ABSTRACT
Acinetobacter baumannii is a Gram-negative bacterium that causes diseases such as pneumonia, bacteremia, and soft tissue infections in hospitalized patients. Relatively little is known about how A. baumannii causes these infections. Thus, we used insertion sequencing (INSeq), a combination of transposon mutagenesis and massively parallel next-generation sequencing, to identify novel virulence factors of A. baumannii. To this end, we generated a random transposon mutant library containing 150,000 unique insertions in A. baumannii strain ATCC 17978. The INSeq analysis identified 453 genes required for growth in rich medium. The library was then used in a murine pneumonia model, and the relative levels of abundance of mutants before and after selection in the mouse were compared. When genes required for growth in rich medium were removed from the analysis, 157 genes were identified as necessary for persistence in the mouse lung. Several of these encode known virulence factors of A. baumannii, such as OmpA and ZnuB, which validated our approach. A large number of the genes identified were predicted to be involved in amino acid and nucleotide metabolism and transport. Other genes were predicted to encode an integration host factor, a transmembrane lipoprotein, and proteins involved in stress response and efflux pumps. Very few genes, when disrupted, resulted in an increase in A. baumannii numbers during host infection. The INSeq approach identified a number of novel virulence determinants of A. baumannii, which are candidate targets for therapeutic interventions.
IMPORTANCE
A. baumannii has emerged as a frequent cause of serious infections in hospitals and community settings. Due to increasing antibiotic resistance, alternative approaches, such as antivirulence strategies, are desperately needed to fight A. baumannii infections. Thorough knowledge of A. baumannii pathogenicity is essential for such approaches but is currently lacking. With the increasingly widespread use of massively parallel sequencing, a class of techniques known as transposon insertion sequencing has been developed to perform comprehensive virulence screens of bacterial genomes in vivo. We have applied one of these approaches (INSeq) to uncover novel virulence factors in A. baumannii. We identified several such factors, including those predicted to encode amino acid and nucleotide metabolism proteins, an integration host factor protein, stress response factors, and efflux pumps. These results greatly expand the number of A. baumannii virulence factors and uncover potential targets for antivirulence treatments.
INTRODUCTION
Acinetobacter baumannii is emerging as a particularly problematic nosocomial pathogen due to the frequency of multidrug-resistant (MDR) strains around the world and its high epidemic potential. Apart from causing a wide range of diseases in hospital settings, including pneumonia, bloodstream infections, soft tissue infections, and meningitis, A. baumannii is a serious concern in wounded military personnel and in community settings (1). Factors contributing to its success as a pathogen include its easy acquisition of antibiotic resistance elements and ability to resist desiccation and common disinfectants (2). Despite its clinical importance, however, relatively little is known about the molecular basis of A. baumannii pathogenicity. The few virulence determinants uncovered in A. baumannii include factors involved in metal acquisition and lipopolysaccharide (LPS) synthesis, as well as membrane-associated proteins, such as outer membrane protein A (OmpA) and two-component regulatory systems (3–9) (see Table S2 in the supplemental material).
High-throughput screens are especially useful for identifying novel genotype-phenotype relationships in understudied bacteria. In recent years, transposon insertion sequencing methods (e.g., insertion sequencing [INSeq], transposon sequencing [Tn-seq], transposon-directed insertion site sequencing [TraDIS], and high-throughput insertion tracking sequencing [HITS]) have emerged as powerful tools to comprehensively screen for gene functions across an entire bacterial genome (10). Using these techniques with animal models of infection, researchers have uncovered novel virulence factors in Haemophilus influenzae, Yersinia pseudotuberculosis, and Pseudomonas aeruginosa (11–13).
We have applied INSeq, one specific method of transposon insertion sequencing, to A. baumannii in a murine lung infection model to identify novel virulence factors important in pneumonia (14). The results not only confirmed several well-established virulence factors of A. baumannii but also uncovered potential novel factors involved in the pathogenicity of this bacterium. Overall, 157 genes were found to be important for effective infection of the host. These high-throughput screen hits can serve as a basis for future research, thus shedding light on the mechanisms employed by A. baumannii to cause pneumonia.
RESULTS AND DISCUSSION
Generation of a transposon insertion library in A. baumannii.
We first generated an A. baumannii transposon insertion library appropriate for the INSeq analysis (14). A transposon vector was modified for use in A. baumannii and designated pJNW684. A library of 150,000 transposon mutants was generated using this vector. Southern blot analysis of a subset of 20 mutants demonstrated that the insertions were random and that each mutant contained a single insertion (see Fig. S1 in the supplemental material).
Adaptation of a mouse model of pneumonia.
Mouse pulmonary infection is an accepted model for the study of A. baumannii pathogenesis (1). We optimized this model for use with the INSeq approach to uncover virulence factors. C57BL/6 mice were infected with 4 × 108 to 8 × 108 CFU of A. baumannii by nasal aspiration, and bacterial burdens were monitored over time. Bacterial CFU increased over the first 24 h but then diverged, as some mice began to clear the infection while others did not (see Fig. S2 in the supplemental material). To further validate this model, we evaluated a mutant with a deletion in the zinc transporter gene znuB (ΔznuB mutant) (3). The ΔznuB mutant persisted less well than the parental strain (data not shown), in agreement with previously published results (3). Together, these findings indicate that the murine pneumonia model is capable of identifying A. baumannii mutants with defects in virulence.
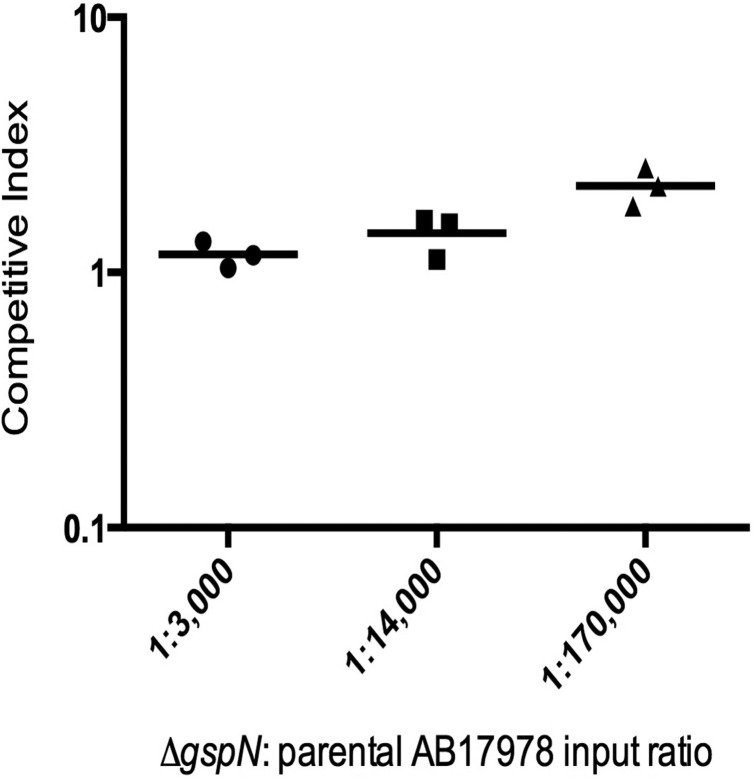
Before carrying out the INSeq selection, we examined whether a bottleneck occurred at this inoculum, which would result in stochastic loss of mutants during selection. We constructed a marked deletion in A1S_0269, a gene predicted to play a role in type II secretion (15) and hereinafter referred to as gspN, for “general secretion pathway protein N.” The mutant did not demonstrate a virulence defect in survival or competition assays with parental A. baumannii in the mouse model of pneumonia (see Fig. S3 in the supplemental material). We therefore concluded that the ΔgspN mutant could be used as a fully virulent but marked A. baumannii strain to assess bottlenecks in the mouse model. The ΔgspN mutant was spiked at various ratios into pools of the parental A. baumannii strain for infection. After 24 h, the bacterial burdens in the mouse lungs were determined. Even when inoculated at a ratio of 1:170,000, the marked ΔgspN mutant could be recovered from the mouse lungs in appropriate numbers (Fig. 1). These results indicated that the entire 150,000-member transposon mutant library could be inoculated into a single mouse without undergoing a bottleneck effect.
FIG 1 .
Recovery of ΔgspN mutant from spiked pools of parental A. baumannii bacteria following inoculation into mouse lungs. Competitive indices were determined with the following formula: CI = (mutant output/parental output)/(mutant input/parental input). Each symbol represents an individual mouse, and bars indicate median values.
Performance of INSeq on A. baumannii.
To carry out the in vivo selection for INSeq, the entire mutant library was first grown in Luria-Bertani (LB) medium. The resulting bacterial culture was split into two halves. One half was designated the input pool of bacteria, and the other half was used to inoculate mice intranasally. After 24 h, lungs from three infected mice were dissected and homogenized to yield the output pool of bacteria. Genomic DNA was extracted from both the input and output pools and processed to capture and amplify transposon insertion site junctions. The amplified DNA was then analyzed by Illumina sequencing. Eight samples were sequenced in all, including three technical replicates of the input pool, three biological replicates of the output pool (i.e., samples from three separate mice), and an additional two technical replicates of one of the output pool biological replicates (i.e., samples from one mouse were processed separately three times).
In total, 174 million sequencing reads were obtained from the eight samples, of which 75% contained the appropriate barcodes. An average of 95% of the barcoded reads in each pool contained the transposon insertion sequence, resulting in 108 million reads, of which 51% aligned to an average of 198,000 TA site flanking sequences in the ATCC 17978 genome. Evaluating the input pools, there were ~61,500 unique insertions within genes in the mutant bank after growth in LB medium. On average, there were 18 insertions per gene and 91 reads per insertion in each pool. Analysis using the ESSENTIALS software (16) indicated that the compositions of the input pool technical replicates and the output pool biological and technical replicates were highly similar (see Fig. S4 in the supplemental material). The reproducibility of these results supports their validity.
A. baumannii genes required for growth in LB medium.
Analysis of the input pool was performed to identify genes that are essential or that substantially affect growth in LB medium. (For simplicity, the word “gene” will be used throughout this discussion to refer to the genetic unit containing the transposon insertion, but it is recognized that polar effects may cause loss of expression of downstream genes within an operon that could account for observed phenotypes.) The number of actual reads for each insertion site in a given gene was compared to the number of expected reads to determine whether the gene was necessary for growth in LB medium. A density plot (log2-transformed fold change in reads versus number of genes) was generated from these data and its local minimum determined to identify the population of genes for which insertion severely impaired growth in LB medium (see Fig. S5A in the supplemental material). A total of 453 genes (13.1% of the total A. baumannii genes) met this criterion (Fig. S6 and Table S1, tab A). This number likely includes genes required for growth, genes necessary for a normal rate of growth, and genes that lie upstream from such genes and therefore exert polar effects on their expression. Another 64 genes could not be evaluated because either the gene is very small or transposon insertion sites could not be unambiguously assigned to a specific gene (Table S1, tab A, NA). Many of these, however, were tRNA or rRNA genes and were also likely required for growth in LB medium. Our value for the number of genes required for growth in medium compares well to that obtained by de Berardinis and colleagues for the closely related species Acinetobacter baylyi. They found 499 genes to be essential by experimentally testing single-gene-deletion mutants for growth in minimal medium (17). In contrast, Kaur and colleagues previously identified only 200 A. baumannii genes as essential (18). However, their analysis used in silico rather than experimental methods, which may account for the identification of a relatively small number of genes. Therefore, our value is a reasonable estimate of the number of genes required for growth in LB medium.
A. baumannii genes required for persistence during pneumonia.
Next, genes that affected the persistence of A. baumannii in the lungs during pneumonia were identified. For the purposes of this paper, persistence of A. baumannii in mouse lungs is defined as the presence of a high bacterial load at 24 h postinfection. All genes necessary for growth in LB medium were first removed from the analysis. For each gene, the number of reads in the output pool was compared to the number of reads in the input pool. In contrast to the analysis of growth in LB medium, the density plot generated for the output pool followed a normal distribution, suggesting that there was not a distinct population of A. baumannii genes dedicated exclusively to lung pathogenesis (see Fig. S5B in the supplemental material). Transposon-containing genes that were underrepresented or overrepresented in the output pool by at least 2-fold (log2 < −1 or log2 > 1) with an adjusted P value of <0.05 were considered to impact A. baumannii persistence in vivo. The INSeq screen uncovered 157 genes (4.5% of predicted A. baumannii genes) that were required for persistence in the lung (Fig. S6 and Table S1, tab B). In contrast, only eight genes were identified that, when disrupted, resulted in significant increases in the corresponding numbers of mutants in the lungs (Fig. S6 and Table S1, tab C). The majority (77.2% of predicted A. baumannii genes) were not associated with significant changes in the numbers of the corresponding mutants in the output pool compared to their numbers in the input pool (Table S1, tab D); the complete data set can be found in Table S1, tab E. Thus, INSeq identified some A. baumannii genes that were associated with increased virulence during pneumonia and others that were associated with decreased virulence.
Functional categorization of genes identified by INSeq.
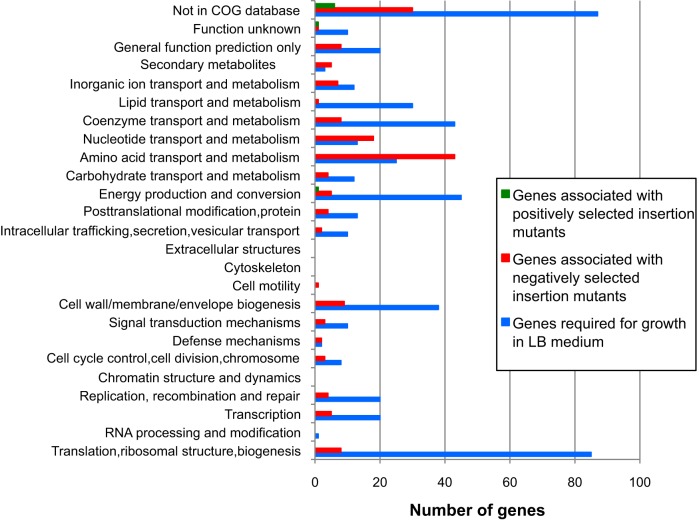
We functionally categorized genes essential for growth in rich medium, as well as genes that positively and negatively impacted persistence in vivo, based on the Clusters of Orthologous Groups (COG) classifications (19). Genes from all three groups were broadly distributed among the COG categories (Fig. 2). As expected, the majority of A. baumannii genes required for growth in LB medium were in the “translation, ribosomal structure, and biogenesis” category. Genes from this group also commonly fell into the categories of “coenzyme transport and metabolism,” “energy production and conversion,” and “cell wall/membrane/envelope biogenesis.” Many genes important for persistence were in the “amino acid transport and metabolism” and “nucleotide transport and metabolism” categories. Surprisingly few virulence genes were found in categories such as “extracellular structures,” “cell motility,” and “defense mechanisms.” These findings suggest that the metabolic attributes of A. baumannii play a major role in its ability to cause pneumonia.
FIG 2 .
Functional categorization of A. baumannii genes identified by INSeq. Each protein sequence was BLAST searched against the COG database, and a COG identification number was assigned to a gene if the best BlastP hit exhibited at least 30% protein sequence similarity over at least 50% of the sequence.
Identification of known A. baumannii virulence genes.
Gratifyingly, INSeq identified several factors previously shown to be important for A. baumannii pathogenesis in vivo (see Table S2 in the supplemental material). For example, disruption of lpsB caused a 15-fold drop in persistence, underscoring the importance of LPS in A. baumannii pathogenesis. Both OmpA and the two-component regulatory protein GacA have been shown to contribute to multiple aspects of A. baumannii infections, perhaps explaining the relatively large (6-fold) decrease in persistence observed with insertions in either gene. The identification of these known virulence factors validated our screen and indicated that several of these factors are relevant in more than one model of infection.
Biofilm formation.
Our screen identified bfmR and bfmS, which encode the response regulator and sensor kinase, respectively, for the two-component regulatory system BfmRS that is essential for biofilm formation and normal cellular morphology (20). Mutants with insertions in bfmR and bfmS were decreased by 40-fold and 2-fold, respectively, in the output pool. Our findings demonstrate that the BfmRS two-component regulatory system also plays an important role in the in vivo pathogenicity of A. baumannii.
LPS and capsule formation.
As with most pathogenic Gram-negative bacteria, LPS and capsule are important aspects of virulence in A. baumannii (6, 7, 21). In addition to lpsB, our INSeq screen identified lpsC (A1S_2900), a previously unstudied gene likely involved in LPS core biosynthesis, as important in virulence. lpsC is predicted to encode a glycosyltransferase similar to LpsB. Mutants with insertions in UDP-glucose 4-epimerase (A1S_0065), which is predicted to play a role in capsule biosynthesis, also showed a significant decrease in persistence in the INSeq screen (−18.6-fold) (22).
Amino acid and nucleotide metabolism.
Disruption of genes involved in amino acid metabolism and transport resulted in decreased bacterial persistence in the mouse pneumonia model. This is perhaps not surprising since the host utilizes amino acid depletion as a defense mechanism against pathogens (23, 24). In response, amino acid biosynthesis genes are induced in these bacteria (25). Tryptophan restriction in the mammalian host is mediated by the tryptophan-catabolizing enzyme indoleamine 2,3-dioxygenase (IDO), which in turn is induced by the gamma interferon (IFN-γ) response to Francisella tularensis and Chlamydia trachomatis infections (23, 24). On the other hand, tryptophan synthesis genes have been shown to be required for virulence in C. trachomatis infection of HeLa cells (25) and in F. tularensis infection of lungs (23). We found that insertions in seven genes involved in tryptophan biosynthesis (A1S_0274, A1S_0688, A1S_2355, A1S_2359, A1S_2360, A1S_2875, and A1S_2876) resulted in about 4- to 5-fold decreased persistence during pneumonia. Thus, it is possible that A. baumannii encounters a similar defense mechanism and mounts an appropriate counterresponse in the lungs during pneumonia. Unlike F. tularensis and C. trachomatis, A. baumannii is thought to be an extracellular pathogen, although recent reports indicate that it can invade the intracellular compartment (26).
Insertions in genes involved in the synthesis of methionine (A1S_0471, A1S_0737, A1S_0778, A1S_1683, A1S_2122, and A1S_2324) and arginine (A1S_0259, A1S_0610, A1S_1980, A1S_2686, A1S_2687, and A1S_3129) caused significantly decreased persistence, ranging from 2- to 6-fold, in the mouse pneumonia model of A. baumannii. Methionine and arginine biosynthetic genes were induced in Saccharomyces cerevisiae and Candida albicans in response to exposure to neutrophils, suggesting that the neutrophil phagosome may be scarce in these amino acids (27). A set of six genes (A1S_0346, A1S_0686, A1S_0688, A1S_3235, A1S_3238, A1S_3245) involved in histidine biosynthesis was also shown to be important for virulence in our screen. Previous proteomic analyses showed that histidine metabolism plays an important role in biofilm formation, which may contribute to the pathogenic potential of A. baumannii (1, 28). These results suggest that the acquisition of amino acids is critical for the persistence of A. baumannii in the pulmonary environment.
Ten genes involved in purine metabolism (A1S_2189, A1S_2251, A1S_2606, A1S_2585, A1S_2605, A1S_2963, A1S_2964, A1S_3425, A1S_2187, and A1S_2188) were identified as important for the persistence of A. baumannii in the murine pneumonia model. These genes were purF, -D, -N, -L, -M, -K, -E, -C, -P, and -O. When they were disrupted, the mutants showed decreases in persistence of between 2- and 12-fold. Typically in bacteria, 11 genes are required for the de novo synthesis of purines. The 11th gene, purB, was not included in our pathogenesis analysis because it was required for growth in LB medium, similar to purB in Escherichia coli (29). Purine biosynthesis genes are critical for optimal pathogenicity in a number of bacteria, including Bacillus anthracis and Streptococcus pneumoniae (30, 31). Again, this likely reflects the scarcity of nutrients, such as nucleotides, in the host environment, thus requiring the bacteria to synthesize these molecules de novo (32).
Stress response.
During an infection, phagocytes produce reactive oxygen species (ROS) through the enzymatic activity of NADPH oxidase as a defense mechanism against bacterial pathogens. A. baumannii is no exception, as NADPH oxidase-deficient mice are more susceptible to A. baumannii acute respiratory infections (33). In response to such oxidative stress, many bacterial species utilize protective glutaredoxins (34). A. baumannii harbors a glutaredoxin (A1S_0529) in which insertions caused significant depletion (2-fold) of the resulting mutants in vivo.
Under stress conditions like those inside an infected host, bacteria require specific proteases that can degrade misfolded proteins and short-lived regulatory proteins in order to survive and propagate efficiently. In E. coli, as well as many other species of bacteria, such proteolysis is carried out by energy-dependent proteases like Lon and members of the Clp family (e.g., ClpAP and ClpXP). Insertions in two genes in an operon that encodes Lon proteases (A1S_1031 and A1S_1030) and one gene that encodes the proteolytic subunit of a Clp protease in A. baumannii (A1S_0476) resulted in significant attenuation of persistence in vivo, by 3-fold, 2-fold, and 7-fold, respectively. Consistent with a role for these factors in A. baumannii pathogenesis is the requirement of their homologs for optimal pathogenicity in a number of bacteria, such as Yersinia pestis, Salmonella enterica, and Staphylococcus aureus (35–37). Both Lon and Clp proteases regulate the expression of the Y. pestis type III secretion system through the controlled proteolysis of a transcriptional repressor, while in S. enterica, the Lon protease regulates the expression of invasion genes encoded on SPI1 (35, 36). In S. aureus, ClpXP regulates host hemoglobin binding and heme-iron extraction, thus playing an important role in virulence during a mouse systemic infection (37). These findings demonstrate that Lon and Clp proteases regulate specific virulence factors, in addition to functioning as universal stress response regulators. Since A. baumannii strain ATCC 17978 expresses a cluster of heme acquisition genes, it is possible that the Clp proteases control virulence through iron acquisition during host infection (38).
Efflux pumps.
A. baumannii produces a large number of efflux pumps, including members of the resistance nodulation division (RND) family, the ATP-binding cassette (ABC) superfamily, and the major facilitator superfamily (MFS) (39). Insertions in genes associated with multiple ABC transporters (A1S_2375, A1S_2378, and A1S_3221) resulted in decreased persistence in the murine pneumonia model. Specifically, one of the ABC transporter genes uncovered by the INSeq screen (A1S_3221) encodes a protein homologous to ChvD, an ABC transporter ATP-binding protein in Agrobacterium tumefaciens that regulates virulence gene expression (40).
Insertions in a four-gene operon (A1S_3103–A1S_3100) annotated as encoding toluene tolerance efflux transporters resulted in highly decreased persistence in the INSeq screen, ranging from 7- to 19-fold. The need for an environmental bacterium like A. baumannii to tolerate toxic organic solvents is understandable, but the implications for such systems in mammalian infections are less clear (1). Toluene tolerance is required for the production of outer membrane vesicles (OMVs) in Pseudomonas putida, and these OMVs contain virulence-associated molecules like LPS (41). Thus, one possibility is that toluene tolerance genes provide a similar function in A. baumannii and allow for the production of OMVs, which are efficient vehicles for known virulence factors like OmpA (42). Alternatively, these genes may perform as-yet-unrecognized virulence functions, such as export of antimicrobial peptides or virulence factors.
Miscellaneous A. baumannii virulence factors uncovered by INSeq.
Our INSeq screen identified several other genes that may play interesting roles in virulence. For example, mutants with insertions in A1S_0622, a gene predicted to encode a VacJ-like transmembrane lipoprotein precursor, were attenuated 15-fold in the mouse pneumonia model. VacJ is important for virulence in Burkholderia pseudomallei and H. influenzae and, more specifically, for intercellular spread during infections caused by Shigella flexneri (11, 43, 44). Integration host factor (IHF) is a DNA-bending protein composed of two subunits, alpha and beta, that is involved in the regulation of virulence gene expression in pathogens such as Brucella abortus and Vibrio cholerae (45, 46). Insertions in the gene that encodes the alpha subunit of IHF, ihfa (A1S_0603), led to a significant decrease (4-fold) in the read frequency in the INSeq screen, suggesting a role for IHF in A. baumannii pathogenesis.
Genes for which insertion led to enrichment in the murine pneumonia model.
The INSeq approach also identified eight genes that, when disrupted, provided a competitive advantage in the lung. This is a relatively small number compared to the results of other studies. For example, Skurnik and colleagues identified 89 genes that, when disrupted, resulted in an increase in the gastrointestinal colonization of P. aeruginosa (47). Four of the genes we identified encode hypothetical proteins, and the associated insertion mutants only showed modest increases in persistence in the lung (2- to 3-fold). The paucity of such genes suggests that evolutionary pressures have maintained virulence genes but selected against genes that decrease pathogenicity in the A. baumannii genome.
Validation of INSeq results through targeted deletions and competition assays.
To validate the INSeq results, we examined five specific mutants with a range of observed virulence levels. These mutants were used in competition assays with the parental A. baumannii strain in the murine pneumonia model to determine whether the INSeq results could be replicated. We chose mutants with disruptions in genes associated with a modest increase in recovery (A1S_0995, +2-fold), unchanged recovery (dotB), a modest decrease in recovery (znuB, −2-fold), and large decreases in recovery (lpsB, decreased −15.0-fold, and A1S_0065, −18.6-fold) in the INSeq screen. Overall, the competitive indices of the targeted deletion mutants in individual 1:1 competition assays followed the same trends as the corresponding mutants in the context of the INSeq experiments (Fig. 3). These findings validate the INSeq results.
FIG 3 .
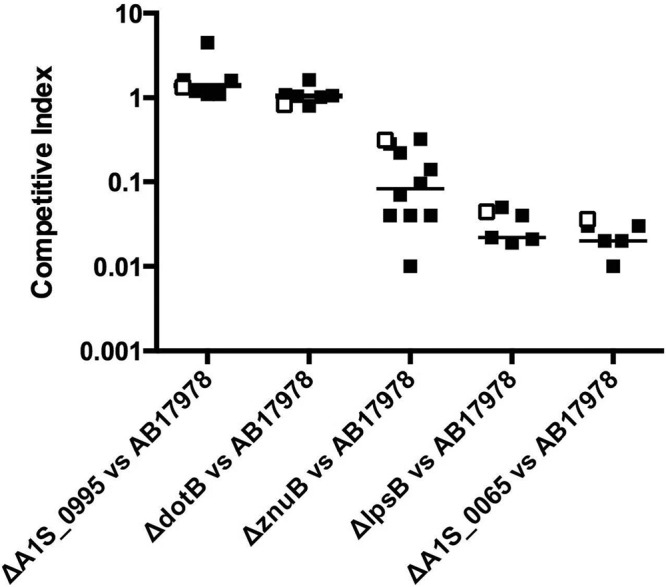
Competitive fitness of representative A. baumannii mutants in the INSeq assay versus individual competition assays. Data show the competitive indices of mutants containing transposon insertions in A1S_0995, dotB, znuB, lpsB, or A1S_0065 in the context of the INSeq screen (□) compared to those of the corresponding targeted deletion mutants in individual 1:1 competition assays with the parental strain (■). The data for the 1:1 competition assays were collected over multiple experiments, with each symbol representing an individual mouse.
We have successfully utilized a high-throughput transposon insertion sequencing technique, INSeq, to find novel virulence factors involved in A. baumannii mouse pneumonia pathogenesis. While we were able to confirm previously characterized virulence factors in this bacterium, many new virulence factors were found as well. We uncovered 157 genes necessary for persistence in the mouse lung and eight genes that, when disrupted, actually lent a fitness advantage to A. baumannii in the lungs. The discovery of these genotype-phenotype relationships will potentially pave the way to further characterization of the pathogenic mechanisms of A. baumannii and the development of novel therapeutics targeting virulence factors for this highly antibiotic-resistant bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. baumannii ATCC 17978 was a generous gift from Paul Dunman (48). E. coli S17.1 λpir [hsdR recA pro RP4-2 (Tc::Mu Km::Tn7) (λpir)] and E. coli β3914 [F− RP4-2-Tc::Mu ΔdapA::(erm-pir) gyrA462 zei-298::Tn10 (ErmR KanR TetR)] were used in matings to introduce cloning or transposon vectors into A. baumannii (49). The ΔznuB and ΔlpsB mutants were a generous gift from Eric Skaar (3).
LB medium was used to culture E. coli and A. baumannii. Vogel-Bonner medium (VBM) agar was used to select for A. baumannii following matings (50). When appropriate, the medium was supplemented with ampicillin (100 µg/ml) or kanamycin (25 to 50 µg/ml).
Generation of A. baumannii transposon mutant library.
To generate the transposon library in A. baumannii, we modified the vector pMarVF1 (J. F. Brooks, M. C. Gyllborg, D. C. Cronin, L. E. H. Markey, A. L. Goodman, and M. J. Mandel, unpublished data). Specifically, the antibiotic resistance cassette was changed to a kanamycin resistance cassette by amplifying this fragment from the pCR-Blunt II-TOPO vector (Invitrogen) with pJNWkan_for and pJNWkan_rev primers (all primers are listed in Table S3 in the supplemental material). Likewise, the promoter used to drive the expression of the transposase gene in pMarVF1was replaced with the σ70 (A1S_2706) promoter from A. baumannii by amplifying the fragment from genomic DNA with rpoD_for and rpoD_rev primers. After appropriate endonuclease restriction digestion, both fragments were ligated into previously digested pMarVF1, resulting in pJNW684. Note that all genomic DNA used in this work was extracted using the DNeasy approach according to the manufacturer’s instructions (Qiagen).
E. coli β3914, which is a diaminopimelic acid (DAP) auxotroph, was transformed with pJNW684 to create the donor strain. We then generated the A. baumannii transposon mutant library through filter mating of E. coli β3914(pJNW684) with A. baumannii (J. F. Brooks et al., unpublished data). Exconjugants were selected on kanamycin-supplemented LB agar plates. A total of 150,000 exconjugants were collected by scraping colonies from mating plates. Colonies were pooled and frozen in 50% glycerol at −80°C.
Murine model of pneumonia.
A. baumannii was grown in 5 ml of medium overnight at 37°C with shaking, diluted into fresh medium, and regrown for 2 to 3 h to achieve the exponential phase of growth. For INSeq selection, an aliquot of the A. baumannii transposon mutant pool was defrosted on ice and grown in 25 ml of medium at 37°C with shaking for 1.25 h. Bacterial cultures were subsequently diluted to an optical density at 600 nm (OD600) of ~1.7 (4 × 108 to 8 × 108 CFU/ml). In the INSeq experiments, a portion of the bacterial culture was saved for genomic DNA extraction (input pool), and the rest was used to inoculate mice. One milliliter of the culture for each mouse infection was collected by centrifugation and resuspended in 50 µl of phosphate-buffered saline (PBS). C57BL/6 female mice (8 to 12 weeks old) were anesthetized with intraperitoneal injections of a mixture of ketamine (100 mg/ml) and xylazine (20 mg/ml) and then infected with 4 × 108 to 8 × 108 CFU of A. baumannii intranasally. For the INSeq experiments, lungs were aseptically removed and homogenized in 5 ml of PBS at 24 h postinfection (output pool). For competition experiments, mice were inoculated with equal mixtures of parental and mutant A. baumannii cells (ratios were confirmed by plating on LB agar with or without kanamycin). Lung homogenates from mice were plated on LB agar with or without kanamycin. Competitive indices (CI) were calculated using the following formula: CI = (mutant output/parental output)/(mutant input/parental input) (51).
Animals were purchased from Harlan Laboratories and housed in the containment ward of the Center for Comparative Medicine at Northwestern University. All experiments were approved by and performed in accordance with the guidelines of the Northwestern University Animal Care and Use Committee.
Preparation of DNA for high-throughput sequencing.
Transposon insertion junctions were amplified from genomic DNA extracted from input and output pools according to the previously published protocol (52). Samples were single-end sequenced using the Illumina HiSeq 2500 platform at the Tufts University Genomics Core Facility.
Sequencing data analysis.
The INSeq data analysis was performed using the online software ESSENTIALS (http://bamics2.cmbi.ru.nl/websoftware/essentials) (16, 52) and the A. baumannii ATCC 17978 genome sequence (GenBank accession number CP000521). Further details are presented in Text S1 in the supplemental material. Raw sequencing reads are available at http://dhcp-165-124-221-156.mimnet.northwestern.edu/~hauserlab/inseq/index.html.
Construction of A. baumannii marked-deletion mutants.
A. baumannii mutants with a marked deletion in the dotB, gspN, A1S_0065, or A1S_0995 gene were generated using the gene replacement method of Schweizer and Hoang (53). Briefly, PCR primers were designed to amplify 300- to 700-bp fragments of the 5′ and 3′ ends of each gene, along with adjacent sequences. The kanamycin resistance cassette was amplified from the pCR-Blunt II-TOPO vector (Invitrogen). Restriction endonuclease sites were included in each of the primer sequences. The appropriate DNA fragments were then either digested and ligated sequentially into previously digested pEX100T (a generous gift from Herbert Schweizer) or pieced together by splicing by overlap extension (SOEing) PCR, digested, and ligated into digested pEX100T (53). After the cloning vectors were confirmed by sequencing, they were transformed into E. coli S17-1 λpir and introduced into A. baumannii via filter mating (54). Selection for vector integration into A. baumannii was achieved by growth on VBM agar supplemented with kanamycin (50 µg/ml). Kanamycin-resistant colonies were then grown in LB liquid for 3 h and plated on LB agar supplemented with sucrose (10% wt/vol) to induce a second recombination event that left behind only the deleted allele of the gene containing the kanamycin resistance cassette. The presence of each of the appropriate deleted alleles within mutants was confirmed by PCR amplification of the allele and subsequent sequencing.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
Southern blot analyses were performed using 20 randomly chosen transposon mutants, the parental wild-type A. baumannii strain (WT AB), and the transposon-containing vector pJNW684 (Tn vector). (A) A probe specific for the transposon was used. (B) A probe specific for the backbone of the transposon-containing vector was used. Molecular weight markers are indicated in kilobases (Kb). Transposon insertions into the A. baumannii genome occurred without backbone integration and with a high frequency of single insertions per bacterium. Download
Bacterial persistence in the lungs following intranasal inoculation of mice with 4 × 108 to 8 × 108 CFU of the parental strain of A. baumannii. Each symbol indicates data collected from a single mouse. Black bars represent medians. Fewer mice were evaluated at the 46-h time point due to drop out from lethality. Download
Disruption of the gspN gene did not result in a defect in virulence in the mouse pneumonia model. (A) Survival curves of mice intranasally infected with 4 × 108 to 8 × 108 CFU of either parental A. baumannii or the ΔgspN mutant. Five mice were used for each group. (B) Competition assays (1:1) between the ΔgspN mutant and the parental strain. Each symbol represents a single mouse. The experiment was performed on two different days, with cumulative results shown. Download
A principal component analysis (PCA) plot of the technical and biological replicates of the INSeq datasets generated by ESSENTIALS. Arrows 1 to 3 represent the three technical replicates of the input pool and arrows 4 to 8 represent the three biological replicates of the output pool, with arrows 5 to 7 representing the three technical replicates for one biological replicate. The proximity of the vectors is an indication of the similarity of the data sets. Locus tags associated with insertion sites are shown in gray. The figure was modified from the output of ESSENTIALS. Download
Density plots of input and output pools. (A) Density plot of the log2-transformed fold changes in measured reads relative to expected reads of the input pools for each gene. The red dot is the fold change value corresponding to the minimum between the peaks of essential and nonessential genes. This value was used as a threshold value for defining genes required for growth in LB medium. (B) Density plot of the log2-transformed fold changes in reads from output pools relative to reads from input pools for each gene. Figures were modified from the output of the ESSENTIALS program. Download
Representation of A. baumannii ATCC 17978 genes required for growth in LB medium (blue lines), genes that when mutated caused at least a 2-fold decrease in persistence in the mouse lung (red lines), and genes that when mutated caused at least a 2-fold increase in persistence in the mouse lung (green lines). The data were plotted using the genome visualization program CGView (http://wishart.biology.ualberta.ca/cgview) [P. Stothard, D. S. Wishart, Bioinformatics 21(4):537–539, 2005, doi:10.1093/bioinformatics/bti054]. Download
Results of the INSeq analysis. A. baumannii genes required for growth in rich medium (tab A), genes required for persistence in lungs of mice (tab B), genes that decrease persistence in lungs of mice (tab C), genes that did not affect persistence in lungs of mice (tab D), and data for all genes compared between the input and output pools (tab E).
Characterized A. baumannii virulence factors confirmed by INSeq screen.
Primers used in this study.
ACKNOWLEDGMENTS
We thank Paul Dunman for providing the A. baumannii ATCC 17978 strain and Eric Skaar for providing the ΔznuB and ΔlpsB strains along with their parental A. baumannii ATCC 17978 strain. We also thank Brittany Mortensen for technical advice on the ΔznuB and ΔlpsB mutants and John Brooks for help with preparing INSeq sequencing samples. We acknowledge Aldert Zomer for valuable technical assistance with ESSENTIALS and Albert Tai for advice on Illumina sequencing.
Support for this work was provided by National Institutes of Health grants AI053674, AI075191, AI099269, AI04831, and AI088286 (A.R.H.) and National Science Foundation grant IOS-0843633 (M.J.M.). This work was supported in part by the Northwestern University Genomics Core Facility.
Footnotes
Citation Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5(3):e01163-14. doi:10.1128/mBio.01163-14.
REFERENCES
- 1. McConnell MJ, Actis L, Pachón J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37:130-155. 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- 2. Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. 2012. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 8:e1003068. 10.1371/journal.ppat.1003068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80:1015–1024. 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC. 2008. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol. 10:309–319. 10.1111/j.1462-5822.2007.01041.x [DOI] [PubMed] [Google Scholar]
- 6. Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, St Michael F, Vinogradov EV, Campagnari AA. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun. 78:2017–2023. 10.1128/IAI.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. 2013. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 81:542–551. 10.1128/IAI.00704-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russo TA, MacDonald U, Beanan JM, Olson R, MacDonald IJ, Sauberan SL, Luke NR, Schultz LW, Umland TC. 2009. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J. Infect. Dis. 199: 513–521. 10.1086/596317 [DOI] [PubMed] [Google Scholar]
- 9. Cerqueira GM, Kostoulias X, Khoo C, Aibinu I, Qu Y, Traven A, Peleg AY. 23 February 2014. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 10.1093/infdis/jiu024 [DOI] [PubMed] [Google Scholar]
- 10. van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11:435–442. 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U. S. A. 106:16422–16427. 10.1073/pnas.0906627106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crimmins GT, Mohammadi S, Green ER, Bergman MA, Isberg RR, Mecsas J. 2012. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS Pathog. 8:e1002828. 10.1371/journal.ppat.1002828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, Pier GB. 2013. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 9:e1003582. 10.1371/journal.ppat.1003582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. 10.1016/j.chom.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78:1952–1962. 10.1128/IAI.00889-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. 2012. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One 7:e43012. 10.1371/journal.pone.0043012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, Durot M, Kreimeyer A, Le Fèvre F, Schächter V, Pezo V, Döring V, Scarpelli C, Médigue C, Cohen GN, Marlière P, Salanoubat M, Weissenbach J. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4:174. 10.1038/msb.2008.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaur N, Khokhar M, Jain V, Bharatam PV, Sandhir R, Tewari R. 2013. Identification of druggable targets for Acinetobacter baumannii via subtractive genomics and plausible inhibitors for MurA and MurB. Appl. Biochem. Biotechnol. 171:417–436. 10.1007/s12010-013-0372-2 [DOI] [PubMed] [Google Scholar]
- 19. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- 21. Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 78: 3993–4000. 10.1128/IAI.00366-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lees-Miller RG, Iwashkiw JA, Scott NE, Seper A, Vinogradov E, Schild S, Feldman MF. 2013. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89:816–830. 10.1111/mmi.12300 [DOI] [PubMed] [Google Scholar]
- 23. Peng K, Monack DM. 2010. Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect. Immun. 78: 2723–2733. 10.1128/IAI.00008-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rapoza PA, Tahija SG, Carlin JP, Miller SL, Padilla ML, Byrne GI. 1991. Effect of interferon on a primary conjunctival epithelial cell model of trachoma. Invest. Ophthalmol. Vis. Sci. 32:2919–2923 [PubMed] [Google Scholar]
- 25. Wood H, Fehlner-Gardner C, Berry J, Fischer E, Graham B, Hackstadt T, Roshick C, McClarty G. 2003. Regulation of tryptophan synthase gene expression in Chlamydia trachomatis. Mol. Microbiol. 49:1347–1359. 10.1046/j.1365-2958.2003.03638.x [DOI] [PubMed] [Google Scholar]
- 26. Choi CH, Lee JS, Lee YC, Park TI, Lee JC. 2008. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 8:216. 10.1186/1471-2180-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 100:11007–11012. 10.1073/pnas.1834481100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou G. 2011. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10:3399–3417. 10.1021/pr101299j [DOI] [PubMed] [Google Scholar]
- 29. Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA. 2008. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog. 4:e37. 10.1371/journal.ppat.0040037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jenkins A, Cote C, Twenhafel N, Merkel T, Bozue J, Welkos S. 2011. Role of purine biosynthesis in Bacillus anthracis pathogenesis and virulence. Infect. Immun. 79:153–166. 10.1128/IAI.00925-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Groisman EA, Ochman H. 1994. How to become a pathogen. Trends Microbiol. 2:289–294. 10.1016/0966-842X(94)90006-X [DOI] [PubMed] [Google Scholar]
- 33. Qiu H, Kuolee R, Harris G, Chen W. 2009. Role of NADPH phagocyte oxidase in host defense against acute respiratory Acinetobacter baumannii infection in mice. Infect. Immun. 77:1015–1021. 10.1128/IAI.01029-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3–8 [PubMed] [Google Scholar]
- 35. Jackson MW, Silva-Herzog E, Plano GV. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54:1364–1378. 10.1111/j.1365-2958.2004.04353.x [DOI] [PubMed] [Google Scholar]
- 36. Takaya A, Tomoyasu T, Tokumitsu A, Morioka M, Yamamoto T. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224–232. 10.1128/JB.184.1.224-232.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farrand AJ, Reniere ML, Ingmer H, Frees D, Skaar EP. 2013. Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J. Bacteriol. 195:5041–5050. 10.1128/JB.00505-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Antunes LC, Imperi F, Towner KJ, Visca P. 2011. Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162:279–284. 10.1016/j.resmic.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 39. Coyne S, Courvalin P, Périchon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55:947–953. 10.1128/AAC.01388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Z, Jacobs M, Schaff DA, McCullen CA, Binns AN. 2001. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J. Bacteriol. 183:3310–3317. 10.1128/JB.183.11.3310-3317.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kobayashi H, Uematsu K, Hirayama H, Horikoshi K. 2000. Novel toluene elimination system in a toluene-tolerant microorganism. J. Bacteriol. 182:6451–6455. 10.1128/JB.182.22.6451-6455.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. 10.1371/journal.pone.0017027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PC, Titball RW, Wren BW. 2007. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun. 75:1186–1195. 10.1128/IAI.01240-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, Sasakawa C. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11:31–41. 10.1111/j.1365-2958.1994.tb00287.x [DOI] [PubMed] [Google Scholar]
- 45. Sieira R, Comerci DJ, Pietrasanta LI, Ugalde RA. 2004. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54:808–822. 10.1111/j.1365-2958.2004.04316.x [DOI] [PubMed] [Google Scholar]
- 46. Stonehouse E, Kovacikova G, Taylor RK, Skorupski K. 2008. Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J. Bacteriol. 190:4736–4748. 10.1128/JB.00089-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Skurnik D, Roux D, Cattoir V, Danilchanka O, Lu X, Yoder-Himes DR, Han K, Guillard T, Jiang D, Gaultier C, Guerin F, Aschard H, Leclercq R, Mekalanos JJ, Lory S, Pier GB. 2013. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 110:20747–20752. 10.1073/pnas.1221552110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Piechaud M, Second L. 1951. Studies of 26 strains of Moraxella Iwoffi. Ann. Inst. Pasteur 80:97–99 [PubMed] [Google Scholar]
- 49. Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73:777–784. 10.1128/AEM.02147-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97–106 [PubMed] [Google Scholar]
- 51. Logsdon LK, Mecsas J. 2003. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect. Immun. 71:4595–4607. 10.1128/IAI.71.8.4595-4607.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protoc. 6:1969–1980. 10.1038/nprot.2011.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schweizer HP, Hoang TT. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22. 10.1016/0378-1119(95)00055-B [DOI] [PubMed] [Google Scholar]
- 54. Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1: 153–161. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
Southern blot analyses were performed using 20 randomly chosen transposon mutants, the parental wild-type A. baumannii strain (WT AB), and the transposon-containing vector pJNW684 (Tn vector). (A) A probe specific for the transposon was used. (B) A probe specific for the backbone of the transposon-containing vector was used. Molecular weight markers are indicated in kilobases (Kb). Transposon insertions into the A. baumannii genome occurred without backbone integration and with a high frequency of single insertions per bacterium. Download
Bacterial persistence in the lungs following intranasal inoculation of mice with 4 × 108 to 8 × 108 CFU of the parental strain of A. baumannii. Each symbol indicates data collected from a single mouse. Black bars represent medians. Fewer mice were evaluated at the 46-h time point due to drop out from lethality. Download
Disruption of the gspN gene did not result in a defect in virulence in the mouse pneumonia model. (A) Survival curves of mice intranasally infected with 4 × 108 to 8 × 108 CFU of either parental A. baumannii or the ΔgspN mutant. Five mice were used for each group. (B) Competition assays (1:1) between the ΔgspN mutant and the parental strain. Each symbol represents a single mouse. The experiment was performed on two different days, with cumulative results shown. Download
A principal component analysis (PCA) plot of the technical and biological replicates of the INSeq datasets generated by ESSENTIALS. Arrows 1 to 3 represent the three technical replicates of the input pool and arrows 4 to 8 represent the three biological replicates of the output pool, with arrows 5 to 7 representing the three technical replicates for one biological replicate. The proximity of the vectors is an indication of the similarity of the data sets. Locus tags associated with insertion sites are shown in gray. The figure was modified from the output of ESSENTIALS. Download
Density plots of input and output pools. (A) Density plot of the log2-transformed fold changes in measured reads relative to expected reads of the input pools for each gene. The red dot is the fold change value corresponding to the minimum between the peaks of essential and nonessential genes. This value was used as a threshold value for defining genes required for growth in LB medium. (B) Density plot of the log2-transformed fold changes in reads from output pools relative to reads from input pools for each gene. Figures were modified from the output of the ESSENTIALS program. Download
Representation of A. baumannii ATCC 17978 genes required for growth in LB medium (blue lines), genes that when mutated caused at least a 2-fold decrease in persistence in the mouse lung (red lines), and genes that when mutated caused at least a 2-fold increase in persistence in the mouse lung (green lines). The data were plotted using the genome visualization program CGView (http://wishart.biology.ualberta.ca/cgview) [P. Stothard, D. S. Wishart, Bioinformatics 21(4):537–539, 2005, doi:10.1093/bioinformatics/bti054]. Download
Results of the INSeq analysis. A. baumannii genes required for growth in rich medium (tab A), genes required for persistence in lungs of mice (tab B), genes that decrease persistence in lungs of mice (tab C), genes that did not affect persistence in lungs of mice (tab D), and data for all genes compared between the input and output pools (tab E).
Characterized A. baumannii virulence factors confirmed by INSeq screen.
Primers used in this study.