ABSTRACT
Since 2009, catfish farming in the southeastern United States has been severely impacted by a highly virulent and clonal population of Aeromonas hydrophila causing motile Aeromonas septicemia (MAS) in catfish. The possible origin of this newly emerged highly virulent A. hydrophila strain is unknown. In this study, we show using whole-genome sequencing and comparative genomics that A. hydrophila isolates from diseased grass carp in China and catfish in the United States have highly similar genomes. Our phylogenomic analyses suggest that U.S. catfish isolates emerged from A. hydrophila populations of Asian origin. Furthermore, we identified an A. hydrophila strain isolated in 2004 from a diseased catfish in Mississippi, prior to the onset of the major epidemic outbreaks in Alabama starting in 2009, with genomic characteristics that are intermediate between those of the Asian and Alabama fish isolates. Investigation of A. hydrophila strain virulence demonstrated that the isolate from the U.S. catfish epidemic is significantly more virulent to both channel catfish and grass carp than is the Chinese carp isolate. This study implicates the importation of fish or fishery products into the United States as the source of highly virulent A. hydrophila that has caused severe epidemic outbreaks in United States-farmed catfish and further demonstrates the potential for invasive animal species to disseminate bacterial pathogens worldwide.
IMPORTANCE
Catfish aquaculture farming in the southeastern United States has been severely affected by the emergence of virulent Aeromonas hydrophila responsible for epidemic disease outbreaks, resulting in the death of over 10 million pounds of catfish. Because the origin of this newly emerged A. hydrophila strain is unknown, this study used a comparative genomics approach to conduct a phylogenomic analysis of A. hydrophila isolates obtained from the United States and Asia. Our results suggest that the virulent isolates from United States-farmed catfish have a recent common ancestor with A. hydrophila isolates from diseased Asian carp. We have also observed that an Asian carp isolate, like recent U.S. catfish isolates, is virulent in catfish. The results from this study suggest that the highly virulent U.S. epidemic isolates emerged from an Asian source and provide another example of the threat that invasive species pose in the dissemination of bacterial pathogens.
Observation
Aeromonas hydrophila is typically an opportunistic bacterial pathogen that is ubiquitous in freshwater environments and is responsible for diseases in different species, including amphibians, reptiles, fish, and mammals (1). Motile Aeromonas septicemia (MAS) caused by mesophilic A. hydrophila affects a wide variety of primarily freshwater fish species, including carp, tilapia, perch, catfish, and salmon (2). Epidemic disease outbreaks in fish caused by A. hydrophila, resulting in millions of dollars of lost revenue, have been reported worldwide but have not occurred in the United States until recently (3, 4). In the summer of 2009, an outbreak of MAS in commercially raised catfish caused by highly virulent A. hydrophila (VAh) began in western Alabama (5). Since the initial outbreak during the summer of 2009, this outbreak has spread to other, adjacent states, including Mississippi and Arkansas (6). To date, this epidemic of MAS outbreaks is responsible for an estimated loss of more than $12 million in catfish aquaculture operations in the southeastern United States. Our comparative and functional genomic analyses demonstrated that A. hydrophila isolates from recent epidemic outbreaks in the United States are highly clonal but genomically distinct from A. hydrophila historical isolates from diseased catfish in the United States, and these epidemic isolates contain a large number of genomic regions predicted to be acquired through lateral genetic transfer (7). Several episodes of epidemic outbreaks caused by A. hydrophila beginning in the late 1980s and within the last 10 years have been reported in farmed carp in China (8, 9). There are striking similarities between the U.S. catfish and Chinese carp epidemics caused by A. hydrophila, including the timing of the epidemics, which primarily occurred during the summer; the extensive mortality observed in seemingly healthy fish; and the highly clonal nature of the epidemic isolates (5, 8).
Nonnative species can endanger the native or endemic species by disturbing the trophic levels of the native fish community (10) and pose a potential threat of the emergence of infectious diseases (11). Because of the globalization of the aquaculture and ornamental fish trade industries, fish transport provides a gateway for the introduction of exotic species that could introduce new pathogens that can devastate populations of indigenous species (12). For instance, spring viremia of carp (SVC), a viral disease historically affecting fish in Europe, the Middle East, and Russia, has been found in carp in the United States since first being reported in 2002 (13). The Chinese origin of the SVC viral isolates in the United States was thought to be mediated through importation of ornamental fish (14). The emergence of several other viral diseases of fish and shrimp reported worldwide has been attributed to the dissemination of infected fish or eggs as a carrier of nonindigenous pathogens (15).
Over the past several decades, Asian carp have been imported into the United States, including grass carp (Ctenopharyngodon idella), bighead carp (Hypophthalmichthys nobilis), silver carp (Hypophthalmichthys molitrix), and black carp (Mylopharyngodon piceus), as well as countless species of live ornamental fish sold in the United States. Grass carp were imported to the United States from east Asia beginning in 1963 by the U.S. Fish and Wildlife Service (16) and have been extensively used in polyculture of catfish and in reservoirs for aquatic weed control. As a result of the Great Flood of 1993, silver carp were released in large numbers into the Mississippi River drainage system and currently threaten the ecological balance of the U.S. Great Lakes region (17).
The origin of the clonal A. hydrophila isolates responsible for the ongoing epidemic MAS outbreak in United States-farmed channel catfish is unknown. We used a phylogenomic approach to study the molecular epidemiology of the bacterial isolates responsible for this epidemic outbreak. Our study clearly demonstrates that the U.S. catfish and Chinese carp isolates have a recent common ancestor.
To determine the evolutionary relationships of the recent virulent A. hydrophila (VAh) isolates, a gyrB-based phylogenetic analysis was conducted using a total of 264 Aeromonas strains downloaded from the Aeromonas multilocus sequence typing (MLST) database (http://www.pubmlst.org/aeromonas) (18) and including other strains of U.S. and non-U.S. origin (see Table S1 in the supplemental material). The A. hydrophila phylogenetic analysis revealed a coherent and well-supported clade that included all VAh strains (see Fig. S1). Interestingly, the only strain retrieved from the GenBank database that was affiliated with VAh strains was A. hydrophila strain ZC1, which was isolated from a diseased grass carp in Guangdong Province, China, from ponds that had experienced an epidemic outbreak of hemorrhagic septicemia (19). In order to identify any other isolates that were affiliated with epidemic strains, we screened A. hydrophila strain collections available in the United States that were isolated from fish and other hosts and identified one additional A. hydrophila strain, S04-690, which was affiliated with VAh strains and strain ZC1 (see Fig. S1). Strain S04-690 was isolated in 2004 from a diseased catfish obtained from a commercial aquaculture pond located in Washington County, Mississippi, which experienced a single event of MAS outbreak that killed thousands of catfish but importantly did not result in a widespread epidemic outbreak in the surrounding regions or subsequent outbreaks in following years within the affected farm.
Since our gyrB-based phylogeny suggests that Chinese carp isolate ZC1 is highly similar to VAh isolates and a 2004 Mississippi isolate of catfish origin, we evaluated the evolutionary relationships of additional Chinese carp isolates of epidemic origin in Hubei Province, China (see Table S1 in the supplemental material) (4). The gyrB phylogeny constructed using the neighbor-joining method revealed that all of the Chinese carp epidemic isolates, including isolate ZC1, consistently group together as a single clade along with the recent U.S. epidemic isolates of catfish origin that included Mississippi isolate S04-690 (see Fig. S1). In contrast, we found that A. hydrophila isolates of nonepidemic origin are widely spread through different clades (see Fig. S1). The gyrB sequences of the epidemic Aeromonas isolates (from Chinese carp or U.S. catfish) formed a monophyletic clade with 100% sequence identities and with 100% bootstrap support for the VAh-associated lineage. This suggests that a clonal A. hydrophila complex descended from a common ancestor responsible for epidemic outbreaks of fish disease in China and the United States.
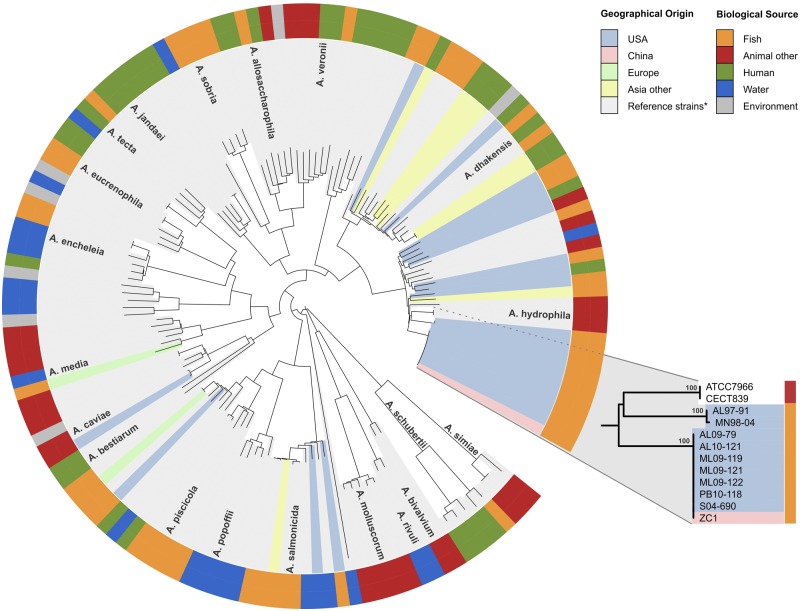
To provide a more refined phylogenetic placement of the U.S. A. hydrophila isolates, we used a multilocus sequence-based phylogeny that included 3,751 bp of concatenated nucleotide sequences from six housekeeping genes: gyrase subunits A and B (gyrA and gyrB), RNA polymerase sigma factor (rpoD), bacterial DNA recombination protein (recA), heat shock protein (dnaJ), and DNA polymerase III tau and gamma subunits (dnaX) (see Table S2 in the supplemental material). Our analysis included A. hydrophila VAh catfish isolates, Mississippi catfish isolate S04-690, Chinese carp isolate ZC1 (19), and nonepidemic Aeromonas isolates of U.S. origin (see Table S1), yielding a phylogenetic tree with strong bootstrap support for the terminal nodes (Fig. 1). This multilocus phylogeny demonstrated that the Chinese carp isolate ZC1 clustered together with the Alabama and Mississippi catfish isolates as a monophyletic clade (Fig. 1); in contrast, the nonepidemic A. hydrophila isolates are highly divergent (Fig. 1). In addition to this multilocus sequence-based phylogeny, we searched the Aeromonas PubMLST database (http://www.pubmlst.org/aeromonas) (18) using gyrB, groL, gltA, metG, ppsA, and recA gene-specific sequences of isolates ML09-119, S04-690, and ZC1, and we found that these three isolates belong to sequence type (ST) ST251, which until this study contained only Aeromonas hydrophila isolate XS91-4-1. Interestingly, this highly virulent Chinese isolate, Aeromonas hydrophila XS91-4-1, was obtained from a diseased silver loweye carp from an epidemic outbreak in 1991 (20). Taken together, these findings demonstrate that Chinese carp isolates XS91-4-1 and ZC1 along with U.S. catfish isolates ML09-119 and S04-690 are clonal. We also constructed a phylogenetic tree using the neighbor-joining method that demonstrates that U.S. catfish isolates ML09-119 and S04-690 and Chinese carp isolates XS91-4-1 and ZC1 cluster together in a monophyletic clade (data not shown).
FIG 1 .
Multilocus sequence-based phylogeny using six different housekeeping genes of 148 Aeromonas strains. The internal color ranges refer to the geographical origin of the isolates, while the external ring indicates the biological source. The cluster containing the epidemic strains is highlighted. *, Aeromonas isolates sequenced by Martinez-Murcia and colleagues (25). The reclassification of Aeromonas aquariorum and Aeromonas hydrophila subsp. dhakensis as Aeromonas dhakensis has been proposed elsewhere (26).
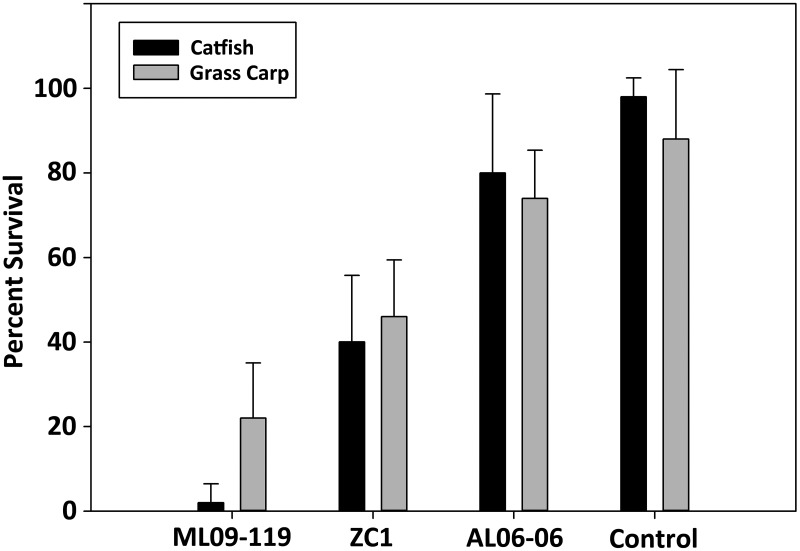
Our multilocus phylogeny based on two different sets of housekeeping genes as well as MLST typing is concordant with the gyrB-based phylogeny that demonstrated that the Chinese carp and U.S. catfish isolates cluster together as a clonal group and share identical alleles of all 10 housekeeping genes. Therefore, these findings strongly support the hypothesis that the recent A. hydrophila isolates responsible for catfish epidemics in the United States emerged due to the rapid spread of a clonal group of pathogenic isolates that share an ancestor with A. hydrophila isolates identified from diseased carp from Asia. These data, while supporting the close phylogenetic affiliation of U.S. catfish and Asian carp isolates, do not provide any indication of the relative timing of the evolutionary changes that have occurred in these A. hydrophila populations, nor did these analyses indicate the relative virulence of these strains within carp or catfish. Channel catfish and grass carp were experimentally challenged to compare the virulence of strains ML09-119, ZC1, and AL06-06, with survival rates ranging from 0.02 ± 0.04 to 0.98 ± 0.04. A. hydrophila isolate AL06-06 was included in the challenge experiment in this study as a reference strain that is typical of A. hydrophila strains that have been historically isolated from stressed fish prior to the advent of the MAS epidemic and has shown reduced mortality (20%) in channel catfish relative to that observed from ML09-119 (7). In this study, we observed that isolate ML09-119 was significantly more virulent than either ZC1 or AL06-06 and that ZC1 was more virulent than AL06-06 (P < 0.0001) (Fig. 2) in both channel catfish and grass carp. Additionally, overall comparisons between fish species show that channel catfish were more susceptible to A. hydrophila than were grass carp (P = 0.0126); however, this observation occurred only for VAh strain ML09-119 (Fig. 2). No interaction effect of the independent variables was observed (P = 0.1002). These data suggest that ML09-119 has evolved increased virulence and that channel catfish appear to be more susceptible.
FIG 2 .
Percent survival of channel catfish or grass carp after challenge with A. hydrophila strain ML09-119, ZC1, or AL06-06. Significant differences in survival were observed for bacterial strains ML09-119, ZC1, and AL06-06 (P < 0.0001) and between fish species in the ML09-119 group (P = 0.0126). No interaction effect between variables of bacterial strains and fish species was observed (Pfish species × bacterial strains = 0.1002). The difference in survival between channel catfish and grass carp injected with ML09-119 was significant (P = 0.0069). No significant differences in survival between channel catfish and grass carp intraperitoneally injected with ZC1 or AL06-06 were observed, but there was a significant difference between strains ZC1 and AL06-06 in both channel catfish (P = 0.0075) and grass carp (P = 0.0089).
One of the phenotypic traits common among all VAh strains is the ability to utilize myo-inositol as a sole carbon source (7). We therefore tested the Chinese carp isolate ZC1 and the Mississippi catfish isolate S04-690 for this phenotype and found that these strains could also utilize myo-inositol as a sole carbon source and had growth curves similar to those observed for VAh strains, whereas isolate AL06-06, used as a negative control, did not utilize myo-inositol as a sole carbon source (data not shown). The comparison of the 17.5-kb myo-inositol utilization cluster of VAh isolates with that of Chinese carp isolate ZC1 (accession no. KF724901) and Mississippi catfish isolate S04-690 (accession no. KF724900) demonstrated that they share identical nucleotide sequences with identical synteny. Previously, we observed a consistent correlation between the presence of an epidemic-specific genetic marker and myo-inositol utilization capacity of all tested VAh strains (7). To our knowledge, the utilization of myo-inositol as a sole carbon source has not been reported for any other A. hydrophila isolates obtained from diseased fish or any other environmental or clinical origin in the United States (1). However, myo-inositol-utilizing Aeromonas isolates are prevalent in East and Southeast Asia (21). This is further evidence in support of an Asian origin for the epidemic A. hydrophila strains in the United States.
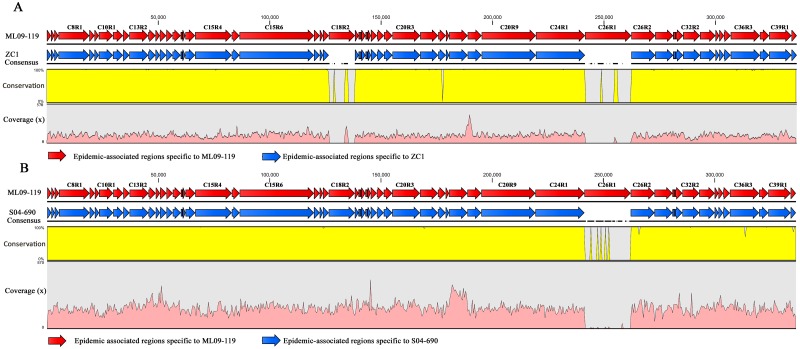
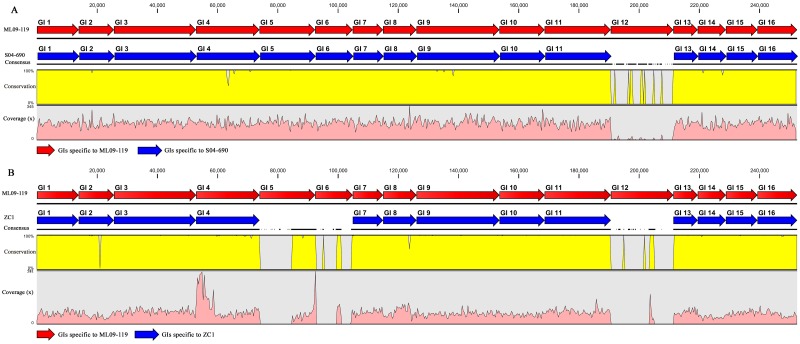
The clonal nature of the Asian carp and U.S. catfish isolates made it impossible to differentiate them based on housekeeping gene divergence. To systematically investigate the emergence of the recent VAh isolates, we sequenced the genomes of the Chinese carp isolate ZC1 and the Mississippi catfish isolate S04-690. The genomic comparison of both of these isolates with the highly virulent recent VAh isolates and other nonepidemic disease isolates was conducted to understand the genomic changes among these strains and provide finer resolution for the phylogenetic relationships among these strains. Reference mappings of the Illumina sequence reads from the genomes of Chinese carp isolate ZC1 and Mississippi catfish isolate S04-690 were conducted against the 55 genomic regions unique among VAh strains (7), demonstrating that the genomes of Chinese carp isolate ZC1 and Mississippi catfish isolate S04-690 share 53 and 54 of these 55 epidemic-associated regions, respectively (Fig. 3). These 55 epidemic-associated genomic regions (336,469 bp in total), present within the genome of all sequenced VAh isolates, are comprised of prophage elements, pathogenicity islands, metabolic islands, fitness islands, and genes of unknown functions (7). These epidemic-associated unique regions are considered horizontally acquired regions of the A. hydrophila genome since they were obtained by excluding core genomes of the 11 A. hydrophila isolates described previously (7). Though it has not been demonstrated experimentally, the analysis of the open reading frames (ORFs) present within these unique regions suggests their potential roles in pathogenesis (7). The C18R2 (11,731 bp) and C26R1 (20,780 bp) epidemic-associated unique regions of a recent VAh isolate are absent in Chinese isolate ZC1 (Fig. 3A). The C26R1 region, the same region absent in ZC1, is the only epidemic-associated unique region of a recent VAh isolate absent in the genome of Mississippi isolate S04-690 (Fig. 3B). In our previous study, we reported that epidemic-associated unique regions C18R2 and C26R1 contained two different genomic islands (GIs), namely, GI6 and GI12, respectively (7), which suggested their possible acquisition through horizontal gene transfer. The comparison of genomic islands present within the genomes of Chinese carp and U.S. catfish isolates demonstrated that most of the genomic islands, except for GI5, GI6, and GI12, which are found consistently within the genomes of VAh isolates are also present within the genome of strain ZC1 (Fig. 4A). We also observed that the majority of the genomic islands, except for GI12, were present in Mississippi isolate S04-690 (Fig. 4B). These shared genomic islands with identical synteny and DNA sequences of high similarity are located in the same positions within the genomes of ZC1, S04-690, and ML09-119 isolates (data not shown). No additional GIs were predicted within the genome of S04-690, whereas 4 additional GIs were predicted within the genome of ZC1 (data not shown). Our analysis of GI12, a VAh-specific GI absent within the genomes of ZC1 and S04-690, predicts the presence of a fitness island with a large number of ORFs involved in DNA modification functions (7). GI5 and GI6 are observed in VAh isolates (7) but absent in ZC1 and are predicted to encode 15 and 8 proteins of unknown functions, respectively. Since genomic islands of bacterial species are highly variable and frequently used to distinguish strains due to their acquisition through horizontal gene transfer (22), the sharing of a large number of identical genomic islands has led to the hypothesis that U.S. VAh isolates originated from Asia and that the Mississippi isolate S04-690 represents an intermediate strain between the Asian carp and Alabama catfish isolates. The commonality of a large number of genomic islands also indicates that the horizontal acquisition of those genomic islands occurred in their common ancestor prior to the diversification of the Chinese carp isolate ZC1, Mississippi catfish isolate S04-690, and Alabama catfish VAh isolates.
FIG 3 .
Reference alignment showing the maps of the orthologous genetic regions (blue arrow) of Chinese carp isolate ZC1 (A) or Mississippi catfish isolate S04-690 (B) specific to the epidemic-associated genetic regions of VAh isolates (red arrow). The scales for the percent conservation and average coverage of the Illumina sequences from both the isolates aligned with epidemic-associated genetic regions of VAh isolates are shown on the left axis. The black horizontal line illustrates the continuous alignment of the sequences, and the discontinuous black line indicates the interruption of the sequence alignment. The average coverages of the ZC1 and S04-690 sequences mapped against epidemic-associated genetic regions of VAh isolates are 87× and 115×, respectively.
FIG 4 .
Reference alignment showing the maps of the orthologous genetic regions (blue arrow) of isolates S04-690 (A) and ZC1 (B) specific to the GIs of VAh isolates (red arrow). The scales for the percent conservation and average coverage of the Illumina sequences from both the isolates aligned with GIs of VAh isolates are shown on the left axis. The black horizontal line illustrates the continuous alignment of the sequences. and the discontinuous black line indicates the interruption of the sequence alignment.
Although the MLST-based phylogeny demonstrated that U.S. catfish and Chinese carp isolates have a common ancestor (Fig. 1), this analysis did not provide sufficient phylogenetic resolution to differentiate between these isolates. To further refine the evolutionary relationships of the U.S. catfish and Chinese carp isolates, we constructed a phylogenetic tree based on the 303,863 bp of concatenated sequences from epidemic-specific regions shared among VAh isolates (7), ZC1, and S04-690. We observed that VAh epidemic isolates form a distinct monophyletic clade that is divergent from the S04-690 strain (see Fig. S2 in the supplemental material). The evolutionary history inferred from this phylogenetic tree suggests a common ancestor between all of these strains, with S04-690 being most closely related to VAh isolates. These findings strongly suggest a stepwise emergence of the U.S. epidemic isolates from Asia with the Mississippi catfish isolate as a representative intermediate in this evolutionary model.
Finally, we conducted a pairwise comparison of the proteomes of 14 A. hydrophila isolates using a BLASTp matrix, an all-against-all BLASTp analysis, to determine the similarity among proteomes based on the number of conserved gene families (23). Similarly to our previous findings (7), these results demonstrated that the VAh strain proteomes are highly clonal and that the proteome of Mississippi catfish isolate S04-690 shows a high degree of similarity to that of VAh strains, with 99.5% similarity to the representative strain ML09-119 (see Fig. S3 in the supplemental material). In contrast, the proteome of the Chinese carp isolate ZC1 shows 97.7% and 97.5% similarity to those of ML09-119 and S04-690, respectively. These findings are concordant with the phylogenomic analysis (see Fig. S2) that suggests that S04-690 is an example of an intermediate strain in the evolutionary emergence of VAh. We observed that the genomes of S04-690, ZC1, and all of the VAh isolates share >97% similarities among themselves. In contrast, none of the genomes of nonepidemic U.S. isolates showed >74% similarity to that of VAh isolates (see Fig. S3). These findings further support the conclusion that the Chinese carp and U.S. catfish VAh and Mississippi isolates are a monophyletic group with a coherent genome and a discriminatory phenotype (i.e., myo-inositol utilization) that clearly differentiate them as clonal, pathogenic strains that are distinct from A. hydrophila isolates that have been previously described in the United States.
This study provides multiple lines of evidence that suggest that highly virulent A. hydrophila isolates responsible for epidemic outbreaks of MAS in catfish in the southeastern United States have an Asian origin. At this point, the exact means of introduction of these virulent isolates in the United States is not clear. There are several possible ways in which Asian isolates might have been introduced in the United States: (i) the introduction of Asian carp into the United States and their extensive use as biological control agents may have introduced an Asian variant of A. hydrophila that served as a precursor for the emergence of highly virulent catfish isolates, (ii) transport and distribution of imported ornamental fish from Asia may have introduced the isolates, or (iii) import of contaminated processed seafood products may have introduced Asian isolates into the United States, since imported seafood is frequently contaminated with A. hydrophila (24). Given the widespread occurrence of this A. hydrophila lineage within different regions of China (Guangdong, Hubei, Jiangsu, and Zhejiang provinces) and the report of myo-inositol-utilizing A. hydrophila strains in eastern and southeastern Asia in the years prior to the U.S. epidemic, the most likely scenario is that this pathogenic A. hydrophila lineage was introduced into the United States from fish imported from Asia. Additionally, highly virulent disease episodes have been documented since the late 1980s in China with A. hydrophila of this genotype (9) and no disease episodes like those observed on commercial catfish farms in the U.S. occurred until 2004, with the onset of epidemic outbreaks occurring in 2009.
We hypothesize that a virulence factor(s) encoded within genomic islands of the epidemic catfish isolates may contribute to the enhanced pathogenicity and/or host specificity of the VAh isolates in catfish. Further studies of the role and mechanisms of this virulence factor(s) in A. hydrophila pathogenesis in catfish and carp will increase our understanding of the emergence of highly virulent bacterial pathogens and the contribution of geographically introduced nonnative species in the pandemic spread of pathogens facilitated by human activities.
Nucleotide sequence accession numbers.
Sequences for the myo-inositol utilization clusters of Chinese carp isolate ZC1 (accession no. KF724901) and Mississippi catfish isolate S04-690 (accession no. KF724900) have been deposited in GenBank. The raw sequence reads for strains ZC1 and S04-690 were deposited in NCBI’s Short Read Archive under accession numbers SRR1031999 and SRR1032027, respectively.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
gyrB phylogeny using sequences from 169 A. hydrophila isolates, including strains of Asian and U.S. origins. Colored ranges refer to the geographical origin of the isolates. The red segments indicate the epidemic strains. The tree figure was generated using the Interactive Tree Of Life web application (http://itol.embl.de) (27). Download
Phylogenetic analysis based on the virulent A. hydrophila (VAh)-specific sequences. The evolutionary history was inferred using the neighbor-joining method. The evolutionary distances are in units of number of base substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 303,863 positions in the final data set. Evolutionary analyses were conducted in MEGA5 with 1,000 repetitions to generate bootstrap support (28). *, isolate PB10-118 was isolated from a diseased fish obtained from Arkansas, USA (7). Download
BLASTp matrix of 14 different A. hydrophila isolates. The proteomes of each of the A. hydrophila isolates were compared pairwise using all-against-all BLASTp as described previously (23). This matrix illustrates the output from the pairwise comparison of conserved protein families of each of the isolates to each other. Green represents the relative percent homology between proteomes, and red represents relative percent homology within the isolate’s own proteome. The dense green pyramid on the right indicates greater homology (>97%) between the proteomes of VAh isolates from U.S. catfish epidemics, Chinese carp isolate ZC1, and Mississippi catfish isolate S04-690. Download
Aeromonas hydrophila isolates used for gyrB phylogeny.
Isolates of Aeromonas species used for MLST analysis.
Footnotes
Citation Hossain MJ, Sun D, McGarey DJ, Wrenn S, Alexander LM, Martino ME, Xing Y, Terhune JS, Liles MR. 2014. An Asian origin of virulent Aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. mBio 5(3):e00848-14. doi:10.1128/mBio.00848-14.
REFERENCES
- 1. Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joseph SW, Carnahan A. 1994. The isolation, identification, and systematics of the motile Aeromonas species. Annu. Rev. Fish Dis. 4:315–343. 10.1016/0959-8030(94)90033-7 [DOI] [Google Scholar]
- 3. da Silva BC, Mouriño JLP, Vieira FN, Jatobá A, Seiffert WQ, Martins ML. 2012. Haemorrhagic septicaemia in the hybrid surubim (Pseudoplatystoma corruscans×Pseudoplatystoma fasciatum) caused by Aeromonas hydrophila. Aquac. Res. 43:908–916. 10.1111/j.1365-2109.2011.02905.x [DOI] [Google Scholar]
- 4. Nielsen ME, Høi L, Schmidt AS, Qian D, Shimada T, Shen JY, Larsen JL. 2001. Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Organ. 46:23–29. 10.3354/dao046023 [DOI] [PubMed] [Google Scholar]
- 5. Hemstreet WB. 2010. An update on Aeromonas hydrophila from a fish health specialist for summer 2010. Catfish J. 24:4 [Google Scholar]
- 6. Pridgeon JW, Klesius PH. 2011. Molecular identification and virulence of three Aeromonas hydrophila isolates cultured from infected channel catfish during a disease outbreak in west Alabama (USA) in 2009. Dis. Aquat. Organ. 94:249–253. 10.3354/dao02332 [DOI] [PubMed] [Google Scholar]
- 7. Hossain MJ, Waldbieser GC, Sun D, Capps NK, Hemstreet WB, Carlisle K, Griffin MJ, Khoo L, Goodwin AE, Sonstegard TS, Schroeder S, Hayden K, Newton JC, Terhune JS, Liles MR. 2013. Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS One 8:e80943. 10.1371/journal.pone.0080943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qian D, Chen Y, Shen J, Shen Z. 1997. Studies on the pathogen of fish bacterial septicemia in Zhejiang Province during 1989-1992: biochemical characteristics, virulence and serogroups of; Aeromonas hydrophila, p 242–245 In Yingqi Z, Fuyuan H, Hongqi Z, He C, Chaoqi Y, Fuhui D, Yi L. (ed), Proceedings of Fourth Asian Fisheries Forum, Beijing, 16 to 20 October 1995. China Ocean Press, Beijing, China [Google Scholar]
- 9. Zhang X-J, Yang W-M, Li T-T, Li A-H. 2013. The genetic diversity and virulence characteristics of Aeromonas hydrophila isolated from fishponds with disease outbreaks in Hubei province. Acta Hydrobiol. Sin. 2013(3):458–466. 10.7541/2013.44 [DOI] [Google Scholar]
- 10. Freedman JA, Butler SE, Wahl DH. 2012. Impacts of invasive Asian carps on native food webs. Illinois-Indiana: Sea Grant College Program, University of Illinois, Urbana, IL [Google Scholar]
- 11. Kennedy D. 2001. Black carp and sick cows. Science 292:169. 10.1126/science.292.5515.169 [DOI] [PubMed] [Google Scholar]
- 12. Naylor RL, Williams SL, Strong DR. 2001. Aquaculture—a gateway for exotic species. Science 294:1655–1656. 10.1126/science.1064875 [DOI] [PubMed] [Google Scholar]
- 13. Phelps NB, Armién AG, Mor SK, Goyal SM, Warg JV, Bhagyam R, Monahan T. 2012. Spring viremia of carp virus in Minnehaha Creek, Minnesota. J. Aquat. Anim. Health 24:232–237. 10.1080/08997659.2012.711267 [DOI] [PubMed] [Google Scholar]
- 14. Miller O, Fuller FJ, Gebreyes WA, Lewbart GA, Shchelkunov IS, Shivappa RB, Joiner C, Woolford G, Stone DM, Dixon PF, Raley ME, Levine JF. 2007. Phylogenetic analysis of spring virema of carp virus reveals distinct subgroups with common origins for recent isolates in North America and the UK. Dis. Aquat. Organ. 76:193–204. 10.3354/dao076193 [DOI] [PubMed] [Google Scholar]
- 15. Walker PJ, Winton JR. 2010. Emerging viral diseases of fish and shrimp. Vet. Res. 41(6):51. 10.1051/vetres/2010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guillory V, Gasaway RD. 1978. Zoogeography of the grass carp in the United States. Trans. Am. Fish. Soc. 107:105–112. [DOI] [Google Scholar]
- 17. Nico LG, Williams JD, Jelks HL. 2005. Black carp: biological synopsis and risk assessment of an introduced fish. American Fisheries Society, Bethesda, MD. [Google Scholar]
- 18. Martino ME, Fasolato L, Montemurro F, Rosteghin M, Manfrin A, Patarnello T, Novelli E, Cardazzo B. 2011. Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes. Appl. Environ. Microbiol. 77:4986–5000. 10.1128/AEM.00708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng G-C, Jiang X-Y, Ye X, Liu M-Z, Xu S-Y, Liu L-H, Bai Y-Q, Luo X. 2009. Isolation, identification and characterization of Aeromonas hydrophila from hemorrhagic grass carp. Microbiol. China 36:1170–1177 [Google Scholar]
- 20. Zhu D, Li A, Wang J, Li M, Cai T, Hu J. 2007. Correlation between the distribution pattern of virulence genes and virulence of Aeromonas hydrophila strains. Front. Biol. China 2:176–179. 10.1007/s11515-007-0024-4 [DOI] [Google Scholar]
- 21. Rahman MM, Somsiri T, Tajima K, Ezura Y. 2004. Distribution of Aeromonas spp. emphasizing on a newly identified species Aeromonas sp. T8 isolated from fish and aquatic animal in South-East Asia. Pak. J. Biol. Sci. 7:258–264. 10.3923/pjbs.2004.258.268 [DOI] [Google Scholar]
- 22. Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics 9:566. 10.1186/1471-2164-9-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friis C, Wassenaar TM, Javed MA, Snipen L, Lagesen K, Hallin PF, Newell DG, Toszeghy M, Ridley A, Manning G, Ussery DW. 2010. Genomic characterization of Campylobacter jejuni strain M1. PLoS One 5:e12253. 10.1371/journal.pone.0012253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsai GJ, Chen TH. 1996. Incidence and toxigenicity of Aeromonas hydrophila in seafood. Int. J. Food Microbiol. 31:121–131. 10.1016/0168-1605(96)00972-5 [DOI] [PubMed] [Google Scholar]
- 25. Martinez-Murcia AJ, Monera A, Saavedra MJ, Oncina R, Lopez-Alvarez M, Lara E, Figueras MJ. 2011. Multilocus phylogenetic analysis of the genus Aeromonas. Syst. Appl. Microbiol. 34:189–199. 10.1016/j.syapm.2010.11.014 [DOI] [PubMed] [Google Scholar]
- 26. Beaz-Hidalgo R, Martinez-Murcia A, Figueras MJ. 2013. Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martinez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 36:171–176. 10.1016/j.syapm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 27. Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
gyrB phylogeny using sequences from 169 A. hydrophila isolates, including strains of Asian and U.S. origins. Colored ranges refer to the geographical origin of the isolates. The red segments indicate the epidemic strains. The tree figure was generated using the Interactive Tree Of Life web application (http://itol.embl.de) (27). Download
Phylogenetic analysis based on the virulent A. hydrophila (VAh)-specific sequences. The evolutionary history was inferred using the neighbor-joining method. The evolutionary distances are in units of number of base substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 303,863 positions in the final data set. Evolutionary analyses were conducted in MEGA5 with 1,000 repetitions to generate bootstrap support (28). *, isolate PB10-118 was isolated from a diseased fish obtained from Arkansas, USA (7). Download
BLASTp matrix of 14 different A. hydrophila isolates. The proteomes of each of the A. hydrophila isolates were compared pairwise using all-against-all BLASTp as described previously (23). This matrix illustrates the output from the pairwise comparison of conserved protein families of each of the isolates to each other. Green represents the relative percent homology between proteomes, and red represents relative percent homology within the isolate’s own proteome. The dense green pyramid on the right indicates greater homology (>97%) between the proteomes of VAh isolates from U.S. catfish epidemics, Chinese carp isolate ZC1, and Mississippi catfish isolate S04-690. Download
Aeromonas hydrophila isolates used for gyrB phylogeny.
Isolates of Aeromonas species used for MLST analysis.