt small RNAs (Supplementary Fig. 5). Therefore, they likely affect small RNA biogenesis, not activity. We examined the impact of PSRs on small RNA biogenesis using transgenic Arabidopsis expressing PSR1 or PSR2 (Supplementary Fig. 6). Each of three independent 35S::PSR1-YFP transgenic Arabidopsis lines had strong morphological phenotypes (Supplementary Fig. 7), including leaf shape changes, small stature and reduced fertility. Some of these phenotypes are also observed in miRNA biogenesis mutants19,20. Indeed, examination of representative miRNAs and endogenous siRNAs showed an across-the-board reduction in their abundance (Fig. 2a and Supplementary Fig. 7). Corresponding to the lower levels of small RNAs, transcripts from a few miRNA and siRNA target genes accumulated to higher levels in PSR1-expressing plants (Supplementary Fig. 8), confirming that PSR1 has a general impact on small RNA biogenesis. As specific small RNAs have been implicated in regulating plant immunity, PSR1 might promote infection by manipulating these defense-regulating small RNAs.
Figure 2.
Effects of PSR1 and PSR2 on small RNA biogenesis in Arabidopsis. (a) RNA blotting showing lower levels of two representative miRNAs, one ta-siRNA (ASR5D8) and one hc-siRNA (SiR1003) in wild-type (Col-0) and three independent PSR1-expressing Arabidopsis lines. (b) Transcript abundance of pri-miRNAs from various miRNA genes in wild-type and PSR1- or PSR2-expressing lines as determined by RT-PCR. Error bars, s.e.m. from two experimental replicates. (c) RNA blotting showing lower levels of pre-miR164b and pre-miR166a in two PSR1-expressing lines. The two bands probably both represent pre-miRNA species. Relative expression levels were analyzed using the prominent upper bands. (d) Accumulation of three representative ta-siRNAs (ASRP255, ASRP1151 and ASR5D8 from TAS1, TAS2 and TAS3 loci, respectively), two miRNAs and one hc-siRNA (SiR1003) in wild-type and three independent PSR2-expressing transgenic lines. Changes in siRNA abundance were not evident in the transgenic line PSR2-18 because of the low expression level of PSR2. U6 served as a loading control. Numbers below the blots represent the relative abundance of the small RNAs with the levels in wild-type plants set to 1.
We further investigated the specific step(s) during miRNA biogenesis that is disrupted by PSR1 by examining the abundance of primiRNAs and pre-miRNAs. Pri-miRNAs are the primary transcripts from miRNA genes, and pre-miRNAs are the products of DICERLIKE1 (DCL1)-mediated processing of pri-miRNAs7. Quantitative RT-PCR showed that the levels of pri-miRNAs were either not affected or were slightly higher for different miRNA genes in PSR1-expressing plants relative to wild-type plants (Fig. 2b). However, a mild but consistent reduction in abundance was observed for two tested pre-miRNAs (Fig. 2c), suggesting that PSR1 inhibits DCL1-mediated processing of pri-miRNAs. Because DCL1 also performs the subsequent biogenesis step to produce miRNAs from pre-miRNAs, the larger reduction in mature miRNA levels is likely due to inhibition of both DCL1-mediated processing steps by PSR1. PSR1 also affects the levels of endogenous siRNAs, which require the activity of DCL2 and DCL4 for their biogenesis. Therefore, PSR1 may target multiple DCLs or common DCL cofactor(s)21,22 that are responsible for both miRNA and siRNA biogenesis.
Obvious abnormalities in developmental phenotypes or alterations in the levels of eight representative miRNAs were not seen for 35S::PSR2-Flag transgenic Arabidopsis plants (Fig. 2d and Supplementary Fig. 9). Further investigation of siRNAs showed that, although PSR2 did not affect 24-nt heterochromatic siRNAs (hc-siRNAs) (Fig. 2d and Supplementary Fig. 9), the levels of two 21-nt trans-acting siRNAs (ta-siRNAs) were lower in PSR2-expressing plants (Fig. 2d). ta-siRNAs are secondary siRNAs generated from miRNA-targeted noncoding transcripts from TAS loci. miRNAs trigger cleavage of TAS transcripts through the endonucleolytic activity of ARGONAUTE1 (AGO1), and the cleavage products then serve as precursors for ta-siRNAs made through the activities of RNA DEPENDENT RNA POLYMERASE6 (RDR6) and DCL4 (ref. 7). Although miR173 levels were not affected, the levels of ASRP255 and ASRP1151 ta-siRNAs, whose biogenesis requires miR173, were lower in PSR2-expressing plants (Fig. 2d). Moreover, PSR2 only affects specific ta-siRNAs, as miR390-mediated TAS3 ta-siRNA accumulation was unaffected (Fig. 2d). We further found that PSR2 does not interfere with the slicer activity of AGO1 at the initial step of ta-siRNA biogenesis (Supplementary Fig. 10); therefore, it likely targets the accumulation of specific ta-siRNA species at a downstream step(s).
In contrast to the broad inhibitory activity of PSR1 on the biogenesis of both miRNAs and siRNAs, the activity of PSR2 specifically controls ta-siRNA accumulation through a different mechanism. Therefore, these effectors could facilitate Phytophthora infection in a synergistic manner. ta-siRNAs were recently found to regulate plant nucleotide binding–leucine-rich repeat (NB-LRR) genes23,24, which are canonical pathogen resistance genes. By suppressing the ta-siRNA pathway, PSR2 might disrupt the expression of NB-LRR and other defense-related genes, leading to host damage and misregulation of defense responses. Because PSR2 is only expressed late in infection (Supplementary Fig. 1), it could also be involved in regulating the transition from biotrophic to necrotrophic growth in Phytophthora by triggering host cell death.
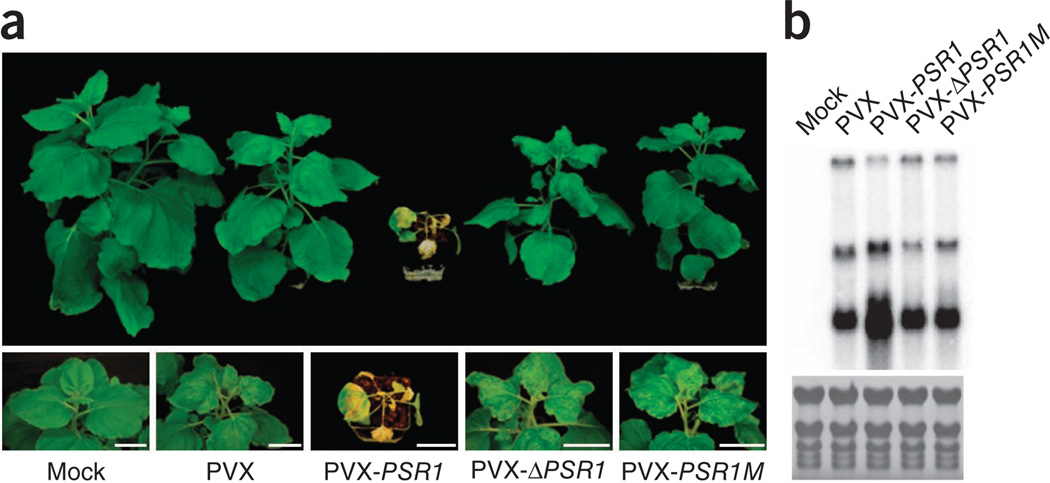
To test whether PSR1 and PSR2 are capable of suppressing siRNA-mediated host immunity, we introduced PSR1 and PSR2 into the potato virus X (PVX) genome and examined their effects on viral virulence. Unlike N. benthamiana plants infected with wild-type PVX, plants infected with PVX-PSR1 showed necrosis on newly emerged organs (leaves and stem) from the shoot apex, which led to plant death (Fig. 3a). Consistent with enhanced disease symptoms, viral RNAs accumulated to a much higher level in PVX-PSR1–infected tissues (Fig. 3b and Supplementary Fig. 11). Furthermore, PVX-PSR1M, which encodes the PSR1 mutant lacking its NLS, caused a weak disease symptom similar to wild-type PVX, suggesting that the RNA silencing suppression activity of PSR1 is responsible for facilitating PVX infection. We also confirmed that the enhanced virulence of PVX-PSR1 involved PSR1 activity as a protein by showing that PVX-ΔPSR1, which carries a stop codon at the beginning of PSR1, exhibited similar virulence to wild-type PVX (Fig. 3b and Supplementary Fig. 11). PVX-PSR2 also enhanced disease symptoms and viral RNA accumulation relative to wild-type PVX (Supplementary Fig. 12), although to a lesser extent than PVX-PSR1. These data suggest that PSRs promote viral infection, likely through their RNA silencing suppression activities.
Figure 3.
PSR1 promotes the infection of N. benthamiana by PVX. (a) Disease symptoms of plants infiltrated with Agrobacterium carrying wild-type PVX or PVX with PSR1, PSR1M or ΔPSR1 expressed from the 35S promoter. Plants infected with wild-type PVX, PVX-PSR1M or PVX-ΔPSR1 had mild disease symptoms, whereas PVX-PSR1 induced severe necrosis on the apex of new tissues (leaves and stem), leading to plant death. Pictures were taken 21 d after Agrobacterium infiltration. (b) RNA blotting showing the accumulation of PVX genomic and subgenomic RNAs 4 d after infection. Uninfected plants (mock) were used as negative controls. This experiment was repeated twice with similar results.
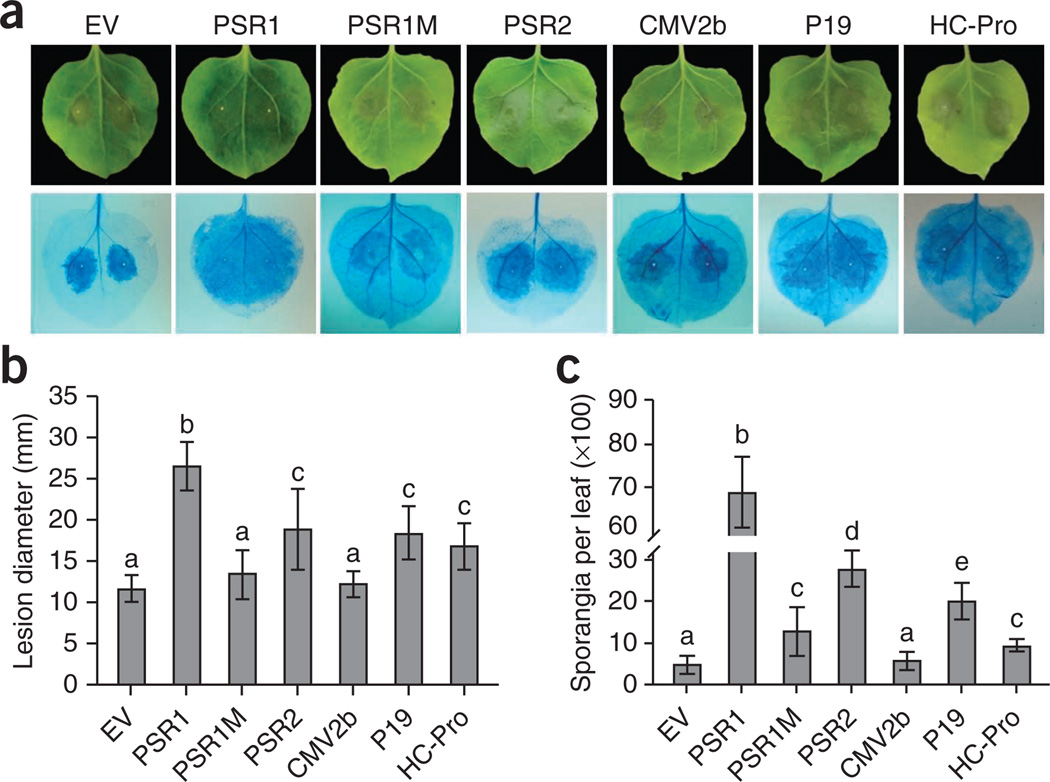
We next determined whether PSRs promote Phytophthora infection by inoculating N. benthamiana leaves expressing PSR1 or PSR2 with P. infestans. The genome sequence of P. infestans contains a homolog of PSR2 (PITG_15152) but no close relative of PSR1. Six days after inoculation by zoospores, we observed enlarged lesions and markedly higher sporangia numbers on leaves expressing PSR1 or PSR2 relative to plants receiving empty vector (Fig. 4 and Supplementary Fig. 13). PSR1-mediated susceptibility relied on its RNA silencing suppression activity because PSR1M had substantially less ability to promote Phytophthora infection than wild-type PSR1. The greater disease promotion activity of PSR1 compared to PSR2 is consistent with its stronger RNA silencing suppression activity but may also be partly due to the presence of the PSR2 homolog in P. infestans.
Figure 4.
Expression of RNA silencing suppressors in N. benthamiana enhances infection by P. infestans. (a) Disease symptoms (top) and lesions visualized using trypan blue staining (bottom) on N. benthamiana leaves expressing Phytophthora or viral effectors and then inoculated with zoospores of P. infestans strain 1306. Empty vector pEG100 (EV) was used as a negative control. Pictures were taken 6 d after P. infestans inoculation. (b) Sizes of lesions caused by P. infestans infection on leaves expressing Phytophthora or viral effectors. (c) Numbers of newly formed P. infestans sporangia on leaves expressing Phytophthora or viral effectors. Error bars in b and c, s.e.m.; different letters indicate values that are statistically different (P < 0.01). These experiments were repeated three times with similar results.
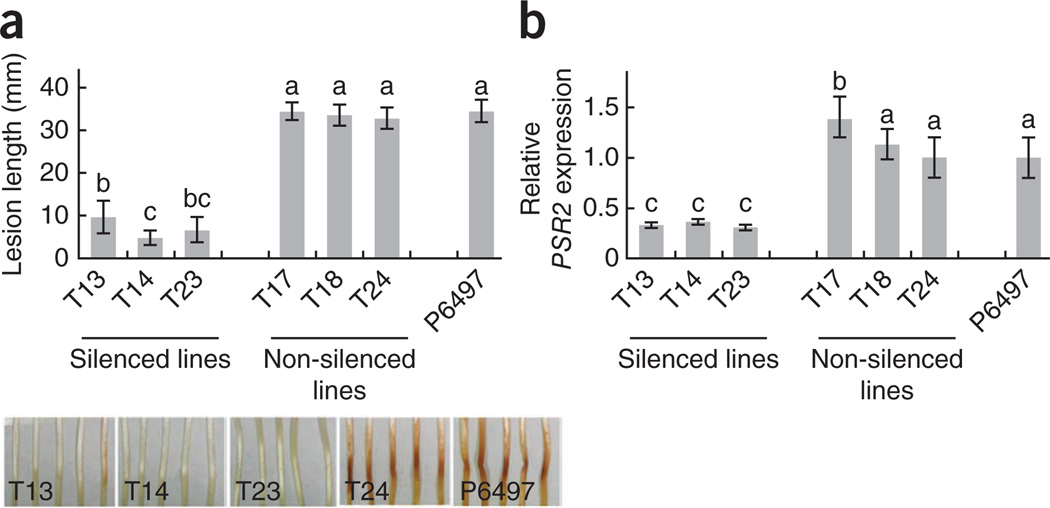
To further confirm that PSRs act as virulence factors during Phytophthora infection, we generated PSR2-silenced P. sojae mutants (we could not silence PSR1, despite numerous trials). Independent PSR2-silenced strains uniformly exhibited significantly decreased virulence when infecting soybean seedlings (Fig. 5). Taken together, our data strongly suggest that PSRs are important virulence factors during Phytophthora infection.
Figure 5.
Silencing of PSR2 in P. sojae impairs virulence in soybean. Hypocotyls from 4-d-old soybean seedlings (cultivar Hefeng47) were infected with P. sojae P6497 (wild type), non-silenced P. sojae transformants and three independent PSR2-silenced lines. (a) Lesion lengths (top) and disease symptoms (bottom) in etiolated hypocotyls 36 h after infection. Means and s.e.m. from at least ten measurements are presented. (b) Relative transcript levels of PSR2 in the silenced lines determined by RT-PCR. The expression of the P. sojae actA gene served as the internal standard. Error bars, s.e.m. from two independent RNA isolations; letters above each column represent statistically significant differences (P < 0.01). This experiment was repeated twice with similar results.
We also investigated the general function of RNA silencing suppression in promoting Phytophthora infection by testing plants expressing three VSRs (CMV2b, P19 from tomato bushy stunt tombusvirus and HC-Pro from potyviruses). Notably, P19 and HC-Pro also enhanced the infection of N. benthamiana by P. infestans (Fig. 4), suggesting that interfering with RNA silencing in general causes greater plant susceptibility to Phytophthora.
Pathogens depend on a multitude of effectors to subvert host immunity. This study shows that oomycete pathogens have evolved effectors to facilitate infection by suppressing host RNA silencing. The fact that the RNA silencing process is targeted by multiple Phytophthora effectors reflects its key role in immunity to oomycete, and this knowledge sets the foundation to enhance resistance against these devastating diseases.
Basic RNA silencing processes are conserved in plant and mammalian systems. Furthermore, a similar host-targeting signal is present in effectors from animal parasites (such as the malaria pathogen Plasmodium spp.), suggesting an evolutionarily conserved means for delivering virulence proteins that affect host immunity4,5,25. Our discovery warrants further efforts to identify and characterize RNA silencing suppressors produced by eukaryotic pathogens that infect mammals.
ONLINE METHODS
Plants, microbial strains and growth conditions
Soybean (Glycine max) and N. benthamiana plants were grown in a temperature-controlled greenhouse. Bacterial and oomycete strains and constructs are listed in Supplementary Table 1. P. sojae strain P6497 was grown on 10% V8 medium at 25 °C in the dark. P. sojae transformants (for PSR2 silencing) were grown on 10% V8 medium supplemented with G418 (10 µg/ml; Sigma). P. infestans isolate 1306 was maintained on rye sucrose agar plates at 18 °C. A. tumefaciens strains26 were grown on LB agar plates supplemented with 50 µg/ml kanamycin, 50 µg/ml rifampicin, 50 µg/ml gentamycin and 5 µg/ml tetracycline when necessary.
RNA silencing suppression assays using Agrobacterium-mediated transient expression in N. benthamiana 16c plants
cDNA fragments, excluding the sequences encoding the N-terminal signal peptides, from 59 RXLR effectors were amplified from P. sojae isolate P6497 (ref. 27) using gene-specific primers (Supplementary Table 2). PCR products were cloned into the Gateway entry vector pENTR1A (Invitrogen) and then into the destination vector pEG100 (ref. 28). Plasmids were transformed into A. tumefaciens strain C58C1 (pCH32), and the resulting strains were used for transient expression in N. benthamiana using previously described protocols29.
Fully expended leaves of N. benthamiana 16c plants at the six-leaf stage were infiltrated with Agrobacterium strains carrying 35S::GFP and individual effecter gene constructs. Green fluorescence was visualized using a handheld long-wavelength UV lamp (Blak-Ray B-100AP, Ultraviolet Products). Agrobacterium carrying the empty vector pEG100 and a construct expressing CMV2b from the 35S promoter30 were used as negative and positive controls, respectively.
GFP mRNA and siRNA were examined 3 d after Agrobacterium infiltration in the infiltrated leaf areas by RNA blotting. The abundance of GFP transcripts was determined using radiolabeled random priming probes. The GFP siRNA probe was generated using the MEGAScript high-yield T7 kit (Ambion) in the presence of [α-32P] UTP. U6 served as a loading control.
Expression of PSRs during P. sojae infection of soybean roots
Soybean seeds (G. max cultivar Williams 82) were pregerminated31 and grown in growth pouches (Mega International) wetted with 10 ml of distilled water. The pouches were kept in a growth chamber with constant 22 °C, 90% humidity and a 16-h photoperiod. Young roots of 8-d-old seedlings were inoculated with hyphal plugs of P. sojae isolate P6497 grown on 10% V8 medium. Transcript levels of PSR1 and PSR2 in the infected tissues were analyzed by semiquantitative RT-PCR using gene-specific primers (Supplementary Table 3) during a time course from 0–24 h after inoculation.
Small RNA binding assays for PSRs
PSR1, PSR2 and P19 genes were cloned into the Escherichia coli expression vector pGEX4T-2 (GE Healthcare Life Science), and glutathione S-transferase (GST)-tagged fusion proteins were purified using immobilized glutathione (Thermo Scientific). Equal amounts of soluble protein were then tested for small RNA binding by electrophoretic mobility shift assays (EMSAs).
Small 21-nt RNA oligonucleotides (Supplementary Table 3) were synthesized and radiolabeled with [γ-32P] ATP. Double-stranded small RNAs were produced by heating a mixture of equimolar complementary single-stranded oligonucleotides in annealing buffer containing 50 mM Tris-HCl (pH 7.5) and 100 mM NaCl at 99 °C for 5 min and cooling to room temperature. We incubated 1 pmol of radiolabeled single-stranded or double-stranded small RNA with 1 µg of purified proteins in binding buffer (20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 300 mM NaCl, 0.1% NP-40 and protease inhibitor cocktail (Roche)) for 40 min at room temperature. Protein-RNA complexes were resolved on 6% native polyacrylamide gels and visualized using a Typhoon phosphorimager.
Subcellular localization of PSR1 in plant cells
Genes encoding PSR1 and PSR1M (a mutant of PSR1 in which all 16 residues of a putative NLS motif were replaced with alanines) were cloned into the vector pEG101 (ref. 28) to generate C-terminal YFP fusion proteins. Fusion proteins were expressed in 3-week-old N. benthamiana leaves by Agrobacterium infiltration, and their localization in plant cells was determined using a Leica SP2 Laser Scanning Confocal Microscope (Leica Microsystems) 48 h after infiltration.
Small RNA analysis in Arabidopsis transgenic lines overexpressing PSRs
A. thaliana eco. Col-0 plants were transformed with A. tumefaciens strain GV3101 carrying pEG101::PSR1 or pEG100::PSR2-Flag by the floral-dip method32. The abundance of miRNAs and siRNAs in three independent transgenic lines (per construct) was examined by RNA blotting. RNA isolation and blotting and the detection of pre-miR166a and pre-miR164b were performed as previously described33–35. cDNA was synthesized from 3 µg of total RNA using reverse transcriptase (Fermentas) and an oligo(dT) primer. RT-PCR was carried out using gene-specific primers (Supplementary Table 3). Quantitative RT-PCR on small RNA target transcripts was performed in triplicate on a Bio-Rad iQ cycler apparatus with iQ SYBR Green Supermix (Bio-Rad).
In vitro transcription and RNA cleavage assays
AGO1 immunoprecipitation from PSR2-expressing or wild-type Arabidopsis plants was performed as described previously36. The TAS1C fragment was amplified by PCR from the genomic DNA of wild-type Arabidopsis using the TAS1CF and TAS1CR primers (Supplementary Table 3). In vitro transcription was performed by incubating 800 ng of DNA in 25-µl reactions with T7 RNA polymerase (Promega) and [α-32P] UTP at 37 °C for 1.5 h. Labeled TAS1C RNA fragment was gel purified and dissolved in 50 µl of nuclease-free water. We added 3 µl of the labeled probe to a 25-µl cleavage reaction mix containing 20 µl of AGO1 immune complexes in RISC buffer (40 mM HEPES (pH 7.4), 100 mM potassium acetate, 5 mM magnesium acetate and 4 mM DTT) supplemented with 1 µl of 25 mM ATP and 1 µl of RNase inhibitor (Fermentas). The reaction mix was incubated at 37 °C for 1.5 h, and RNAs were resolved on an 8 M urea/5% polyacrylamide gel and detected using a Typhoon phosphorimager.
PVX infection assays
PCR products of PSR1, PSR1M, Δ PSR1 (with point mutations generating a stop codon three amino acids into the PSR1 ORF), PSR2 and Δ PSR2 (with point mutations generating a stop codon eight amino acids into the PSR2 ORF) were ligated into the pGR106 vector37, which carries the full PVX genome. Recombinant plasmids were transformed into A. tumefaciens strain GV3101, and the resulting strains were then used to infiltrate 3-week-old wild-type N. benthamiana plants. Total RNA was extracted 4 d after inoculation from plants infected with PVX, PVX-PSR1, PVX-PSR1M or PVX-Δ PSR1 and 21 d after inoculation from plants infected with PVX, PVX-PSR2 or PVX-Δ PSR2. Viral RNAs were detected by probes corresponding to the PVX coat protein–encoding gene (CP).
P. infestans infection assays
PSR1, PSR1M, PSR2, CMV2b, P19 and HC-Pro were transiently expressed in wild-type N. benthamiana plants. Twenty-four hours after Agrobacterium infiltration, infiltrated leaves were detached from the plants and inoculated with 30 µl of zoospores suspension (containing approximately 1,000 zoospores) of P. infestans isolate 1306. Inoculated leaves were incubated in a growth chamber at 18 °C for 6 d before disease progression was analyzed. Newly formed sporangia were washed from each inoculated leaf and counted under a microscope. Lesions were visualized after trypan blue staining38, and the size of the lesion on each leaf was measured. None of the effectors caused visible tissue damage when expressed in N. benthamiana without the subsequent P. infestans infection (Supplementary Fig. 14), excluding the possibility that enhanced susceptibility could be due to potential cytotoxic effects from these effectors. The experiment was repeated three times. In each experiment, leaves from 6–10 plants were analyzed for each treatment.
Construction and characterization of PSR2-silenced P. sojae mutants
A 160-nt region within the PSR2 gene was PCR amplified using gene-specific primers (Supplementary Table 3). The PCR product was used as a template to synthesize double-stranded RNA in vitro using the MEGAscript RNAi kit (Invitrogen). PSR2-targeting double-stranded RNA was then introduced into P. sojae isolate P6497 using a previously described transformation protocol39. The pTH209 vector40 was used as a helper plasmid. We analyzed 32 transformants for silencing efficiency by determining the transcript levels of PSR2 using RT-PCR. PSR2-silenced lines, selected non-silenced transformants (as negative controls) and wild-type strain P6497 were further analyzed for virulence in soybean seedlings.
For soybean infection, hypocotyls from 4-d-old seedlings of the susceptible cultivar Hefeng47 were inoculated with zoospores of P. sojae strains. Soybean plants were grown in vermiculite at 25 °C for 4 d in the dark. P. sojae was grown for 3 d in Petri dishes containing 10% V8 medium at 25 °C. Mycelia were rinsed twice and then flooded with sterile distilled water overnight at 25 °C to release the zoospores. Hypocotyls removed from the 4-d-old soybean seedlings were inoculated with 10 µl of zoospore suspensions (containing approximately 100 zoospores). Inoculated plants were maintained in the dark at 25 °C and at high humidity for 36 h before lesion lengths were analyzed. The experiment was repeated twice with similar results. At least five seedlings were analyzed for each treatment.
Statistical analyses
Statistical analyses were performed using JMP 8.0 (SAS Institute).
Supplementary Material
Acknowledgments
We thank B. Tyler (Oregon State University) for providing soybean seeds and ten effector clones. S. Kamoun (The Sainsbury Laboratory) and M. Coffey (University of California, Riverside) kindly provided pGR106 and P. sojae strain P6497, respectively. We are indebted to S-W. Ding for sharing viral suppressor constructs and thoughtful input. This work was supported by funds from the University of California, Riverside, to W.M. and X.C., National Science Foundation (NSF) grant IOS-0847870 to W.M. and US Department of Agriculture–National Institute of Food and Agriculture (USDA-NIFA) grants 2010-04209 and 2008-00694 to X.C. and H.S.J., respectively. L.L. was supported by a fellowship from the China Scholarship Council.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
W.M. and X.C. developed the concept. W.M., X.C., Y.Q., Y.W. and H.S.J. designed the experiments. Y.Q., L.L., Q. Xiong, C.F., J.W., J.S., X.W., X.L., Q. Xiang, S.J. and F.Z. performed the experiments. W.M., X.C. and Y.W. analyzed the data. W.M., X.C., H.S.J., Y.Q., L.L. and Y.W. wrote the manuscript. W.M. conceived, directed and coordinated the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 2.Thines M, Kamoun S. Oomycete-plant coevolution: recent advances and future prospects. Curr. Opin. Plant Biol. 2010;13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 4.Kale SD. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Whisson SC. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 6.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 2012;15:483–492. doi: 10.1016/j.pbi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Chen X. Small RNAs—secrets and surprises of the genome. Plant J. 2010;61:941–958. doi: 10.1111/j.1365-313X.2009.04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 9.Ding SW. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 10.Vance V, Vaucheret H. RNA silencing in plants—defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 11.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 13.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brigneti G, et al. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Brosnan CA, Voinnet O. Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr. Opin. Plant Biol. 2011;14:580–587. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 18.Díaz-Pendon JA, Ding SW. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 2008;46:303–326. doi: 10.1146/annurev.phyto.46.081407.104746. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prigge MJ, Wagner DR. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren G, et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, et al. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haldar K, Kamoun S, Hiller NL, Bhattacharje S, van Ooij C. Common infection strategies of pathogenic eukaryotes. Nat. Rev. Microbiol. 2006;4:922–931. doi: 10.1038/nrmicro1549. [DOI] [PubMed] [Google Scholar]
- 26.Wroblewski T, Tomczak A, Michelmore R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 2005;3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- 27.Jiang RH, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earley KW. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H, et al. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe. 2011;9:177–186. doi: 10.1016/j.chom.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Lu R, et al. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA. 2004;101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou H, Morgan RL, Guttman DS, Ma W. Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol. Plant Microbe Interact. 2009;22:176–189. doi: 10.1094/MPMI-22-2-0176. [DOI] [PubMed] [Google Scholar]
- 32.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 33.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji L, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones L, et al. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dou D, et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell. 2008;20:1118–1133. doi: 10.1105/tpc.107.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Judelson HS, Tyler BM, Michelmore RW. Transformation of the oomycete pathogen Phytophthora infestans. Mol. Plant Microbe Interact. 1991;4:602–607. doi: 10.1094/mpmi-4-602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.