Abstract
Genomics and proteomics have emerged as key technologies in biomedical research, resulting in a surge of interest in training by investigators keen to incorporate these technologies into their research. At least two types of training can be envisioned in order to produce meaningful results, quality publications and successful grant applications: (1) immediate short-term training workshops and (2) long-term graduate education or visiting scientist programs. We aimed to fill the former need by providing a comprehensive hands-on training course in genomics, proteomics and informatics in a coherent, experimentally-based framework. This was accomplished through a National Heart, Lung, and Blood Institute (NHLBI)-sponsored 10-day Genomics and Proteomics Hands-on Workshop held at National Jewish Health (NJH) and the University of Colorado School of Medicine (UCD). The course content included comprehensive lectures and laboratories in mass spectrometry and genomics technologies, extensive hands-on experience with instrumentation and software, video demonstrations, optional workshops, online sessions, invited keynote speakers, and local and national guest faculty. Here we describe the detailed curriculum and present the results of short- and long-term evaluations from course attendees. Our educational program consistently received positive reviews from participants and had a substantial impact on grant writing and review, manuscript submissions and publications.
Keywords: Proteomics, Mass spectrometry, Genomics, Bioinformatics, Hands-on, Workshops
Introduction
In recent years, genomics and proteomics have emerged as key technologies in biomedical research programs. These types of analyses are now critical to performing effective clinical research aimed at developing a better understanding of disease pathology, as well as designing new approaches for clinical treatment [1]. For example, as genomic technologies have matured, so have a growing number of examples of genomic profiles directly being used to create individualized treatment plans [2]. These include the use of relatively simple single nucleotide polymorphism (SNP) genotyping for determining an appropriate treatment plan or for predicting the chemotherapeutic regimen most likely to benefit a breast cancer patient [3–5]. In addition, mass spectrometry, metabolomics and proteomics are becoming essential for providing sensitive methods of characterizing and analyzing both small molecules and proteins [6–12]. Both genomics and proteomics have proven so successful in these regards that demand has far exceeded the supply of trained personnel equipped to exploit the available technologies. This has resulted in a surge of interest in training by investigators keen to utilize these technologies in their research. The need for researchers trained in omics technologies has been described by several groups, including working groups from the National Institutes of Health (NIH) [13]. In our own experience, the demand for training has been demonstrated at local, national and international levels. For example, in the past 8 years, previously established hands-on workshops in Metabolomics and Proteomics at National Jewish Health (NJH) and the University of Colorado School of Medicine (UCD) have been attended by over 400 individuals from around the world.
To support technology-dependent clinical research studies, at least two types of training in omics technologies can be envisioned: (1) immediate short-term training in the form of workshops and (2) long-term instruction in the form of graduate education and visiting science programs. We aimed to fill the former need by providing an intensive and comprehensive hands-on training course in genomics, proteomics and bioinformatics in a coherent experimentally-based framework. Information gleaned from former participants’ surveys enabled us to determine that there is still a large need for introductory courses which focus on biomedical research such as the one described herein.
Several recent publications have focused on the need for experimental standards in omics research, especially when applied to human disease [14–17] and education is one means of lessening experimental error. To be of maximum benefit, education should ideally occur before experimental planning has begun, in order to decrease the possibility that the wrong platform, technology, experimental design or data analysis is chosen. For example, a first step toward utilizing omics technologies is the development of a well-defined hypothesis which clearly outlines the research question under investigation. If a researcher is interested in global expression patterns and/or biomarkers, he/she will need to determine if genomics, metabolomics or proteomics is more appropriate. Alternatively, an investigator may determine that a combination of approaches is necessary. Even after determining a general approach, the researcher must choose which specific platform and sample preparation strategies to utilize and this requires an understanding of the technologies available and the strengths and weaknesses of each approach. Given the plethora of equipment and protocols currently available, a new investigator could conceivably become overwhelmed by alternatives and subsequently make a poor choice that could lead to downstream difficulties in data analysis and interpretation. After determining an appropriate platform in which to test the hypothesis, careful experimental design and quality control measures are also critical to the success of the project as well as for interpreting the results. Given the large datasets generated by these technologies, knowledge of data analysis workflows and options, biostatistics and bioinformatics are vital and these are areas in which many investigators require additional training, specifically in the context of omics research. Finally, interpretation of omics results is dependent on a broad understanding of biochemistry and molecular biology, often presented within a knowledge-based pathway framework. Our training course exposed participants to each of these areas and, more specifically, enabled them to make informed decisions regarding the critical choice of platform for their particular research question.
To this end, we assembled a group of educators and scientists who understand the need for training and have experience in omics technologies and hands-on workshops. NJH is a premier hospital and research institution with a reputation for high quality clinical and basic research, effective professional education and extensive collaborations. The mass spectrometry facility is well established and supports over 100 national investigators involved in NIH-funded research in lung and immune diseases, diabetes, nutrition/obesity and others [18–25]. Similarly, the Program Directors at Colorado State University (CSU) and UCD have established genomics and proteomics research and support projects in pulmonary diseases, cancer and women’s health, and have programs in nutrition, environmental and microbial science [26–38]. The Directors of the workshop have developed and taught courses and workshops covering the subjects of metabolomics, genomics, proteomics, bioinformatics and biostatistics. In general, we accomplished the following: (1) built a curriculum focusing on outcome-structured learning in genomics, proteomics and bioinformatics technologies, which enabled investigators to immediately apply their knowledge to existing research studies; (2) provided novel, multidisciplinary training experiences using hands-on techniques and an established model by experienced investigators and educators; (3) disseminated educational materials such that knowledge and information could be used to benefit a wide variety of audiences; and (4) provided a portal for collaborative opportunities for both young and established investigators, thereby encouraging formation of interdisciplinary teams.
The intensive course schedule covered 10 consecutive days and was offered annually from 2007 to 2010 in the summer months. The NHLBI provided funding through a T15 award (N. Reisdorph, Principal Investigator) which covered faculty salary, invited keynote speakers, transportation between institutions and hotels, administrative support and laboratory module consumables. One salient feature of the course was the inclusion of over 20 faculty members with an array of experience spanning genomics, proteomics, bioinformatics, biostatistics, mass spectrometry, genetics, molecular biology, biochemistry, lung and heart disease, clinical studies, human research, and mouse and cell models. In addition, a number of vendors (Agilent Technologies, Affymetrix, Biorad and Pierce) provided laboratory module consumables as well as beverages, lunches and keynote speaker dinners for participants. Participants paid for their travel and housing expenses and a moderate $1000 tuition fee was charged to cover items not covered through the T15 award, including catering and keynote receptions. Scholarships were available each year to cover all costs for up to four participants.
Course details
The genomics and proteomics workshop structure combined hands-on training with lecture-based learning. It was carefully designed to allow students to experience an application-based workflow from sample preparation to data analysis. Classes were composed of no more than 22 students, who were split into 2 groups. Following an introductory session on the day of arrival, Group 1 attended proteomics training for days 2–4, then optional workshops for day 5, then genomics training for days 6–8 and finally bioinformatics for days 8–10 (Table 1 showing general schedule). Conversely, Group 2 attended genomics training for days 2–4, then optional workshops for day 5, then proteomics training for days 6–8 and finally bioinformatics for days 8–10. This allowed for small class size and one-on-one contact during instruction, while enabling all participants to interact with each other. Individual curricula for genomics, proteomics, bioinformatics and optional workshops are described below and detailed schedules are included in the online supplementary information. Information on group events is also described.
Table 1.
General course layout
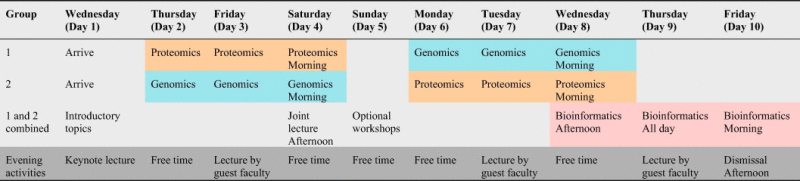 |
Note: Group 1 attended proteomics training for days 2–4, then optional workshops for day 5, then genomics training for days 6–8 and finally bioinformatics for days 8–10. Conversely, Group 2 attended genomics training for days 2–4, then optional workshops for day 5, then proteomics training for days 6–8 and finally bioinformatics for days 8–10.
Proteomics curriculum
A copy of the course curriculum is included in the supplementary information (Supplementary File 1), with proteomics spanning days 2–4. The proteomics curriculum was carefully designed to maximize the retention of information and minimize student fatigue. Training material was presented in both lecture and laboratory session modules. Lectures and laboratory sessions were strategically interspersed throughout the 3 day period. Thus student training time consistently alternated between lecture and hands-on activities, thereby reducing mental fatigue and helping to keep students engaged. Lectures and laboratory sessions were ordered logically to reflect a typical workflow, i.e., sample preparation > data acquisition > data analysis. Lecture topics included introduction to mass spectrometry and database searching, quantitative proteomics, advanced technologies and clinical proteomics. During hands-on lab sections, students were divided into groups of three, making the student-to-instructor ratio 3:1. The composition of these groups was predetermined by experience level and remained the same throughout the entire course. This provided an element of consistency that helped students feel comfortable with the learning environment. It also enabled the laboratory instructor to tailor the session to meet the needs of the individual participants. For example, the instructor may spend additional time describing the individual components of a mass spectrometer to one group, but may discuss specific method parameters with another more experienced group. Laboratory sessions were divided into three stations and organized in a rotation-based format. Each group of three students spent 60–75 min per rotation at the following stations: sample preparation, instrumentation, informatics and data analysis. Over the course of the workshop, students returned several times to specific lab stations to continue and complete various aspects of sample preparation and instruction on mass spectrometry instruments and software. This rotation style format exposes students to a wide variety of proteomics and mass spectrometry concepts, while maintaining the continuity required to effectively experience an application-based workflow from sample preparation to data analysis.
During laboratories, students were taken through typical proteomics work flows from sample preparation to data acquisition and from data analysis to protein identification. Each student was responsible for preparing their own digested protein sample and students contributed to creating mass spectrometry data acquisition methods. Students identified the protein(s) in their sample during the database search laboratory session, which was the final session of the 3-day proteomics course. In the days leading up to this final session, students were reminded that they would discover the identity of the protein(s) in their sample and there was a friendly competition to see who would have the highest search result score. This was designed to build anticipation and help keep students engaged.
Genomics curriculum
A copy of the course curriculum is included in the Supplementary File 1, with genomics spanning days 6–8. The genomics curriculum schedule interweaved hands-on bench work with lectures and demonstration videos. For the hands-on section, the participants in 5 groups of 2 converted total RNA into biotinylated cDNA using the standard protocol for running on an Affymetrix gene expression microarray. Quality control of the intermediate and final products was assessed using the Agilent Bioanalyzer. During incubation times, lectures and demonstration videos were presented. The lecture topics included: principles and applications of DNA microarray technology, a comparison of multiple gene expression platforms, genomic DNA SNP analysis using microarrays, epigenetic applications, application of microarray technology to advancing medicine and translational research, using publicly available datasets in your own research projects, troubleshooting quality control issues and data analysis, and integrating proteomic and gene expression datasets. In the final year’s course offering (2010), lectures in next-generation sequencing methods and applications were expanded. Various hands-on components typically completed by genomic core personnel were presented by video demonstrations, including microarray sample loading, washing, staining and scanning.
Bioinformatics curriculum
A copy of the course curriculum is included in the Supplementary File 1, with bioinformatics spanning days 8–10. All bioinformatics instructions were provided in the computational lab and synchronized with the laboratory sessions for a unique hands-on approach. Lecture topics included experimental design, power analysis, data pre-processing, quality control, exploratory data analysis, class discovery, differential expression and class prediction. The BRB-Array Tools software, based on an Excel interface of R BioConductor packages, was used for demonstrations by the instructors followed by hands-on sessions by the participants. This software was selected since it allowed participants with a diverse computational background to learn the fundamental data analysis steps in a user-friendly environment. Additional bioinformatics instruction was provided on the final days of the workshop and covered additional online resources such as Genome Browsers, Gene Set Enrichment Analysis, Promoter Analysis, Gene Ontology Analysis, Database Resources for Genetic Variations and Disorders and in later years, an introduction to Next-generation Sequencing Data Analysis.
Keynote and special lectures, optional workshops and on-line training
In addition to the genomics, proteomics and informatics curricula, the course consisted of 4 keynote lectures, one half-day of optional workshops, and pre- and post-course online training. Table 2 lists keynote lecturers and their affiliations. Special lectures that were presented by local speakers are also listed in Table 2.
Table 2.
Keynote and special lecturers
| Speaker | Institution/affiliation | Title | Year(s) presented |
|---|---|---|---|
| Natalie Ahn, PhD | University of Colorado, Boulder | Phosphoproteomics | 2010 |
| Keith Baggerly, PhD | MD Anderson Cancer Center | Evolution of forensic bioinformatics | 2009, 2010 |
| Pierre Chaurand, PhD | University of Montreal | Mass spectrometry imaging | 2009 |
| David Erle, MD | University of California, San Francisco | Microarray pearls and pitfalls | 2007–2010 |
| Leroy Hood, MD/PhD | Institute for Systems Biology | Systems biology and systems medicine | 2007 |
| Alexy Nesvizhskii, PhD | University of Michigan | Bioinformatics in proteomic applications of heart, lung and blood disorders | 2007–2010 |
| Kevin Schey, PhD | Vanderbilt University | Spatially-resolved proteomics: MALDI tissue imaging and beyond | 2007, 2008, 2010 |
| David Schwartz, MD | National Jewish Health | Environmental genomics and human health | 2008 |
| Michael Edwards, PhD | University of Colorado, AMC | Integrating publicly-available microarray data into your analysis | 2007–2010 |
| Mark Geraci, PhD | University of Colorado, AMC | Advancing translational medicine with DNA microarrays | 2007–2010 |
| Kirk Hansen, PhD | University of Colorado, AMC | Practical proteomics and spectral interpretation | 2007–2010 |
| Sonia Leach, PhD | National Jewish Health | SNP analysis with F-SNP, VarioWatch and SequenceVariantAnalyzer | 2010 |
| David Riches, PhD | National Jewish Health | Introduction to model systems | 2008–2010 |
| Ivana Yang, PhD | University of Colorado, AMC | Genomic studies of DNA methylation patterns | 2009, 2010 |
Note: Keynote speakers are highlighted in bold.
Optional workshops varied each year depending on participants’ interest. Optional workshops were held on Sunday morning and included the following topics: Additional Hands-on Experience with Mass Spectrometers, Two-dimensional Gel Electrophoresis, Introduction to Grant Writing, Biostatistics, Alternate Microarray Platform, Bioinformatics, MicroRNA Analysis and Literature Reviews.
Online training was held via live and recorded sessions and consisted of introductory material for the proteomics sessions. A schedule with topics is listed in Supplementary File 2. In addition, a 4-session online workshop was held in January 2012 to provide updated information to participants (Supplementary File 3). The fields of genomics, proteomics and bioinformatics are constantly changing and one challenge of omics training is consistently providing updated and relevant information. This was accomplished both through annual review and revision of material, and through the final online session. All sessions were recorded and are available by contacting the corresponding author.
Participant demographics and statistics
The workshop was aimed at junior and senior investigators but was also open to postdoctoral and clinical fellows and senior graduate students. In all, we provided hands-on and web-based training to 87 individuals from 55 institutions, all of whom rated the training they received as outstanding at the end of the course. Over 120 individuals applied for training and, as the course was NIH-NHLBI funded, priority was given to individuals with funding in an area related to heart, lung or blood diseases. Participants were chosen based on recommendations from a selection committee composed of course directors, instructors and advisors. Educational experience of participants from the four annual course offerings included: BS/BA (13%), PhD (33%), MD (48%) and MD/PhD (6%) (Figure 1A). Participants were primarily (48% of total) faculty members including Assistant Professor (26%), Associate Professor (6%), Professor (7%), Research Assistant Professor (3%) and Instructor (3%), although postdocs and fellows made up 30% of the participants (Figure 1B). Just over 10% of participants were graduate students and 2 proteomics core managers attended the course. Generally speaking, only 5% of the participants stated that they had significant training in omics and considered themselves experts, with the vast majority not having any prior training. Information gleaned from these participants’ statistics enabled us to determine that there is still a large need for introductory courses which focus on biomedical research. While advanced course work is certainly necessary, our course filled the significant niche for introductory and mid-level investigators.
Figure 1.
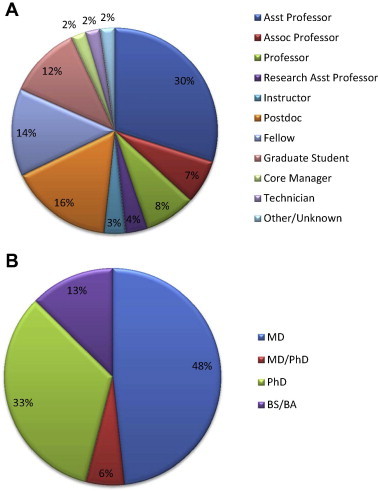
Participant demographics Shown are professional positions (A) and educational background (B) of course participants. Participants were mainly PhDs and MDs with faculty positions and included 2 proteomics core facility managers.
Short-term evaluations
Following each section, participants were asked to fill out a web-based evaluation form which was developed in collaboration with Dr. Carol Hodgson, an Associate Dean for Curriculum and Evaluation and Director of the Educational Development and Research Office at the University of Colorado, Denver. The form required rating various aspects of the workshop from poor, fair, good, very good and excellent, with corresponding numerical scores of 1 (poor)–5 (excellent). Median scores were tabulated for each laboratory and lecture and for administrative and overall course ratings. Composite scores for courses are shown in Figure 2. Scores for “overall quality” of the workshops in 2007, 2008, 2009 and 2010 were 4.76, 4.80, 4.84 and 4.63, respectively (far right of bar graph).
Figure 2.
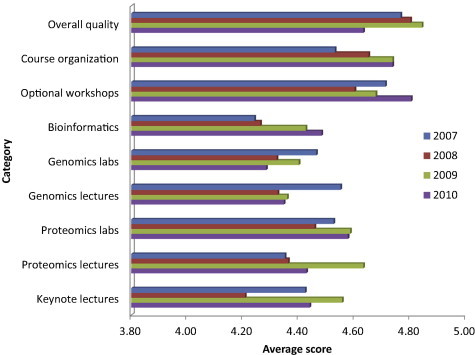
Composite scores Scores are shown for courses held in 2007–2010. The numbers indicate average scores for all lectures and laboratories within a category.
Course evaluations were taken seriously and were critical to shaping and improving the following years’ offering. For example, a number of participants suggested video demonstrations to replace hands-on genomics sample preparation, which was implemented in 2008. In addition, bioinformatics sessions originally spanned entire days; however these were split into half-days and interspersed with hands-on laboratory sessions after participant feedback suggested there were too many hours spent in the computer lab. Lectures and labs that received relatively poor ratings (greater than 10% fair ratings or that had less than 40% excellent ratings) were revised where possible in subsequent years.
Long-term evaluations
Approximately 3 months following the 2010 course, a follow-up evaluation was distributed to former NHLBI 10-day omics course attendees. Supplementary File 4 lists the questions that were asked to participants. Of particular note is that of the 49 responders, 98% rated the course very good to excellent and 100% indicated that they would recommend the course to others. Ninety-eight percent of responders rated the quality of the workshop as very good to excellent. Impressively, 44% said that the workshop had had high impact on their research. As shown in Figure 3, responders who were frequently using specific techniques in their own laboratories, or through a core facility, more than quadrupled following the course. For example, only 10% frequently used genomics informatics techniques prior to the course, but over 50% frequently used these techniques following the course. This could indicate that education helped overcome a significant barrier in learning by exposing individuals to the techniques in a hands-on fashion. In addition to increased utilization of techniques, training apparently helped participants in other aspects of their work. Figure 4 shows the results to the question “please indicate if you have used knowledge gained in the course in any of the following scenarios”. Almost all participants indicated that they had used knowledge gained in the workshop in reading journal articles (96%) and attending scientific meetings/seminars (94%). An impressive 76% indicated that the course had benefited them in terms of manuscript review, and almost half indicated the course had helped with non-NIH grant review and thesis review. This indicates that the course not only helped with research programs, but the knowledge gained was applied in a myriad of other ways, all of which have a direct positive impact on NIH-funded omics research.
Figure 3.
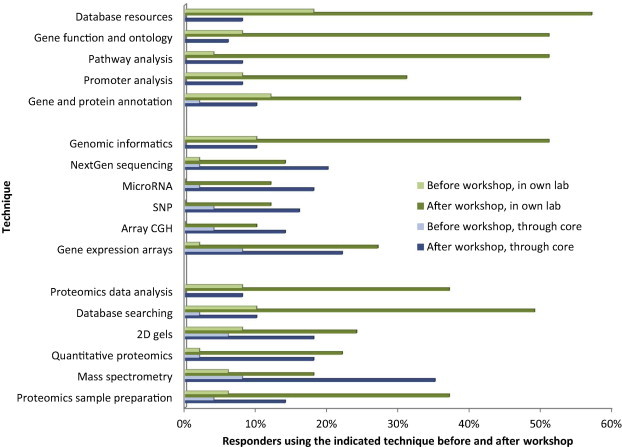
Usage of techniques The percentage of responders who frequently use a technique in their own laboratory (green bars) or through a core facility (blue bars) is shown. A total of 49 individuals responded to this series of questions. In all cases the number of individuals using a specific technique increased following participation in the course. Percentage of participants utilized the listed techniques in their own lab before the workshop and after the workshop was indicated in light green bars and dark green bars, respectively, while percentage of participants utilized the listed techniques through core facility before the workshop and after the workshop was indicated in light blue bars and dark blue bars, respectively.
Figure 4.

Value of workshop beyond research Respondents’ answers to the question “please indicate if you have used knowledge gained in the NHLBI omics course in any of the following scenarios” were analyzed. This chart demonstrates the utility of hands-on training beyond direct usage in research projects.
Participant comments
In addition to ranking specific course elements, participants were asked to provide written feedback regarding the course. This enabled participants to make specific requests regarding the curriculum. For example, in the first year the curriculum included a “mock grant proposal” which entailed working in groups to develop short grant proposals that included genomics and proteomics experiments. Groups were balanced to include junior and senior scientists and were given time for 2 group discussions and 1 presentation. In addition to the discussion periods, participants were also encouraged to discuss the mock proposals during non-workshop hours as a means of facilitating collaborations after the workshop. While over half of the participants rated the mock grant proposal modules as very good to excellent, their written comments demonstrated that the inclusion of this module was too time-consuming and challenging and it was thus eliminated in subsequent workshops.
Participant feedback also resulted in developing videos to complement the genomics modules, including alternate genomics strategies (SNP, next-generation sequencing), including more proteomics informatics sessions, expanding quantitative proteomics sessions and decreasing the amount of material covered in the bioinformatics sessions. A short compilation of written comments submitted by workshop attendees is included in Supplementary File 5.
Technical details and additional information
A search of PubMed and educational journals demonstrated a lack of publications related to omics-based education, in spite of the availability of several courses at educational and industrial institutions. To our knowledge, the described course constituted the only hands-on multi-omics course of its kind. As mentioned, the NHLBI provided significant funding to enable the presentation of the courses from 2007 to 2010 and hence no further courses are currently being held. Therefore, we have included additional information that could be of use to others in the field of omics education and training.
Advertising
The course was advertised through several portals including local institutions, institutions with primarily-underserved members, vendor contact lists, proteomics and genomics core laboratories and websites. Scientists receiving funding from NHLBI for proteomics or genomics research were identified through the NIH Research Portfolio Online Reporting Tools (RePORT) website and contacted through email. Course brochures were also sent to prospective and previous participants and course instructors for distribution.
Course demand
In the first year 43 individuals applied and 22 were admitted. In the second year, there were 48 applicants and 23 were admitted. In the third year, 27 applied and 20 were admitted into the course. In the fourth year, 28 applied and 22 were admitted. Due to the hands-on nature of the course, the course directors determined that 20–22 participants allowed individuals to receive experience on instrumentation while providing substantial one-on-one time with instructors. While lecture-only based courses could conceivably reach a larger audience, by definition they lack hands-on experience. The drop in demand from the last 2 years was attributed to several circumstances including a declining national economy that resulted in a decrease in overall scientific budgets. In addition, the course necessarily recruited investigators whose research focused on heart, lung and blood studies and our applicant pool was therefore limited. Members of industry were also not allowed to attend the course. Based on the number of inquiries between 2007 and 2010 from non-NHLBI investigators, and based on full capacity for current metabolomics and proteomics courses at NJH and UCD, it is the opinion of the authors that there is still a high demand for these types of hands-on courses.
Summary and accessing course material
Course evaluations and participant comments have shown that our intensive hands-on workshop formats are an effective model for presenting essential lecture and laboratory information in a relatively short amount of time. This curriculum could be the framework for long-term training strategies and can be conducted in any number of laboratories. While intensive, 10 days seemed an appropriate length of time for the course and allowed full-time researchers, physician-scientists, post-docs and graduate students to attend. For copies of the videotaped lectures, copies of course evaluations or access to the web-based introductory material, please contact the corresponding author.
Authors’ contributions
NR was responsible for the overall development of the workshop, directing the proteomics modules, managing the grant, compiling evaluations and for the production of the manuscript. CC and RS were responsible for the genomics modules, NR, RR and JP were responsible for proteomics modules including instructing and curriculum development, while KK and TLP were responsible for the bioinformatics modules. DJE, KS, AN and MG assisted with curriculum development, participant selection and presented lectures in the workshop. TLP prepared figures for the manuscript. All authors reviewed the manuscript, read and approved the final manuscript.
Competing interests
None of the authors has any competing interests to declare.
Acknowledgements
This work was supported by a grant through the National Institutes of Health, National Heart, Lung, and Blood Institute to Dr. Nichole Reisdorph (Grant No. T15HL086386).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Appendix Supplementary. material
Detailed schedule from 2009 course.
Web-based introduction schedule (pre-requisite).
Web-based update schedule.
Evaluation questions for workshop participants.
Written comments by participants.
References
- 1.Chadwell K. Clinical practice on the horizon: personalized medicine. Clin Nurse Spec. 2013;27:36–43. doi: 10.1097/NUR.0b013e318277703c. [DOI] [PubMed] [Google Scholar]
- 2.Venegas M., Brahm J., Villanueva R.A. Genomic determinants of hepatitis C virus antiviral therapy outcomes: toward individualized treatment. Ann Hepatol. 2012;11:827–837. [PubMed] [Google Scholar]
- 3.Dandona S., Roberts R. Personalized cardiovascular medicine: status in 2012. Can J Cardiol. 2012;28:693–699. doi: 10.1016/j.cjca.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Miller D.B., O’Callaghan J.P. Personalized medicine in major depressive disorder – opportunities and pitfalls. Metabolism. 2013;62:S34–S39. doi: 10.1016/j.metabol.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez de Castro D., Clarke P.A., Al-Lazikani B., Workman P. Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clin Pharmacol Ther. 2013;93:252–259. doi: 10.1038/clpt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker E.S., Liu T., Petyuk V.A., Burnum-Johnson K.E., Ibrahim Y.M., Anderson G.A. Mass spectrometry for translational proteomics: progress and clinical implications. Genome Med. 2012;4:63. doi: 10.1186/gm364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung L., Baxter R.C. Breast cancer biomarkers: proteomic discovery and translation to clinically relevant assays. Expert Rev Proteomics. 2012;9:599–614. doi: 10.1586/epr.12.62. [DOI] [PubMed] [Google Scholar]
- 8.Indovina P., Marcelli E., Pentimalli F., Tanganelli P., Tarro G., Giordano A. Mass spectrometry-based proteomics: the road to lung cancer biomarker discovery. Mass Spectrom Rev. 2013;32:129–142. doi: 10.1002/mas.21355. [DOI] [PubMed] [Google Scholar]
- 9.Lin J.L., Bonnichsen M.H., Nogeh E.U., Raftery M.J., Thomas P.S. Proteomics in detection and monitoring of asthma and smoking-related lung diseases. Expert Rev Proteomics. 2010;7:361–372. doi: 10.1586/epr.10.9. [DOI] [PubMed] [Google Scholar]
- 10.McDonnell L.A., Corthals G.L., Willems S.M., van Remoortere A., van Zeijl R.J., Deelder A.M. Peptide and protein imaging mass spectrometry in cancer research. J Proteomics. 2010;10:1921–1944. doi: 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Ozben T. Expanded newborn screening and confirmatory follow-up testing for inborn errors of metabolism detected by tandem mass spectrometry. Clin Chem Lab Med. 2013;51:157–176. doi: 10.1515/cclm-2012-0472. [DOI] [PubMed] [Google Scholar]
- 12.Sharma N., Martin A., McCabe C.J. Mining the proteome: the application of tandem mass spectrometry to endocrine cancer research. Endocr Relat Cancer. 2012;19:R149–R161. doi: 10.1530/ERC-12-0036. [DOI] [PubMed] [Google Scholar]
- 13.Participants NCFW . NIH Common Fund; Bethesda, MD: April 6, 2011. Metabolomics and translational research videoconference. Available from: < http://commonfund.nih.gov/pdf/Metabolomics_Workshop.pdf>. [Google Scholar]
- 14.Baggerly K. Disclose all data in publications. Nature. 2010;467:401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
- 15.Baggerly K.A., Coombes K.R. What information should be required to support clinical “omics” publications? Clin Chem. 2011;57:688–690. doi: 10.1373/clinchem.2010.158618. [DOI] [PubMed] [Google Scholar]
- 16.Leek J.T., Scharpf R.B., Bravo H.C., Simcha D., Langmead B., Johnson W.E. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orchard S., Binz P.A., Borchers C., Gilson M.K., Jones A.R., Nicola G. Ten years of standardizing proteomic data: a report on the HUPO-PSI Spring workshop: April 12–14th, 2012, San Diego, USA. Proteomics. 2012;12:2767–2772. doi: 10.1002/pmic.201270126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armstrong M., Jonscher K., Reisdorph N.A. Analysis of 25 underivatized amino acids in human plasma using ion-pairing reversed-phase liquid chromatography/time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:2717–2726. doi: 10.1002/rcm.3124. [DOI] [PubMed] [Google Scholar]
- 19.Bowler R., Reisdorph N. Mining the liver proteome for drug targets for sepsis. Crit Care Med. 2007;35:2443–2444. doi: 10.1097/01.CCM.0000285451.40021.26. [DOI] [PubMed] [Google Scholar]
- 20.Chu H.W., Gally F., Thaikoottathil J., Janssen-Heininger Y.M., Wu Q., Zhang G. SPLUNC1 regulation in airway epithelial cells: role of Toll-like receptor 2 signaling. Respir Res. 2010;11:155. doi: 10.1186/1465-9921-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delong T., Baker R.L., Reisdorph N., Reisdorph R., Powell R.L., Armstrong M. Islet amyloid polypeptide is a target antigen for diabetogenic CD4+ T cells. Diabetes. 2011;60:2325–2330. doi: 10.2337/db11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munks M.W., McKee A.S., Macleod M.K., Powell R.L., Degen J.L., Reisdorph N.A. Aluminum adjuvants elicit fibrin-dependent extracellular traps in vivo. Blood. 2010;116:5191–5199. doi: 10.1182/blood-2010-03-275529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabinovitch N., Reisdorph N., Silveira L., Gelfand E.W. Urinary leukotriene E4 levels identify children with tobacco smoke exposure at risk for asthma exacerbation. J Allergy Clin Immunol. 2011;128:323–327. doi: 10.1016/j.jaci.2011.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadinski B.D., Delong T., Reisdorph N., Reisdorph R., Powell R.L., Armstrong M. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol. 2010;11:225–231. doi: 10.1038/ni.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong M., Liu A.H., Harbeck R., Reisdorph R., Rabinovitch N., Reisdorph N. Leukotriene-E4 in human urine: comparison of on-line purification and liquid chromatography–tandem mass spectrometry to affinity purification followed by enzyme immunoassay. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3169–3174. doi: 10.1016/j.jchromb.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett B., Saba L.M., Hornbaker C.K., Kechris K.J., Hoffman P., Tabakoff B. Genetical genomic analysis of complex phenotypes using the PhenoGen website. Behav Genet. 2011;41:625–628. doi: 10.1007/s10519-010-9427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto S., Seebart C., Guastafierro T., Prenni J., Caiafa P., Zlatanova J. Proteome analysis of protein partners to nucleosomes containing canonical H2A or the variant histones H2A.Z or H2A.X. Biol Chem. 2012;393:47–61. doi: 10.1515/BC-2011-216. [DOI] [PubMed] [Google Scholar]
- 28.Goldstrohm D.A., Broeckling C.D., Prenni J.E., Curthoys N.P. Importance of manual validation for the identification of phosphopeptides using a linear ion trap mass spectrometer. J Biomol Tech. 2011;22:10–20. [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman P.L., Bennett B., Saba L.M., Bhave S.V., Carosone-Link P.J., Hornbaker C.K. Using the phenogen website for ‘in silico’ analysis of morphine-induced analgesia: identifying candidate genes. Addict Biol. 2011;16:393–404. doi: 10.1111/j.1369-1600.2010.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kechris K.J., Biehs B., Kornberg T.B. Generalizing moving averages for tiling arrays using combined p-value statistics. Stat Appl Genet Mol Biol. 2010;9 doi: 10.2202/1544-6115.1434. Article 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruh N.A., Troudt J., Izzo A., Prenni J., Dobos K.M. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS One. 2010;5:e13938. doi: 10.1371/journal.pone.0013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehaffy C., Hess A., Prenni J.E., Mathema B., Kreiswirth B., Dobos K.M. Descriptive proteomic analysis shows protein variability between closely related clinical isolates of Mycobacterium tuberculosis. Proteomics. 2010;10:1966–1984. doi: 10.1002/pmic.200900836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller N.H., Walters M.S., Marcus R.A., Graf L.L., Prenni J., Gilden D. Identification of phosphorylated residues on varicella-zoster virus immediate-early protein ORF63. J Gen Virol. 2010;91:1133–1137. doi: 10.1099/vir.0.019067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollock D.D., de Koning A.P., Kim H., Castoe T.A., Churchill M.E., Kechris K.J. Bayesian analysis of high-throughput quantitative measurement of protein–DNA interactions. PLoS One. 2011;6:e26105. doi: 10.1371/journal.pone.0026105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan E.P., Heuberger A.L., Weir T.L., Barnett B., Broeckling C.D., Prenni J.E. Rice bran fermented with Saccharomyces boulardii generates novel metabolite profiles with bioactivity. J Agric Food Chem. 2011;59:1862–1870. doi: 10.1021/jf1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saba L.M., Bennett B., Hoffman P.L., Barcomb K., Ishii T., Kechris K. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology. 2011;60:1269–1280. doi: 10.1016/j.neuropharm.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serkova N.J., Niemann C.U. Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev Mol Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 38.Serkova N.J., Standiford T.J., Stringer K.A. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184:647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed schedule from 2009 course.
Web-based introduction schedule (pre-requisite).
Web-based update schedule.
Evaluation questions for workshop participants.
Written comments by participants.



