ABSTRACT
Cryptococcus spp. cause life-threatening fungal infection of the central nervous system (CNS), predominantly in patients with a compromised immune system. Why Cryptococcus neoformans has this remarkable tropism for the CNS is not clear. Recent research on cerebral pathogenesis of C. neoformans revealed a predominantly transcellular migration of cryptococci across the brain endothelium; however, the identities of key fungal virulence factors that function specifically to invade the CNS remain unresolved. Here we found that a novel, secreted metalloprotease (Mpr1) that we identified in the extracellular proteome of C. neoformans (CnMpr1) is required for establishing fungal disease in the CNS. Mpr1 belongs to a poorly characterized M36 class of fungalysins that are expressed in only some fungal species. A strain of C. neoformans lacking the gene encoding Mpr1 (mpr1Δ) failed to breach the endothelium in an in vitro model of the human blood-brain barrier (BBB). A mammalian host infected with the mpr1Δ null strain demonstrated significant improvement in survival due to a reduced brain fungal burden and lacked the brain pathology commonly associated with cryptococcal disease. The in vivo studies further indicate that Mpr1 is not required for fungal dissemination and Mpr1 likely targets the brain endothelium specifically. Remarkably, the sole expression of CnMPR1 in Saccharomyces cerevisiae resulted in a robust migration of yeast cells across the brain endothelium, demonstrating Mpr1’s specific activity in breaching the BBB and suggesting that Mpr1 may function independently of the hyaluronic acid-CD44 pathway. This distinct role for Mpr1 may develop into innovative treatment options and facilitate a brain-specific drug delivery platform.
IMPORTANCE
Cryptococcus neoformans is a medically relevant fungal pathogen causing significant morbidity and mortality, particularly in immunocompromised individuals. An intriguing feature is its strong neurotropism, and consequently the hallmark of cryptococcal disease is a brain infection, cryptococcal meningoencephalitis. For C. neoformans to penetrate the central nervous system (CNS), it first breaches the blood-brain barrier via a transcellular pathway; however, the identities of fungal factors required for this transmigration remain largely unknown. In an effort to identify extracellular fungal proteins that could mediate interactions with the brain endothelium, we undertook a proteomic analysis of the extracellular proteome and identified a secreted metalloprotease (Mpr1) belonging to the M36 class of fungalysins. Here we found that Mpr1 promotes migration of C. neoformans across the brain endothelium and into the CNS by facilitating attachment of cryptococci to the endothelium surface, thus underscoring the critical role of M36 proteases in fungal pathogenesis.
INTRODUCTION
Cryptococcus neoformans is an opportunistic fungal pathogen that causes life-threatening meningitis, most commonly in populations with impaired immunity (1). The number of cases of cryptococcosis worldwide is estimated at 1 million, with more than 600,000 deaths per year (2). The morbidity and mortality associated with cryptococcal meningitis are unacceptably high, especially in HIV-infected populations in resource-poor settings (3). Without rapid intervention, cryptococcal meningitis is universally fatal regardless of the immune status of the host (1).
Humans are frequently in contact with C. neoformans through the inhalation of fungal spores or desiccated cells originating in environments such as soil and pigeon dwellings (4, 5). In the healthy population, the host defense can prevent fungal cells in the lung from causing symptomatic infection (1). However, in patients with impaired cellular immunity, C. neoformans can establish pulmonary infection and subsequently spread to other organs of the body, especially the central nervous system (CNS), where it causes life-threatening meningitis (1).
The mechanism responsible for C. neoformans’ remarkable neurotropism has yet to be fully resolved. In order for C. neoformans to invade the CNS, it must first breach the blood-brain barrier (BBB), which serves to protect the organism from circulating infectious and toxic agents (6, 7). At the cellular level, the BBB consists of specialized brain microvascular endothelial cells with robust tight junctions and few pinocytic vesicles (6). Work by various groups has provided strong evidence for C. neoformans’ ability to cross the BBB in vitro (8–10) and in vivo (11–14). Three major pathways have been proposed for the movement of C. neoformans across the BBB (15). Paracellular crossing of the BBB involves the loss of BBB integrity as a result of compromised tight junctions or injury to the brain endothelium, both of which can lead to opening of the BBB to facilitate C. neoformans’ entry into the CNS (9, 11, 13, 16–18). Crossing the BBB via the “Trojan horse” pathway involves the hijacking of phagocytic cells as a vehicle for C. neoformans to enter the CNS (14, 19, 20). Histological sections show the presence of cryptococci within macrophage-like cells either within or outside the capillaries in infected brain sections (14), suggesting that C. neoformans could have been transported inside circulating phagocytes. Accumulated evidence to date, however, suggests that C. neoformans crosses the BBB primarily through the transcellular pathway (8, 10, 15, 21). This mode of transmigration requires that cryptococci adhere to and are internalized by the brain endothelium from the luminal surface (blood side). Subsequently, the yeast cells transmigrate through the endothelial cytoplasm and exit on the abluminal side (brain side) of the BBB (8, 10, 12). Transcellular crossing has been observed in many invasive bacterial pathogens (22–25) and in Candida albicans (26) and has also been suggested for C. neoformans based on in vitro (8–10, 27) and in vivo (8, 12, 21, 28, 29) studies.
Compelling evidence in the literature supports the involvement of C. neoformans’ cell surface and secreted proteins in CNS invasion (17, 29–35). For example, the expression of extracellular phospholipase B (29, 35), urease (11, 12), and laccase (34) contributes to the ability of C. neoformans to cause brain infection. In addition, a ligand-receptor interaction involving cryptococcal surface-bound hyaluronic acid (a product of the CPS1 gene) and the CD44 host receptor has been amply demonstrated to take place during the adhesion of cryptococci to the brain endothelium in vitro (36–40). Based on the importance of the extracellular proteome of C. neoformans in CNS infection, we used a proteomic approach to isolate cryptococcal cell surface proteins and identified an assortment of proteases and free radical-inducing proteins with possible roles in CNS invasion (41). One of the candidate proteins was an uncharacterized fungalysin metalloprotease belonging to the M36 peptidase class of proteases that are unique to some fungi (41). Previous work by Steen et al. (42) also identified this metalloprotease as being encoded by a gene that is highly upregulated during CNS infection in a rabbit model of cryptococcal meningitis. C. neoformans metalloprotease is structurally similar to a neutral metalloprotease of Aspergillus fumigatus (43, 44) that is capable of breaking down elastin (45), a protein within connective tissue that is particularly abundant in lung and arteries. Accordingly, immunogold localization studies demonstrated that A. fumigatus secretes a fungalysin metalloprotease into invading lung tissue (45). The M36 fungalysin class of metalloprotease appears to mediate some aspects of the host-fungus interaction in A. fumigatus and some basidiomycetes, but the identities of their substrates remain unknown (46).
In this study, we identified a novel metalloprotease (Mpr1) through a screen for mutants of C. neoformans defective in crossing the brain endothelium in an in vitro model of the human BBB. We found that Mpr1 facilitates the migration of C. neoformans across the in vitro BBB and the subsequent invasion of cryptococci into the CNS. Our work indicated that the inability of a strain of C. neoformans lacking MPR1 (mpr1Δ) to cross the BBB was due to its failure to adhere to the brain endothelium. As expected, the mpr1Δ null strain was significantly less virulent than the wild-type C. neoformans strain in both the murine inhalation and tail-vein models of infection due to a significant reduction in brain fungal burden and brain pathology. This study fills a significant gap in our understanding of the role of a secreted protease in promoting the invasion of C. neoformans into the CNS and ultimately in the development of cryptococcal meningitis. Given the significant contribution of Mpr1 to C. neoformans’ CNS pathogenesis, this study may lead the way to the development of novel therapeutic interventions, and it provides a keen understanding of the fundamental mechanism of this important M36 class of fungalysins.
RESULTS
A newly identified metalloprotease (Mpr1) in C. neoformans.
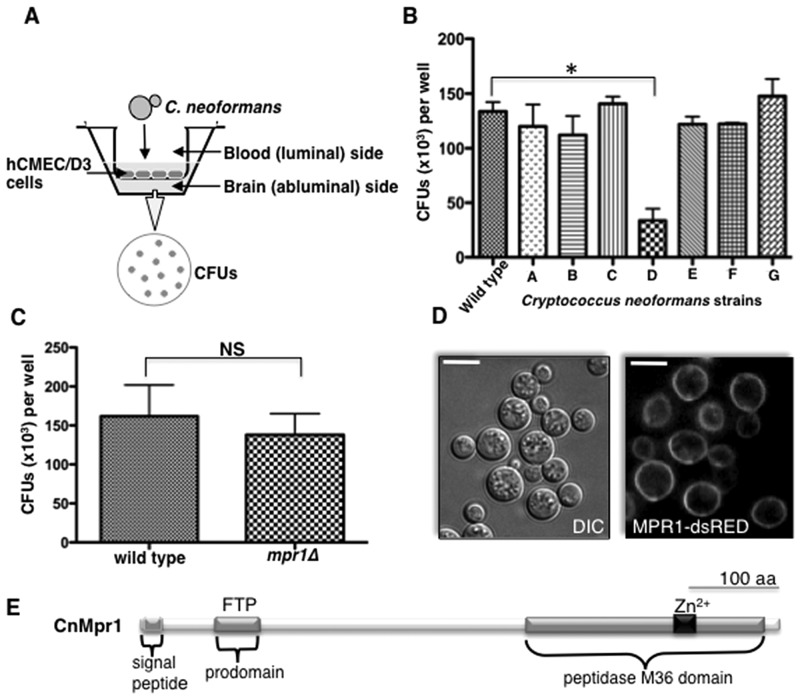
To determine whether extracellular proteases of C. neoformans promoted fungal disease of the CNS, individual strains of C. neoformans lacking known and predicted proteases were examined in an in vitro model of the BBB (Fig. 1A) (27, 47). In this model, human cerebral microvascular endothelial cells (hCMEC/D3 cells) were grown as a monolayer on a transwell insert submerged in medium specific for brain endothelial cells such that the luminal and abluminal sides represented the blood and brain sides, respectively. Cryptococci were added to the luminal side and collected from the abluminal side approximately 18 h later for CFU determination (Fig. 1A). Interestingly, a strain of C. neoformans lacking the newly identified metalloprotease Mpr1 (mpr1Δ) was defective in crossing the endothelium compared to a wild-type strain of C. neoformans (H99) (Fig. 1B). The inability of the mpr1Δ null strain to migrate across brain endothelial cells in the in vitro model of the BBB was not due to medium effects or an inability to tolerate 5% CO2 and 37°C, since both H99 and the mpr1Δ strain behaved similarly (Fig. 1C; see also Fig. S1A, B, and C in the supplemental material).
FIG 1 .
A novel metalloprotease (Mpr1) is required for the migration of Cryptococcus neoformans across the brain endothelium. (A) A schematic representation of the in vitro model of the BBB. A monolayer of hCMEC/D3 cells grown on transwell inserts submerged in medium such that the luminal and abluminal sides represented the blood and brain sides, respectively. Cryptococci were added to the luminal side and collected from the abluminal side approximately 18 h later for CFU determination. (B) Strains of C. neoformans lacking the genes encoding various proteases (H99 serotype A background; see Materials and Methods) were tested for their ability to transmigrate the brain endothelium in the in vitro model of the BBB. The strain D (mpr1Δ null strain, H99 serotype A background) was defective in crossing the endothelial barrier (*, P = 0.009) (n = 9). (C) CFUs representing cryptococci of the wild-type (H99) and mpr1Δ strains, equally able to proliferate in hCMEC/D3 growth medium and conditions in the absence of endothelial cells (n = 9). (D) Mpr1 is an extracellular protein. An MPR1-dsRED fusion protein was expressed in the mpr1Δ background strain to assess its localization. Confocal microscopy of an MPR1-dsRED fusion protein under the control of an ACTIN promoter revealed a surface localization (scale bar = 10 µm). (E) A schematic diagram of the predicted structure of Mpr1 illustrates the N-terminal signal sequence, the predicted prodomain (FTP region, Pfam), and the peptidase M36 domain with the conserved Zn2+-binding site.
Confocal microscopy demonstrated a surface localization of Mpr1 in Cryptococcus, indicating that Mpr1 is an extracellular protein (Fig. 1D). This result was further supported by the presence of the predicted N-terminal signal peptide in Mpr1 and by our proteomic analysis, where we identified Mpr1 as a secreted protease (48). Further in silico analysis of Mpr1 revealed a predicted prodomain and a peptidase M36 catalytic domain with a highly conserved Zn2+ binding motif (Fig. 1E).
Mpr1 mediates attachment of cryptococci to the brain endothelium.
The inability of the mpr1Δ strain to cross the endothelium suggested that Mpr1 promoted transcellular migration by facilitating an aspect of a multistep process that included adhesion, invasion, transmigration, or extrusion from the brain endothelium (9, 37, 49).
To resolve this further, we examined whether Mpr1 promoted the attachment of cryptococcal cells to the endothelium using an assay with the in vitro model of the BBB (8). To collect cells of C. neoformans that had initially associated with endothelial cells, Millipore water was added to the top transwell insert, resulting in the rupture of the endothelial cells (hCMEC/D3). The attached cryptococcal cells were recovered and plated to determine CFUs. The CFUs revealed how much of the initial inoculum of the mpr1Δ strain associated with the endothelium compared to results for H99 and the reconstituted strain (mpr1Δ::MPR1).
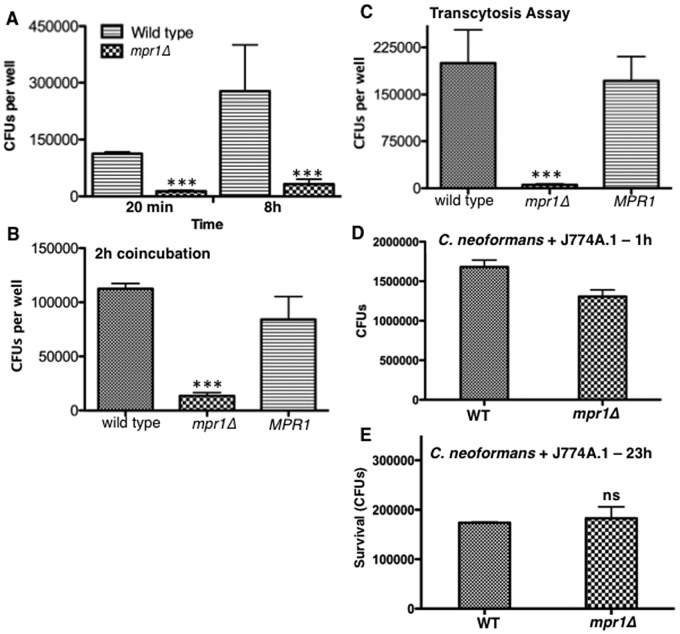
We found that Mpr1 was required for associating cryptococci with brain endothelial cells. This was demonstrated by the low CFU counts for the mpr1Δ strain compared to those for H99 and the reconstituted strains (mpr1Δ::MPR1) following either a 20-min (Fig. 2A) or 2-h coincubation (Fig. 2B) between cryptococci and the endothelium. Increasing the incubation time of the mpr1Δ strain with the brain endothelium to 8 h did not enhance its attachment (Fig. 2A). After a 24-h coincubation, significantly fewer mpr1Δ cryptococci crossed the endothelium, in stark contrast to results for H99 and the reconstituted strains, suggesting that cryptococci could not migrate across brain endothelial cells in the absence of Mpr1 (Fig. 2C).
FIG 2 .
Mpr1 promotes the adherence of Cryptococcus neoformans to brain endothelial cells. Shown are CFUs representing the number of cryptococci adhering to brain endothelial cells following 20 min or 8 h of coincubation (A) or a 2-h incubation (B) of H99 or the MPR1 (mpr1Δ::MPR1; reconstituted strain) and mpr1Δ mutant strains with the endothelium in the in vitro model of the BBB. The mpr1Δ mutant strain did not adhere to brain endothelial cells (***, P = 0.002) (n = 9). (C) The CFUs representing cryptococci that transmigrated to the abluminal (brain) side of the endothelium following a prolonged incubation with the brain endothelium revealed that the mpr1Δ mutant strain did not transmigrate. (D and E) Wild-type (H99) and mpr1Δ cells were incubated with the macrophage cell line J774A.1 to analyze intracellular survival. Following a 1-h coincubation at 37°C in 5% CO2, extracellular cryptococci were removed and the cocultures were either lysed and plated for CFU analysis (D) or replenished with fresh medium and incubated for an additional 23 h (overnight) at 37°C in 5% CO2 to determine intracellular viability of cryptococci (E). Following either a 1-h or overnight coincubation, cryptococci were collected from macrophages, plated on YPD medium, and incubated at 30°C for 2 days for CFU determination. The two strains were similar in their ability to survive within macrophages (n = 9). Statistical significance was determined by an unpaired t test with Welch’s correction (GraphPad Prism5 software).
Taken together, the results suggested that Mpr1 was likely required for attaching cryptococci to the surface of the brain endothelium; however, we could not rule out the possibility that Mpr1 might instead be required for survival within brain endothelial cells given that the cell association assay could not distinguish between adherence-defective cryptococci and those that might be inviable in endothelial cells. To address this issue, we performed intracellular survival assays using a macrophage-like cell line (J774A.1). The J774A.1 lung macrophages were challenged with either H99 or the mpr1Δ null strain of C. neoformans. The mpr1Δ null strain did not display any significant difference in its ability to survive within macrophages compared to the H99 strain, indicating that Mpr1 is not required for intracellular survival (Fig. 2D and E). Thus, these data further support the notion that Mpr1 mediates the attachment of cryptococci to the brain endothelium.
S. cerevisiae gains the ability to migrate across the brain endothelium upon the sole expression of CnMpr1.
Given that Saccharomyces cerevisiae does not express metalloproteases from the M36 fungalysin class, we sought to examine whether the expression of C. neoformans MPR1 (CnMPR1) in S. cerevisiae might generate a transformed strain capable of attaching to and penetrating the brain endothelium. A positive outcome with this approach would provide further support for the role of Mpr1 in transmigration.
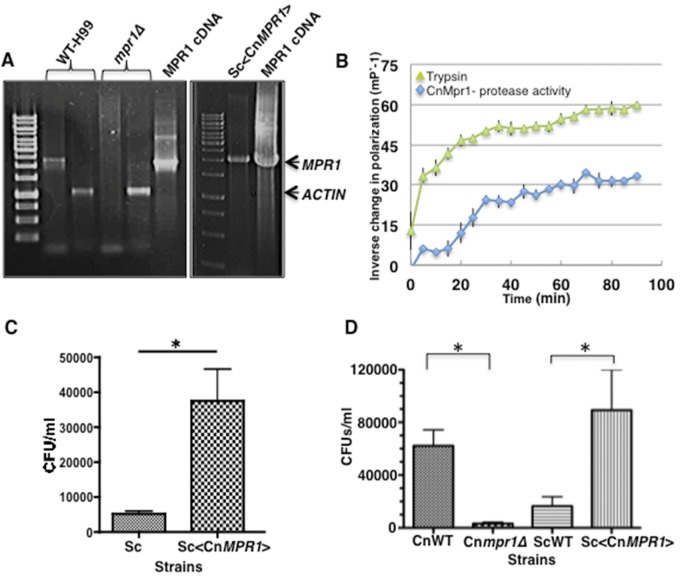
In order to test this, we subcloned the cDNA of CnMPR1 adjacent to the GPD promoter of an episomal 2µ plasmid and expressed it in a wild-type strain of S. cerevisiae. Reverse transcriptase PCR confirmed expression of the CnMPR1 transcript in S. cerevisiae (Fig. 3A). The MPR1 transcript was not detected in S. cerevisiae alone or in the mpr1Δ null strain, as expected. To examine whether Mpr1 retained proteolytic activity when expressed in S. cerevisiae, the recombinant His-tagged Mpr1 protein was isolated from S. cerevisiae and purified by nickel affinity chromatography. Mpr1 activity was measured by using a soluble fluorescein isothiocyanate (FITC)-labeled casein enzymatic assay (50). The assay revealed that the Mpr1 recombinant protein maintained proteolytic activity comparable to that of the trypsin control (Fig. 3B). Remarkably, the sole expression of CnMpr1 protein in S. cerevisiae resulted in the attachment (Fig. 3C) and migration of yeast cells across brain endothelial cells in the in vitro model of the BBB, strongly supporting a primary role for Mpr1 in attaching and penetrating the brain endothelium (Fig. 3D).
FIG 3 .
Expression of CnMPR1 cDNA in Saccharomyces cerevisiae (Sc) resulted in the attachment to and migration of S. cerevisiae across the brain endothelium. (A) Reverse transcriptase PCR confirmed the expression of the CnMPR1 transcript in S. cerevisiae. The band corresponding to the MPR1 transcript is absent in the mpr1Δ and S. cerevisiae (wild-type) strains, as expected. Actin is shown as an internal control for H99 given the typical faint appearance of the MPR1 transcript in H99. cDNA of CnMPR1 was subcloned adjacent to a GDP promoter in a yeast episomal 2µ expression vector. (B) A recombinant CnMpr1-His-tagged protein isolated via nickel chromatography from a yeast S. cerevisiae strain transformed with CnMPR1-HIS cDNA (Sc<CnMPR1-HIS>) retained proteolytic activity (n = 3). Trypsin activity is shown as a positive control for the proteolytic assay. (C) A 4-h coincubation of the Sc<CnMPR1> strain with brain endothelial cells in the in vitro BBB model resulted in a significant increase in the ability of yeast cells to associate with the surface of endothelial cells following a 4-h coincubation (*, P = 0.0294; n = 9). (D) The Sc<CnMPR1> strain migrated across the brain endothelial cells following an overnight coincubation. Transformed yeast cells (Sc<CnMPR1>) were added to the luminal side of the in vitro model of the BBB and collected from the abluminal side for CFU determination (*, P = 0.009; n = 9). Shown are C. neoformans strains (CnWT and Cnmpr1Δ) and S. cerevisiae strains (ScWT and Sc<CnMPR1>: yeast strain transformed with cDNA of MPR1 from C. neoformans).
Extracellular changes at cryptococcus-brain endothelium interface in absence of Mpr1.
We questioned whether Mpr1 might directly impact the fungus-brain endothelium extracellular environment such that the lack of MPR1 might result in specific physical changes to this interface. Scanning electron microscopy (SEM) was used to examine the surface structure of both cryptococci and brain endothelial cells. SEM micrographs revealed surface changes both to cryptococci lacking Mpr1 and to the brain endothelium (Fig. 4). Upon examination of C. neoformans in the absence of brain endothelial cells, the mpr1Δ null strain appeared to have a more porous surface than that of H99 cells (Fig. 4A and B). This was not due to a lack of capsule production, since both mpr1Δ and H99 strains displayed India ink-stained capsule (data not shown). Upon coincubation of H99 with brain endothelial cells, microvillus-like structures were observed on the surface of the endothelium (indicated by arrows in the figure); however, the endothelium alone did not produce these structures, thus supporting the notion that these protrusions likely promote transmigration of cryptococci, as previously reported (Fig. 4C, E, and G) (8, 26, 27). Interestingly in the presence of mpr1Δ strain, the microvillus-like structures appeared less abundant (Fig. 4D and F). Taken together, the data suggest that Mpr1 activity appears to alter the extracellular landscape of the fungus-brain endothelium interface (23, 24).
FIG 4 .
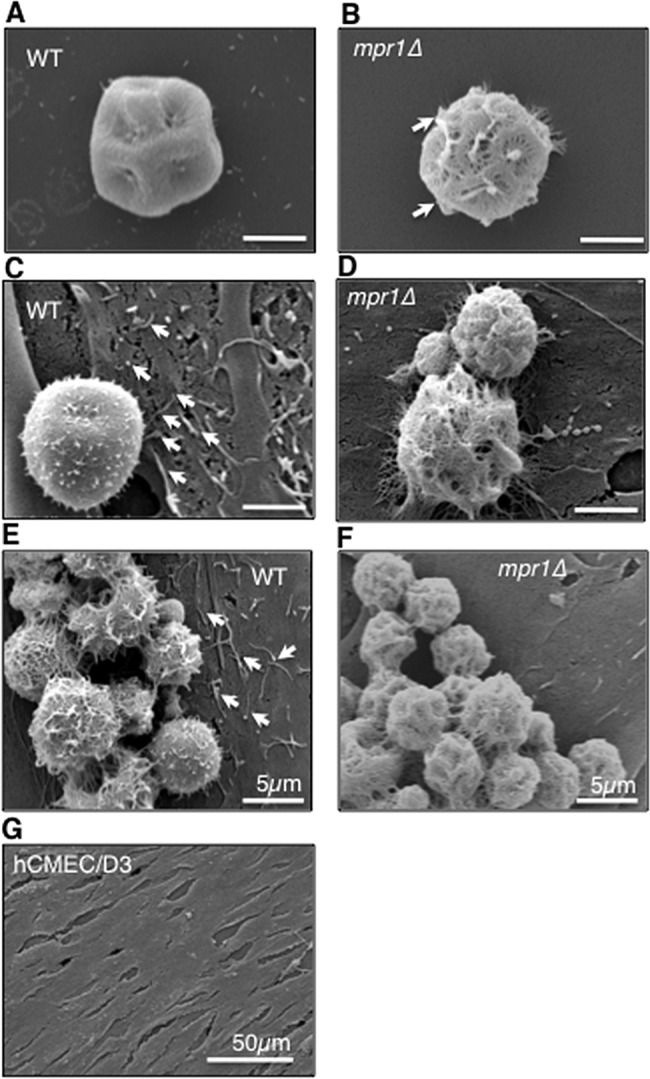
Mpr1 alters the extracellular environment of the fungus-endothelium interface. Scanning electron micrographs of cryptococcal cells from H99 and mpr1Δ strains reveal physical differences at the brain endothelium-fungus interface. In the absence (A and B) or presence (C to F) of hCMED/D3 cells (brain endothelial cells), the mpr1Δ null cells displayed a more porous surface than H99 cells (indicated by arrows). (C and E) Exposure of hCMED/D3 cells to H99 revealed extensive microvillus-like structures that appeared to be less abundant upon coincubation with mpr1Δ cells (D and F) (indicated by arrows). The microvillus-like protrusions were not observed on the surface of hCMED/D3 in the absence of C. neoformans (G) (scale bar = 2 µm unless noted differently in the figure).
Mpr1 promotes pathogenesis of C. neoformans in the inhalation murine model.
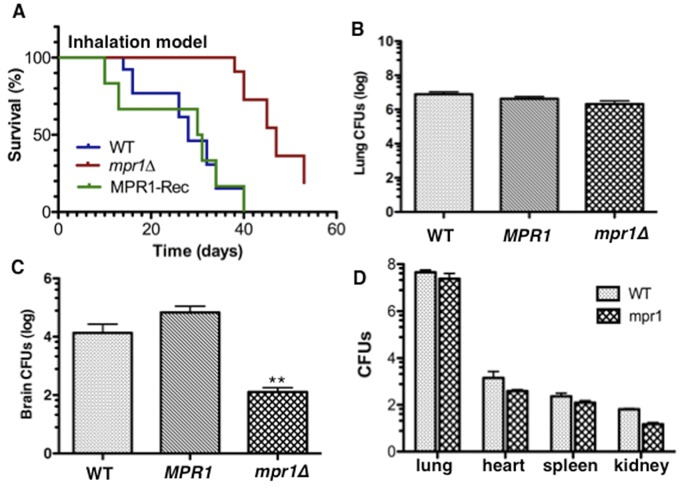
We next determined the contribution of Mpr1 to the virulence of C. neoformans in terms of survival and CNS invasion after pulmonary infection in a mammalian host. Mice were inoculated via the intranasal route to mimic the natural path of infection. By day 37 postinfection (p.i.), no mortality had occurred in mpr1Δ strain-infected mice, whereas 85% of H99-infected mice died (Fig. 5A) (P = 0.001), and at day 52 p.i., 15% of mpr1Δ strain-infected mice remained alive. These results demonstrated that the lack of Mpr1 significantly prolonged survival, indicating its major role in the pathogenesis of C. neoformans.
FIG 5 .
The murine inhalation model inoculated with a strain of Cryptococcus neoformans lacking Mpr1 showed significant improvement in survival due to a lack of fungal burden in the brain. (A) Shown is percent survival of mice inoculated via the nares, mimicking the natural route of infection, with the H99 (wild type), mpr1Δ, and MPR1 (mpr1Δ::MPR1; reconstituted strain) strains. Mpr1Δ strain-infected mice showed a significant improvement in survival (P = 0.001, Wilcoxon test, sample size determined by power analysis [GPower3.1]) (B and C) CFUs representing the fungal burden at the time of death in lungs (B) or brains (C) of mice inoculated with H99, the mpr1Δ mutant, and the MPR1-reconstituted strains revealed significantly less fungal burden in brains from mice infected with the mpr1Δ mutant strain (**, P = 0.009). (D) Following 10 days p.i. (postinfection), mpr1Δ strain-infected mice and H99-infected mice showed similar fungal burdens in lungs, kidneys, spleen, and heart, suggesting that Mpr1 is not required for dissemination out of the lung and Mpr1 does not play a role in breaching other vascular endothelia (P > 0.05 for all tissues).
Total-lung CFU analysis at the time of death (Fig. 5A) and 10 days p.i. (Fig. 5D) revealed a high cryptococcal burden in mpr1Δ strain-infected mice, similar to CFU in lungs of mice infected with H99 (Fig. 5D). These data indicated that improved survival of mpr1Δ strain-infected mice was not due to enhanced clearance of the mpr1Δ strain-induced infection from the lungs, the primary site of infection. However, lack of MPR1 from C. neoformans significantly reduced the fungal burden in the brain following pulmonary inoculation (Fig. 5C) (P = 0.009, CFU measured at time of death), suggesting a crucial role for Mpr1 in the transmigration of C. neoformans into the CNS. Examination of the fungal burden in the heart, spleen, and kidney following pulmonary infection further demonstrated that the mpr1Δ strain colonized other organs similarly to colonization by H99, suggesting that cryptococci do not require Mpr1 to exit the lung nor to invade other vascular endothelia (Fig. 5D) (P > 0.05 for all tissues).
Mpr1 promotes migration of C. neoformans across the brain endothelium.
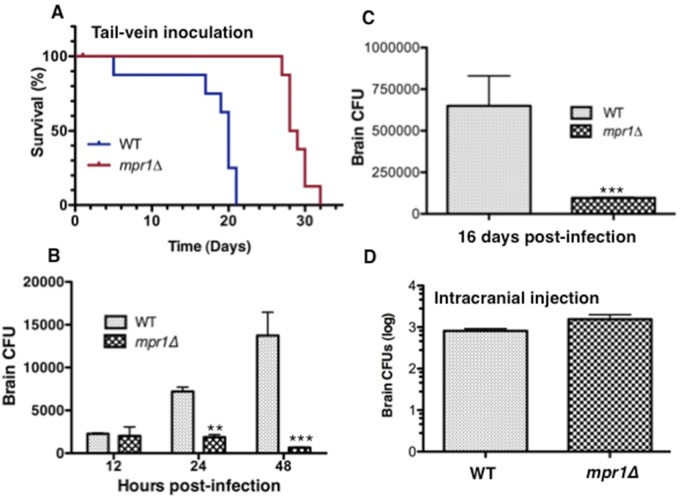
When mice were inoculated via a more direct route to the brain (tail vein injection), we found that by day 21 p.i., 100% of mpr1Δ strain-infected mice were alive and 100% of H99-infected mice had died, suggesting that the lack of MPR1 in the pathogen significantly prolonged survival of the host (Fig. 6A) (P = 0.0002). Upon examination of fungal burden at 48 h p.i. in whole brains of mice infected with the mpr1Δ strain and H99, numbers of CFU revealed a significant increase in cryptococci in whole brains of mice infected with H99 but dramatically less fungal burden in the brains from mpr1Δ strain-infected mice (Fig. 6B) (P < 0.0001). By 16 days p.i., the brain fungal burden in mice inoculated with the mpr1Δ strain remained significantly lower (Fig. 6C). Given that our histological analysis (Fig. 7) revealed no pathology in brains of mice infected with the mpr1Δ null strain, it is very likely that the CFU observed at 12 h postinfection are a reflection of cryptococci that have been trapped within small microvessels because they are unable to adhere to and cross the blood-brain barrier. This is reminiscent of findings of a previous study that showed that prior to actively transmigrating through the brain endothelium, C. neoformans can become mechanically trapped in small brain capillaries independent of rolling and tethering along the surface of the endothelium (12).
FIG 6 .
Association of cryptococci with the brain endothelium requires Mpr1 in the tail-vein murine model. (A) The percent survival of mice inoculated via a more direct route to the brain (tail vein) with the H99 and mpr1Δ strains revealed a dramatic improvement in survival of mice infected with the mpr1Δ strain. (B and C) CFUs representing the fungal burden in brains of mice following 12 h, 24 h, and 48 h (B) or 16 days (C) postinfection with the H99 and mpr1Δ strains revealed significantly less fungal burden in brains of mice infected by the mpr1Δ null strain. (D) The number of cryptococci that survived within brain parenchyma of mice following inoculation by direct intracranial injection of the wild-type and mpr1Δ strains revealed no significant differences between strains. (Asterisks correspond to P values. **, P = 0.008; ***, P < 0.001.)
FIG 7 .
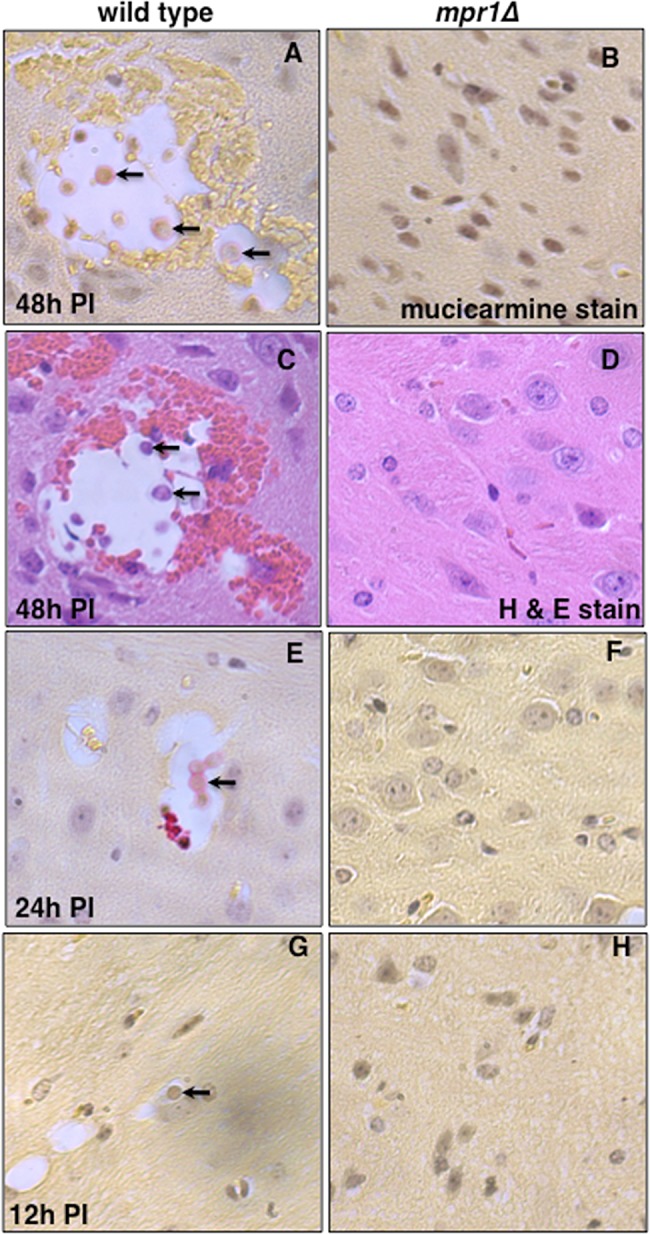
Brains from mice inoculated via the tail vein with the strain lacking Mpr1 revealed a complete lack of brain pathology, suggesting that Mpr1 is required for fungal invasion of the CNS. Mice were inoculated by tail-vein injection, and brains were examined at 12 h, 24 h, and 48 h p.i. by taking 20 consecutive, longitudinal brain sections and staining with mucicarmine (specific for cryptococci) or H&E. Histological analysis of brain slices from brains of mice inoculated with the H99 (A, C, E, and G) revealed lesions filled with cryptococci at various stages of cell division (arrows point to cryptococci within lesions), consistent with fungal disease of the CNS (n = 12). However, brain sections from mice infected with the mpr1Δ mutant strain (B, D, F, and H) at 48 h p.i. (A and D), 24 h p.i. (F), or 12 h p.i. (H) revealed a complete absence of cryptococcus-filled lesions, indicating no brain pathology (n = 12). Brain sections were stained with mucicarmine (A, B, E, F, G, and H) or H&E (C and D).
Taken together, the in vivo data strongly suggested that the mpr1Δ fungal strain was defective in crossing the brain endothelium and invading the brain parenchyma. However, we could not rule out the possibility that the significantly lower CFU counts detected in whole brains of mice infected with the mprΔ1 strain might instead reflect a requirement for Mpr1 in intraparenchyma brain survival. To address this, mice were directly inoculated with cryptococci (H99 or the mpr1Δ strain) by intracranial injections into the brain, and survival of the strains within the brain parenchyma was assessed by determining CFU counts for isolated brains (51). The results show that the mpr1Δ strain and H99 were equally able to proliferate within the brain (Fig. 6D). Thus, we concluded that Mpr1 is not required for the growth and survival of C. neoformans within the brain; instead, Mpr1 functions primarily to attach cryptococci to the surface of the brain endothelium.
Mpr1 contributes to brain pathology in a murine model.
We determined the extent of brain pathology in mpr1Δ strain-infected mice by histochemical analysis of mice inoculated via the tail vein by examining at least 12 brain slices from mice infected with either H99 or the mpr1Δ strain after 12 h, 24 h, and 48 h p.i. Cryptococcal filled lesions were consistently observed in the brain parenchyma of H99-infected mice 48 h p.i., often in close proximity to microvessels. In a representative image shown in Fig. 7A, cryptococci at various stages of division were easily identified within cysts that occurred throughout the brain tissue. Similarly, cryptococci were also observed within lesions in brains from mice at 12 h and 24 h p.i. (Fig. 7E and G). In stark contrast, cryptococcus-filled cysts were not detected in brains from mice inoculated with the mpr1Δ strain at 12 h, 24 h, or 48 h p.i. Remarkably here we observed intact brain tissue with healthy microvessels that were devoid of cryptococci (Fig. 7B, D, F, and H).
DISCUSSION
Although it has been known that C. neoformans expresses several different types of proteases, this is the first clear demonstration of a specific role for a secreted protease in CNS invasion. We first identified the metalloprotease (Mpr1) in a proteomic study aimed at isolating and identifying extracellular proteins of C. neoformans (41). Among the extracellular proteome, we found that Mpr1 belonged to a relatively uncharacterized, new class of secreted fungalysins (the peptidase M36 family) whose substrates have yet to be fully characterized (41, 46). This class of metalloproteases is most closely related to the bacterial thermolysin family, whose members have been implicated as key factors in the pathogenesis of many diseases (46). The fungal M36 class was first described for Aspergillus fumigatus, where the metalloprotease MEP42 was shown to target elastin in human lungs (45, 52). Mounting evidence further supports the notion that fungalysins promote disease by targeting factors in both mammalian and plant hosts, and they have also been implicated in fungal disease in amphibians and insects, but the substrates in most cases have not been resolved (52–55).
Genome sequence analysis has revealed that not all fungi express fungalysins; for example Saccharomyces cerevisiae and Neurospora crassa have none, while Coprinopsis cinerea has eight predicted fungalysins (46). In the case of C. neoformans, only one fungalysin is present in its genome (46). Here we demonstrated that this fungalysin, which we refer to as Mpr1, facilitates the migration of C. neoformans across the brain endothelium and the subsequent invasion into the CNS. Our study proposes that the inability of the mpr1Δ null strain to cross the brain endothelium was due to its failure to adhere to the brain endothelium, and as expected, mice infected with the mpr1Δ null strain showed significant improvement in survival due to a reduced brain fungal burden, and the brain pathology that is commonly seen in cryptococcal meningoencephalitis disease was absent.
The inability of the mpr1Δ strain to migrate across the brain endothelium suggests that Mpr1 promotes the transcellular crossing of cryptococci by facilitating an aspect of a complex series of events that include attachment to the endothelium luminal surface, internalization, transmigration and extrusion from the abluminal side of the brain endothelium (9, 37, 49). Despite the limitations of the attachment assay performed here with the in vitro model of the brain endothelium, three separate observations strongly suggest that Mpr1 promotes the attachment of cryptococci to the endothelium and does not play a role in the intracellular survival of cryptococci. First, significantly fewer cells of the mpr1Δ strain associated with the brain endothelium following only a 20-min coincubation, before most cryptococci had been internalized. Second, Mpr1 was not required for the intracellular survival of cryptococci, as demonstrated by the assays performed with the macrophage cell line. Given that the mpr1Δ strain can survive as well as H99 in a professional phagocytic cell like an activated macrophage, it would seem probable that mpr1Δ cells likely survive within brain endothelial cells. Third, the sole expression of CnMPR1 in S. cerevisiae resulted in a significant migration of the transformed yeast strain across the brain endothelium, demonstrating that the strain likely attached to the endothelial surface prior to crossing. Last, direct intracranial inoculations in the murine model revealed that the mpr1Δ strain was as viable as H99 within the brain parenchyma, and although this may not reflect survival within host cells, it does indicate that survival within a harsh and restrictive environment like the brain is independent of Mpr1. Taken together, these results strongly support the notion that Mpr1 mediates that attachment and/or association of C. neoformans with the surface of the brain endothelium.
In C. neoformans, surface-bound hyaluronic acid serves as a ligand for the CD44 host receptor and mediates adherence of cryptococci to the brain endothelium in vitro (36–40). Interestingly, it has been shown that S. cerevisiae does not naturally express hyaluronic acid but can be engineered to do so upon expression of a hyaluronic acid synthase gene (CPS1). We found that the sole expression of CnMPR1 in a wild-type strain of S. cerevisiae resulted in a robust migration of yeast cells across the brain endothelium, but given the lack of hyaluronic acid in yeast, the CnMPR1-transformed S. cerevisiae strain is likely crossing the brain endothelium independently of the CD44 pathway. Given this observation, CnMpr1 likely targets other host components that may alter endothelium permeability and thus facilitate cryptococcus migration into the CNS.
Consistent with these observations and previous reports, the movement of C. neoformans across the brain endothelium observed in the in vitro BBB model was indicative of a transcellular mechanism whereby cryptococci crossed the endothelium with little or no disruption of the intercellular tight junctions; thus, protein complexes that constitute the tight junctions of the brain endothelial cells are likely not substrates of Mpr1 activity. This observation is further supported by previous reports demonstrating that cryptococci breach the endothelium through an active process via a receptor-mediated event involving the formation of microvillus-like structures on the surface of endothelial cells that likely facilitate the transcellular movement (8, 11, 12, 14, 27). We found that Mpr1 appears to alter the physical characteristics of the extracellular environment of the cryptococcus-endothelium interface as noted in the SEM micrographs, but how these structural changes relate to the targets of Mpr1 activity remains to be seen.
The in vivo studies clearly demonstrate that Mpr1 plays a significant role in the virulence of C. neoformans, and several observations suggest that its role is specific to the pathogenesis of the CNS. First, the significant reduction in the fungal burden in the brain of mice infected with the mpr1Δ strain, the ability of the mpr1Δ null strain to survive within brain parenchyma, and the lack of brain pathology all suggest that Mpr1 is required for attaching cryptococci to the surface of the brain endothelium in vivo, consistent with the in vitro data. Second, the ability of the mpr1Δ strain to disseminate out of the lungs and into other vascular beds point to a role for Mpr1 that could be specific for the brain endothelium. Interestingly, brain sections were devoid of immune cells, as demonstrated by the lack of neutrophils in brain sections stained with hematoxylin and eosin (H&E). The lack of immune cells in the histochemical analysis of brain tissue is consistent with findings of previous work that demonstrated that both in humans and in mice, inflammation in the brain tissue was limited, presumably due to late production of low concentrations of tumor necrosis factor (TNF) alpha and interleukin-6 in brains of infected mice (14, 56, 57). In addition, only low concentrations of these cytokines were found in the cerebrospinal fluid (CSF) of patients with AIDS who suffered from cryptococcal meningitis (14, 56, 57).
This study further supports the notion that multiple virulence factors in addition to Mpr1 likely come into play during invasion of the CNS. In our murine studies, at the time of death, some cryptococcal cells could be detected in brains from mpr1Δ strain-infected mice, indicating that other virulence factors (and other mechanisms, such as “Trojan horse”: phagocyte-associated invasion) may be involved in promoting CNS fungal disease (11, 20). For example, urease has been shown to promote the sequestration of cryptococcal cells within the brain microvasculature (11). Increasing evidence about cryptococcal virulence factors in CNS disease would suggest that a combination of factors, rather than a single one, is most likely involved in the complex, multistep process of cryptococcal dissemination and CNS invasion. In addition, prolonged exposure of the brain endothelium to cryptococci results in disruption of brain endothelial cells and the eventual breakdown of the barrier, thus allowing the passive movement of cryptococci into the brain (12, 14, 16, 58, 59).
In conclusion, we propose a targeted role for Mpr1 in establishing fungal disease of the CNS by promoting the attachment of C. neoformans to the blood-brain barrier. Since Mpr1 represents a major tactic used by cryptococci to invade the CNS, blocking its function may promote development of a novel therapeutic intervention, and coupling it to nanocarriers may give rise to a brain-specific drug delivery system.
MATERIALS AND METHODS
Transcytosis screen for mutants defective at crossing the in vitro BBB model.
The in vitro model of the BBB was established here as previously described (27). Briefly, the in vitro BBB model consists of a transwell apparatus separating the luminal (blood) and abluminal (brain) side of the BBB. hCMEC/D3 cells were grown in the top transwell chamber on a collagen-coated microporous membrane (8 µm). Fungal cells (1 × 105 cells) were added to the top chamber and collected subsequently from the bottom chamber for CFU determination. The Cryptococcus neoformans null strains used in the transcytosis assays shown in Fig. 1 are as follows: wild type, H99; A, putative disintegrin; B, putative protease; C, putative secreted hydrolase; D, Mpr1; E, serine protease; F, IgA1 protease; G, propyl endopeptidase. The strains lacking each gene were obtained from a library of ~1,200 deletion strains of Cryptococcus neoformans (MATα, serotype A, var. grubii) (ATCC CNKO-PS).
Adhesion/cell association assay of C. neoformans with hCMEC/D3 cells.
hCMEC/D3 cells were grown to confluence on collagen-coated transwells as discussed above. An inoculum of 1 × 106 C. neoformans cells in a 50-µl phosphate-buffered saline (PBS) volume was added to the endothelial barrier at the start of the experiment. After the indicated amount of incubation time, the endothelial barrier was washed 3 times with 1× PBS and either lysed with water to determine the number of yeast cells that adhere to and associate with the barrier or incubated overnight to assess the number of yeast cells that transmigrate across the barrier after the initial adhesion step. This last transcytosis step was done to show that a transcytosis defect follows an initial adhesion defect.
Expression of CnMPR1 cDNA in Saccharomyces cerevisiae and measurement of proteolytic activity of Mpr1.
The CnMPR1 cDNA containing a C-terminal 6×-HIS tag was cloned into the EcoRI and HindIII sites of an episomal yeast expression vector under the constitutive GDP promoter with the following primers: forward, GTAGAATTCATGCGCTCCTCCGCGCTCATC; and reverse, TACAAGCTTTCAAGATTTGGTCGGCCTGAA. The plasmid was transformed into a wild-type strain (W303) of Saccharomyces cerevisiae via electroporation (it should be noted that S. cerevisiae does not express any homologues of the M36 fungalysin class of metalloproteases, including MPR1). Expression was confirmed by reverse transcriptase PCR. Large quantities of CnMpr1 was extracted from 500-ml cultures of S. cerevisiae expressing CnMPR1. Yeast cells were grown overnight at 30°C in yeast nitrogen base (YNB) medium lacking uracil. Cultures were centrifuged for 5 min at 5,251 × g in 250-ml Falcon tubes to pellet yeast. Pellets were resuspended in 4 ml morpholineethanesulfonic acid (MES)-buffered saline (pH 6.8, containing 1% Nonidet P-40, 300 mM NaCl, 20 mM imidazole, and 120 µl yeast protease inhibitor cocktail [Sigma-Aldrich, St. Louis MO]) with the addition of acid-washed glass beads (0.5 mm; Sigma-Aldrich). The yeast-bead mixture was vortexed and iced in altering 1-min cycles at 4°C Lysates were separated from the beads by centrifugation at 5,251 × g for 5 min at 4°C and then centrifuged again to remove debris. The protein Mpr1 was isolated from the crude lysate by passage through gravity-flow 1.5-ml nickel-nitrilotriacetic acid (Ni-NTA) columns (Qiagen, Valencia, CA) according to the directions of the kit (note that buffer composition was changed from using 50 mM phosphate buffer at pH 7.2 to using 50 mM MES buffer at pH 6.8) and eluted into a 10-ml volume. Eluate was concentrated by centrifugation on 30,000-molecular-weight-cutoff (MWCO) Amicon Ultra centrifuge columns (Millipore, Billerica, MA) for 15 min at 4°C at 5,251 × g. Concentrates were washed twice with MES-buffered saline, pH 6.8 (50 mM MES and 150 mM NaCl). Final protein concentrations were determined by a Bradford assay (Bio-Rad). The protein activity assay was performed by serially diluting Mpr1 in 2-fold dilutions, ranging from 100 µg/ml to 0.78 µg/ml. The activity was assayed using a Pierce fluorescent protease activity assay kit (Thermo Scientific), according to manufacturer’s directions. Fluorescence polarization (Excitation, 488 nm; emission, 535 nm) was measured every 5 min at 30°C on a SpectraMax M5 microtiter plate reader (Molecular Devices, Sunnyvale, CA) and analyzed using the Excel 2011 software program (Microsoft, Redmond, WA).
SEM.
hCMEC/D3 cells were grown on transwell filters (8 µm; BD Biosciences) under the same conditions used for the transcytosis assays. Approximately 2 × 106 cells of a wild-type strain of C. neoformans (H99) were added to the top chamber for 1 h. The transwell filters were then washed with 1× PBS to remove nonspecific adhering yeast cells, fixed in Karnovsky’s fixative, and sent to an on-campus scanning electron microscopy (SEM) facility for sample processing (Electron Microscopy Lab, University of California, Davis [UC Davis]). For SEM preparation of C. neoformans in the absence of an endothelial barrier, the fungal cells were grown overnight in yeast extract-peptone-dextrose (YPD), washed in PBS, spun down, and fixed in Karnovsky’s fixative.
Macrophage intracellular survival assay.
J774A.1 (ATCC TIB-67, kindly provided by R. Tsolis, UC Davis) is a macrophage-like cell line from a BALB/c, haplotype H-2d, reticulum sarcoma used in the assay. Macrophages were grown at 37°C with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) (11995; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (16140-071; Invitrogen), 1% nonessential amino acids (21340; Gibco), 50 µg/ml penicillin-streptomycin (Pen/Strep) (15070-063; Gibco), and 10% NCTC-109 medium (11140; Gibco). J774A.1 was grown in 25-cm2 or 75-cm2 flasks (136196 or 156499; Thermo Sci Nunc) and used between passages 4 and 15.
For the assay, 2.5 × 105 J774A.1 cells were counted using a hemacytometer (1475; Hausser Scientific) and grown in 96-well culture plates (353072; BD Falcon) overnight. C. neoformans was cultured overnight in YPD plus 2% glucose at 30°C with agitation. Cultures were then synchronized and normalized based on the absorbance value measured (optical density at 600 nm [OD600]) and allowed to grow overnight. Using a hemacytometer, 1 × 106 C. neoformans cells were added to macrophages supplemented with 100 U/ml mouse gamma interferon (IFN) (RM200120; Thermo Sci Pierce Protein Research products), 0.3 µg/ml lipopolysaccharide (LPS) (L4391; Sigma), and 1 µg/ml mAb18B7 (kindly provided by A. Casadevall, Yeshiva University). To obtain a CFU count for C. neoformans following incubation with macrophages, J774A.1 cells were lysed using 100 µl of ice-cold 0.05% SDS. Lysed J774A.1 cells were then serially diluted and plated on YPD plus Pen/Strep plates for 48 h at 30°C for CFU determination
Two experiments were done in parallel: experiment 1, to assess the association of the mpr1Δ strain versus that of C. neoformans H99 cells after an initial 1-h incubation with the macrophage cell line J774A1, and experiment 2, to assess the intracellular survival of the mpr1Δ strain versus that of H99 cells in macrophages 23 h following the 1-h initial incubation period. In experiment 2, nonadherent cryptococcal cells were washed away at 1 h (3 times with 1× PBS), and the infected macrophage culture was replenished with fresh medium (DMEM plus supplement) and allowed to continue for another 23 h. Therefore, the CFU obtained from experiment 1 represented the yeast “inoculum” used in experiment 2. The advantage of this experimental design was that the “inoculum” as defined represented only the number of yeast cells that adhered to or was internalized by macrophages at 1 h of infection. This reduces any variations due to growth differences between mpr1Δ cells and H99 cells in DMEM.
Inhalation model (survival study and CFU analysis). (i) Experimental parameters.
Yeast cells (1 × 107) in a 50-ml volume were used to inoculate mice. Mice (A/J strain, 000646; Jackson Laboratories) were 5 weeks old, inbred, and housed with 3 to 4 animals per cage (n = 7 animals per group: wild type [WT], reconstituted, and mutant).
(ii) Preparation of yeast inoculum.
Frozen stocks of the 3 yeast strains used in the study (H99, the mpr1Δ strain, and Rec1) were streaked on YPD plates and incubated overnight at 30°C. Colonies were then grown in liquid YPD overnight at 30°C until log phase (approximately 16 h). Cultures were regrown a second time (to log phase) by adding 100 µl of liquid culture into fresh YPD to ensure that yeast colonies were robust. Ten milliliters of each cultured strain was cleaned with sterile 1× PBS (4,000 rpm, 4 min, at 25°C) and resuspended in YPD. The concentration of the original culture was determined by hemacytometer count (Brite-Line) and was diluted in 1× sterile PBS to a final concentration of 2 × 108 cells/ml (equates to 1 × 107 cells/50-µl volume).
(iii) Inoculation (survival assay).
After arrival at our animal facilities, the 4-week-old mice were given 1 week to acclimate and given food and water ad libitum. Experiments were conducted in a separate surgical room. Mice were placed under 5% isoflurane anesthesia (2.5% maintenance dose) and inoculated with 1 × 107 cells in 50-µl volume through the nares. The animals were suspended by their top incisors in order to extend their neck, thus ensuring that the inoculum was inhaled smoothly with minimal obstruction. After applying the inoculum, mice were suspended for another 30 s to ensure that all of the inoculum was inhaled. Changes in the health status of the animals were monitored, and body weight was recorded daily until they showed signs of sickness due to the infection. Animals were sacrificed when a loss of 15% of peak body weight was observed or when a loss of 1 g of body weight was observed for 3 consecutive days. Other signs of sickness were monitored, including lethargy and lack of activity and grooming (indicated by ruffled fur). Mice were sacrificed by an overdose of pentobarbital (Fatal Plus; MWI Veterinary Supply) at 150 mg/kg body weight. Survival data were collected and analyzed using the GraphPad-Prism 5.0 software program.
(iv) Tissue homogenization (CFU analysis).
Lungs and brains (and other organs) were collected for homogenization immediately after the animals were sacrificed. Lungs were cut into several pieces, mashed dry with a mortar and pestle for 1 min, and suspended in 5 ml of 1× PBS. Brains were mashed dry for 30 s and suspended in 1× PBS. Original homogenized tissue was considered 1×. Lung tissue was serially diluted with 1× PBS to 100×, 1,000×, and 10,000× dilutions; brain tissue was diluted to 10×, 100×, and 1,000× dilutions; heart, spleen, and kidney were diluted to 10× and 100×. One hundred microliters of each respective dilution was spread on YPD plates containing penicillin-streptomycin and incubated for 2 days at 30°C. Total CFUs were calculated based on the number of colonies counted on the plate, plating volume (100 µl), dilution (from 1× to 10,000×), and the original volume of the homogenized tissue (5 ml). With a few exceptions, we tried to count plates with at least 100 colonies to maximize accuracy.
Intravenous model (survival study and CFU analysis). (i) Experimental parameters.
Yeast cells, 1.2 × 106 in a 200-µl volume, were used to inoculate mice. Mice (A/J strain, 000646; Jackson Laboratories) were 8 weeks old, inbred, and housed with 3 to 4 animals per cage (n = 8 animals per group: WT, reconstituted, and mutant).
(ii) Preparation of yeast inoculum.
The inoculum was prepared as described above. The concentration of the original culture was diluted in YPD to a final concentration of 6 × 106 cells/ml (equates to 1.2 × 106 cells/200 µl total volume).
(iii) Inoculation (survival assay).
Upon arrival at our animal facilities, 7-week-old mice were given 1 week to acclimate and given food and water ad libitum. Experiments were conducted in a separate surgical room. A/J mice, under 3% isoflurane anesthesia (1% maintenance dose as needed), were inoculated with 1.2 × 106 cells in a 200-µl total volume through the lateral tail vein. Mice were held briefly in a restrainer (Kent Scientific) during anesthesia and injection. Once the tail vein became dilated by exposure to a heat lamp, it was disinfected with alcohol swabs (Becton, Dickinson). Thirty-gauge, 1/2-in. needles (Becton, Dickinson) and 1-ml tuberculin syringes (Kendall) were used to deliver the inoculum. After injection was completed, mice were given fresh oxygen until they awoke. Mice were monitored as discussed above and sacrificed with an overdose of pentobarbital (Fatal Plus; MWI Veterinary Supply) at 150 mg/kg body weight.
(iv) CFU analysis.
Brains were collected for homogenization immediately after the animals were sacrificed. Brains were mashed on a mortar and pestle, dry, for 30 s and resuspended in 5 ml of 1× PBS. Original homogenized tissue was considered 1×. Brain tissue was serially diluted with 1× PBS to 10×, 100×, and 1,000× dilutions (1 ml of homogenate in 9 ml 1× PBS). Ten milliliters of each respective dilution was spread on YPD containing penicillin-streptomycin and incubated for 2 days at 30°C. Total CFU were calculated based on the number of colonies counted on the plate, plating volume (100 µl), dilution (from 1× to 1,000×), and the original volume of the homogenized tissue (5 ml).
Intravenous model: brain burden time course (CFU analysis and histology). (i) Experimental parameters.
Yeast cells (5 × 105) in a 200-µl volume were used to inoculate mice. Mice (A/J strain, 000646; Jackson Laboratories) were 8 weeks old, inbred, and housed with 3 to 4 animals per cage. The number of animals used per groups (WT and mutant) was 4 per time point (12 h, 48 h, and 72 h). Three of the animals per group were used for CFU analysis, and one was saved for histological analysis.
(ii) Preparation of yeast inoculum.
The inoculum was prepared as described above. The concentration of the original culture was diluted in 1× sterile PBS to a final concentration of 2.5 × 106 cells/ml (equates to 5 × 105 cells/200 ml total volume).
(iii) Inoculation (CFU analysis).
Injection protocols were the same as discussed above. Mice were sacrificed at 12 h, 24 h, and 72 h using Pentobarbital (150 mg/kg of body weight). Brains, extracted immediately following euthanasia, were mashed dry for 30 s and suspended in 3 ml YPD (designated 1×). The homogenate was serially diluted to 10× and 100× (200 µl of homogenate diluted in 1,800 µl YPD). Two hundred microliters of each respective dilution was spread on YPD plates containing P/S and incubated for 2 days at 30°C. Total CFUs were calculated based on the number of colonies counted on the plate, plating volume (200 µl), dilution (from 1× to 100×), and the original volume of the homogenized tissue (3 ml).
(iv) Histology.
Brains were fixed in 4% paraformaldehyde prior to sectioning. Sectioning was carried out by the UC Davis Pathology Laboratory. Briefly, 20 consecutive, longitudinal sections (50 µm apart) were taken and stained with mucicarmine or H&E (hematoxylin and eosin) stain.
Intraparenchymal survival study (CFU analysis only). (i) Experimental parameters.
Yeast cells (1 × 104) in a 60-µl volume were used to inoculate mice. Mice (A/J strain, 000646; Jackson Laboratories) were 10 weeks old, inbred, and housed with 3 to 4 animals per cage (n = 6 animals used per group, WT and mutant).
(ii) Preparation of yeast inoculum.
Inoculum was prepared as described above. The concentration of the original culture was diluted in 1× sterile PBS to a final concentration of 1.67 × 106 cells/ml (equates to 1 × 104 cells/60 ml, total volume).
(iii) Inoculation (CFU analysis).
Mice were anesthetized using isoflurane anesthesia and randomized for intracranial inoculation of C. neoformans strains H99. Inoculation was delivered as a 60-µl volume through a 27-gauge needle fastened to a tuberculin syringe with a cuff to prevent penetration of more than 1 mm. A midline puncture through the cranial vault approximately 6 mm posterior to the orbit was made, and the inoculum was injected, after which the mice were allowed to recover. The euthanasia protocol was the same as that described above. Mice were sacrificed at 48 h using pentobarbital (150 mg/kg of body weight). Brains, extracted as soon as possible following euthanasia, were mashed dry for 30 s and suspended in 3 ml of YPD (considered 1×). The homogenate was serially diluted to 10× and 100× dilutions (200 µl of homogenate was diluted in 1,800 µl YPD). Two hundred milliliters of each respective dilution was spread on YPD plus Pen/Strep and incubated for 2 days at 30°C. Total CFUs were calculated based on the number of colonies counted on the plate, plating volume (200 µl), dilution (from 1× to 100×), and the original volume of the homogenized tissue (3 ml).
All experiments involving vertebrates were in agreement with ethical regulations established by the UC Davis Institutional Animal Care and Use Committee (IACUC).
Statistical analysis.
For the vertebrate studies, sample size was determined by power analysis (G*Power3.1 software program). Significance of survival curves for the vertebrate studies was determined by using the Wilcoxon test (GraphPad-Prism 5 software). The statistical significance of the data presented in Fig. 1, 2, and 3 was determined by an unpaired t test with Welch’s correction (unequal variance) or by one-way analysis of variance (ANOVA) (GraphPad-Prism 5).
SUPPLEMENTAL MATERIAL
(A, B, and C) Wild type (WT; H99), null (mpr1Δ), and reconstituted (mpr1Δ::MPR1; Rec1) strains have comparable growth in minimal medium (synthetic dextrose-SD) at 30°C and comparable growth in rich medium (YPD) at both 30°C and 37°C. Also, the mpr1Δ strain retained all inherent virulence traits, such as capsule and melanin formation and urease and laccase production (data not shown). Three reconstituted strains were selected for analysis, and strain 1 was selected for all subsequent studies based on its growth, comparable to that of both the wild-type and mpr1Δ strains. (D) Genomic DNA from the WT and mpr1Δ strains was digested with AatII and AgeI, while DNA from the Rec1 (reconstituted) strain (mpr1Δ::MPR1) was digested with two sets of enzymes, AatII and AgeI (Rec1a) or AatII and PmlI (Rec1b). Digested DNA was fractionated on agarose gel, transferred to a nylon membrane, and incubated with a digoxigenin-labeled probe (primer F, ACTTCCAGGCTTACAATTTCAG; primer R, AGATTTGGTCGGCCTGAATGATCG), spanning a region of MPR1 that was deleted from the mpr1Δ strain. A single band was observed for the Rec1 strain, while no bands were detected for the mpr1Δ strain. The presence of one band for the Rec1 strain indicated that only one integration of a wild-type copy of MPR1 took place during the biolistic transformation process. Download
ACKNOWLEDGMENT
This work was supported by Public Health Service grants from the National Institutes of Health (NIAID and NINDS).
Footnotes
Citation Vu K, Tham R, Uhrig JP, Thompson GR, III, Na Pombejra S, Jamklang M, Bautos JM, Gelli A. 2014. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. mBio 5(3):e01101-14. doi:10.1128/mBio.01101-14.
REFERENCES
- 1. Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. 2010. Cryptococcus: from human pathogen to model yeast, 1st ed, vol 1, p 620 American Society for Microbiology, Washington, DC [Google Scholar]
- 2. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 3. Ma H, May RC. 2009. Virulence in cryptococcus species. Adv. Appl. Microbiol. 67:131–190. 10.1016/S0065-2164(08)01005-8 [DOI] [PubMed] [Google Scholar]
- 4. Ellis DH, Pfeiffer TJ. 1990. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336:923–925. 10.1016/0140-6736(90)92283-N [DOI] [PubMed] [Google Scholar]
- 5. Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77:4345–4355. 10.1128/IAI.00542-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weiss N, Miller F, Cazaubon S, Couraud PO. 2009. The blood-brain barrier in brain homeostasis and neurological diseases. Biochim. Biophys. Acta 1788:842–857. 10.1016/j.bbamem.2008.10.022 [DOI] [PubMed] [Google Scholar]
- 7. Kim KS. 2008. Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 6:625–634. 10.1038/nrmicro1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang YC, Stins MF, McCaffery MJ, Miller GF, Pare DR, Dam T, Paul-Satyaseela M, Kim KS, Kwon-Chung KJ. 2004. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect. Immun. 72:4985–4995. 10.1128/IAI.72.9.4985-4995.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen SH, Stins MF, Huang SH, Chen YH, Kwon-Chung KJ, Chang Y, Kim KS, Suzuki K, Jong AY. 2003. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J. Med. Microbiol. 52:961–970. 10.1099/jmm.0.05230-0 [DOI] [PubMed] [Google Scholar]
- 10. Sabiiti W, May RC. 2012. Capsule independent uptake of the fungal pathogen Cryptococcus neoformans into brain microvascular endothelial cells. PLoS One 7:e35455. 10.1371/journal.pone.0035455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, Huffnagle GB. 2004. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am. J. Pathol. 164:1761–1771. 10.1016/S0002-9440(10)63734-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, Mody CH. 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Invest. 120:1683–1693. 10.1172/JCI41963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charlier C, Chrétien F, Baudrimont M, Mordelet E, Lortholary O, Dromer F. 2005. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 166:421–432. 10.1016/S0002-9440(10)62265-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chrétien F, Lortholary O, Kansau I, Neuville S, Gray F, Dromer F. 2002. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J. Infect. Dis. 186:522–530. 10.1086/341564 [DOI] [PubMed] [Google Scholar]
- 15. Liu TB, Perlin DS, Xue C. 2012. Molecular mechanisms of cryptococcal meningitis. Virulence 3:173–181. 10.4161/viru.18685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ibrahim AS, Filler SG, Alcouloumre MS, Kozel TR, Edwards JE, Jr, Ghannoum MA. 1995. Adherence to and damage of endothelial cells by Cryptococcus neoformans in vitro: role of the capsule. Infect. Immun. 63:4368–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu CY, Zhu HM, Wu JH, Wen H, Liu CJ. 2014. Increased permeability of blood-brain barrier is mediated by serine protease during Cryptococcus meningitis. J. Int. Med. Res. 42:85–92. 10.1177/0300060513504365 [DOI] [PubMed] [Google Scholar]
- 18. Stie J, Fox D. 2012. Blood-brain barrier invasion by Cryptococcus neoformans is enhanced by functional interactions with plasmin. Microbiology (Reading, England) 158:240–258. 10.1099/mic.0.051524-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santangelo R, Zoellner H, Sorrell T, Wilson C, Donald C, Djordjevic J, Shounan Y, Wright L. 2004. Role of extracellular phospholipases and mononuclear phagocytes in dissemination of cryptococcosis in a murine model. Infect. Immun. 72:2229–2239. 10.1128/IAI.72.4.2229-2239.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. 2009. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect. Immun. 77:120–127. 10.1128/IAI.01065-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jong A, Wu CH, Gonzales-Gomez I, Kwon-Chung KJ, Chang YC, Tseng HK, Cho WL, Huang SH. 2012. Hyaluronic acid receptor CD44 deficiency is associated with decreased Cryptococcus neoformans brain infection. J. Biol. Chem. 287:15298–15306. 10.1074/jbc.M112.353375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim KS. 2000. E. coli invasion of brain microvascular endothelial cells as a pathogenetic basis of meningitis. Subcell. Biochem. 33:47–59. 10.1007/978-1-4757-4580-1_3 [DOI] [PubMed] [Google Scholar]
- 23. Doran KS, Engelson EJ, Khosravi A, Maisey HC, Fedtke I, Equils O, Michelsen KS, Arditi M, Peschel A, Nizet V. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J. Clin. Invest. 115:2499–2507. 10.1172/JCI23829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Badger JL, Stins MF, Kim KS. 1999. Citrobacter freundii invades and replicates in human brain microvascular endothelial cells. Infect. Immun. 67:4208–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pujol C, Eugène E, de Saint Martin L, Nassif X. 1997. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 65:4836–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jong AY, Stins MF, Huang SH, Chen SH, Kim KS. 2001. Traversal of Candida albicans across human blood-brain barrier in vitro. Infect. Immun. 69:4536–4544. 10.1128/IAI.69.7.4536-4544.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vu K, Weksler B, Romero I, Couraud PO, Gelli A. 2009. Immortalized human brain endothelial cell line HCMEC/D3 as a model of the blood-brain barrier facilitates in vitro studies of central nervous system infection by Cryptococcus neoformans. Eukaryot. Cell 8:1803–1807. 10.1128/EC.00240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tseng HK, Liu CP, Price MS, Jong AY, Chang JC, Toffaletti DL, Betancourt-Quiroz M, Frazzitta AE, Cho WL, Perfect JR. 2012. Identification of genes from the fungal pathogen Cryptococcus neoformans related to transmigration into the central nervous system. PLoS One 7:e45083. 10.1371/journal.pone.0045083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maruvada R, Zhu L, Pearce D, Zheng Y, Perfect J, Kwon-Chung KJ, Kim KS. 2012. Cryptococcus neoformans phospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cell. Microbiol. 14:1544–1553. 10.1111/j.1462-5822.2012.01819.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang SH, Wu CH, Chang YC, Kwon-Chung KJ, Brown RJ, Jong A. 2012. Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS One 7:e48570. 10.1371/journal.pone.0048570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen SC, Wright LC, Santangelo RT, Muller M, Moran VR, Kuchel PW, Sorrell TC. 1997. Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect. Immun. 65:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chayakulkeeree M, Johnston SA, Oei JB, Lev S, Williamson PR, Wilson CF, Zuo X, Leal AL, Vainstein MH, Meyer W, Sorrell TC, May RC, Djordjevic JT. 2011. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Mol. Microbiol. 80:1088–1101. 10.1111/j.1365-2958.2011.07632.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shea JM, Kechichian TB, Luberto C, Del Poeta M. 2006. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74:5977–5988. 10.1128/IAI.00768-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu Y, Davis MJ, Dayrit JK, Hadd Z, Meister DL, Osterholzer JJ, Williamson PR, Olszewski MA. 2012. Immune modulation mediated by cryptococcal laccase promotes pulmonary growth and brain dissemination of virulent Cryptococcus neoformans in mice. PLoS One 7:e47853. 10.1371/journal.pone.0047853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39:166–175. 10.1046/j.1365-2958.2001.02236.x [DOI] [PubMed] [Google Scholar]
- 36. Jong A, Wu CH, Chen HM, Luo F, Kwon-Chung KJ, Chang YC, Lamunyon CW, Plaas A, Huang SH. 2007. Identification and characterization of CPS1 as a hyaluronic acid synthase contributing to the pathogenesis of Cryptococcus neoformans infection. Eukaryot. Cell 6:1486–1496. 10.1128/EC.00120-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jong A, Wu CH, Shackleford GM, Kwon-Chung KJ, Chang YC, Chen HM, Ouyang Y, Huang SH. 2008. Involvement of human CD44 during Cryptococcus neoformans infection of brain microvascular endothelial cells. Cell. Microbiol. 10:1313–1326. 10.1111/j.1462-5822.2008.01128.x [DOI] [PubMed] [Google Scholar]
- 38. Long M, Huang SH, Wu CH, Shackleford GM, Jong A. 2012. Lipid raft/caveolae signaling is required for Cryptococcus neoformans invasion into human brain microvascular endothelial cells. J. Biomed. Sci. 19:19. 10.1186/1423-0127-19-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang SH, Long M, Wu CH, Kwon-Chung KJ, Chang YC, Chi F, Lee S, Jong A. 2011. Invasion of Cryptococcus neoformans into human brain microvascular endothelial cells is mediated through the lipid rafts-endocytic pathway via the dual specificity tyrosine phosphorylation-regulated kinase 3 (DYRK3). J. Biol. Chem. 286:34761–34769. 10.1074/jbc.M111.219378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim JC, Crary B, Chang YC, Kwon-Chung KJ, Kim KJ. 2012. Cryptococcus neoformans activates RhoGTPase proteins followed by protein kinase C, focal adhesion kinase, and ezrin to promote traversal across the blood-brain barrier. J. Biol. Chem. 287:36147–36157. 10.1074/jbc.M112.389676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, Gelli A. 2007. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 7:499–510. 10.1111/j.1567-1364.2006.00198.x [DOI] [PubMed] [Google Scholar]
- 42. Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJ, Perfect JR, Kronstad J. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336–1349. 10.1128/EC.2.6.1336-1349.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Monod M, Paris S, Sanglard D, Jaton-Ogay K, Bille J, Latgé JP. 1993. Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect. Immun. 61:4099–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fernández D, Russi S, Vendrell J, Monod M, Pallarès I. 2013. A functional and structural study of the major metalloprotease secreted by the pathogenic fungus Aspergillus fumigatus. Acta Crystallogr. D Biol. Crystallogr. 69:1946–1957. 10.1107/S0907444913017642 [DOI] [PubMed] [Google Scholar]
- 45. Markaryan A, Morozova I, Yu H, Kolattukudy PE. 1994. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 62:2149–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lilly WW, Stajich JE, Pukkila PJ, Wilke SK, Inoguchi N, Gathman AC. 2008. An expanded family of fungalysin extracellular metallopeptidases of Coprinopsis cinerea. Mycol. Res. 112:389–398. 10.1016/j.mycres.2007.11.013 [DOI] [PubMed] [Google Scholar]
- 47. Weksler BB, Subileau EA, Perrière N, Charneau P, Holloway K, Leveque M, Tricoire-Leignel H, Nicotra A, Bourdoulous S, Turowski P, Male DK, Roux F, Greenwood J, Romero IA, Couraud PO. 2005. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 19:1872–1874. 10.1096/fj.04-3458fje [DOI] [PubMed] [Google Scholar]
- 48. Allendoerfer R, Marquis AJ, Rinaldi MG, Graybill JR. 1991. Combined therapy with fluconazole and flucytosine in murine cryptococcal meningitis. Antimicrob. Agents Chemother. 35:726–729. 10.1128/AAC.35.4.726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jong A, Wu CH, Prasadarao NV, Kwon-Chung KJ, Chang YC, Ouyang Y, Shackleford GM, Huang SH. 2008. Invasion of Cryptococcus neoformans into human brain microvascular endothelial cells requires protein kinase C-alpha activation. Cell. Microbiol. 10:1854–1865. 10.1111/j.1462-5822.2008.01172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Twining SS. 1984. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 143:30–34. 10.1016/0003-2697(84)90553-0 [DOI] [PubMed] [Google Scholar]
- 51. Najvar LK, Bocanegra R, Graybill JR. 1999. An alternative animal model for comparison of treatments for cryptococcal meningitis. Antimicrob. Agents Chemother. 43:413–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Naumann TA, Wicklow DT, Price NP. 2011. Identification of a chitinase-modifying protein from Fusarium verticillioides: truncation of a host resistance protein by a fungalysin metalloprotease. J. Biol. Chem. 286:35358–35366. 10.1074/jbc.M111.279646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brouta F, Descamps F, Monod M, Vermout S, Losson B, Mignon B. 2002. Secreted metalloprotease gene family of Microsporum canis. Infect. Immun. 70:5676–5683. 10.1128/IAI.70.10.5676-5683.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu J, Baldwin D, Kindrachuk C, Hegedus DD. 2006. Serine proteases and metalloproteases associated with pathogenesis but not host specificity in the Entomophthoralean fungus Zoophthora radicans. Can. J. Microbiol. 52:550–559. 10.1139/w06-004 [DOI] [PubMed] [Google Scholar]
- 55. Rosenblum EB, Stajich JE, Maddox N, Eisen MB. 2008. Global gene expression profiles for life stages of the deadly amphibian pathogen Batrachochytrium dendrobatidis. Proc. Natl. Acad. Sci. U. S. A. 105:17034–17039. 10.1073/pnas.0804173105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lortholary O, Improvisi L, Rayhane N, Gray F, Fitting C, Cavaillon JM, Dromer F. 1999. Cytokine profiles of AIDS patients are similar to those of mice with disseminated Cryptococcus neoformans infection. Infect. Immun. 67:6314–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lortholary O, Dromer F, Mathoulin-Pélissier S, Fitting C, Improvisi L, Cavaillon JM, Dupont B, French, Cryptococcosis Study Group 2001. Immune mediators in cerebrospinal fluid during cryptococcosis are influenced by meningeal involvement and human immunodeficiency virus serostatus. J. Infect. Dis. 183:294–302. 10.1086/317937 [DOI] [PubMed] [Google Scholar]
- 58. Roseff SA, Levitz SM. 1993. Effect of endothelial cells on phagocyte-mediated anticryptococcal activity. Infect. Immun. 61:3818–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vu K, Eigenheer RA, Phinney BS, Gelli A. 2013. Cryptococcus neoformans promotes its transmigration into the central nervous system by inducing molecular and cellular changes in brain endothelial cells. Infect. Immun. 81:3139–3147. 10.1128/IAI.00554-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B, and C) Wild type (WT; H99), null (mpr1Δ), and reconstituted (mpr1Δ::MPR1; Rec1) strains have comparable growth in minimal medium (synthetic dextrose-SD) at 30°C and comparable growth in rich medium (YPD) at both 30°C and 37°C. Also, the mpr1Δ strain retained all inherent virulence traits, such as capsule and melanin formation and urease and laccase production (data not shown). Three reconstituted strains were selected for analysis, and strain 1 was selected for all subsequent studies based on its growth, comparable to that of both the wild-type and mpr1Δ strains. (D) Genomic DNA from the WT and mpr1Δ strains was digested with AatII and AgeI, while DNA from the Rec1 (reconstituted) strain (mpr1Δ::MPR1) was digested with two sets of enzymes, AatII and AgeI (Rec1a) or AatII and PmlI (Rec1b). Digested DNA was fractionated on agarose gel, transferred to a nylon membrane, and incubated with a digoxigenin-labeled probe (primer F, ACTTCCAGGCTTACAATTTCAG; primer R, AGATTTGGTCGGCCTGAATGATCG), spanning a region of MPR1 that was deleted from the mpr1Δ strain. A single band was observed for the Rec1 strain, while no bands were detected for the mpr1Δ strain. The presence of one band for the Rec1 strain indicated that only one integration of a wild-type copy of MPR1 took place during the biolistic transformation process. Download