Abstract
In the December 28, 2001 issue of Cell, Hoang and Ferré-D’Amaré report the structure of a tRNA pseudouridine synthase, showing the target uridine flipped out from the tRNA and confirming that base flipping is not limited to enzymes that act on DNA.
Base flipping is a phenomenon first discovered less than a decade ago, when the structure of a DNA cytosine 5-methyltransferase bound to substrate DNA was solved [1]. Though the methylated C5 atom is exposed in the DNA major groove, the major groove surface cannot fit into the concave catalytic pocket typical of methyltransferases and other enzymes. The elegant solution to this problem is that the target cytosine swings ~180° out of the B-form helix to fit into the methyltransferase catalytic pocket, while other parts of the protein fill the gap left by the flipped base. Since then, base flipping has been found in many cases and predicted in others, where an enzyme needs access to an individual DNA base [2]. For example, many DNA glycosylases use this mechanism in removing abnormal bases.
There have been suggestions that base flipping might also be widespread in the RNA world. Base flipping may have arisen before the appearance of DNA as the genetic material for use on double-stranded (ds) RNA molecules [2]. Certainly the same basic needs exist for any enzyme requiring access to a single base, whether dsRNA or dsDNA is the substrate. Linear dsRNA is now relatively rare, though some phages and viruses have linear dsRNA genomes, but tRNAs, rRNAs, and small nuclear (sn) RNAs all have double-stranded regions to which various enzymes need access. Experimental demonstrations of base flipping in RNA that do not directly involve proteins have included a hairpin ribozyme [3], VS ribozyme [4], and the 30S ribosomal subunit complexed with the antibiotic paromomycin [5]. Paromomycin binds the major groove of helix H44 in 16S RNA and flips out the functionally important adenines 1492 and 1493; the flipped bases point directly into the ribosomal A site and interact with the minor groove of the codon-anticodon duplex (see Figure, panel A, left).
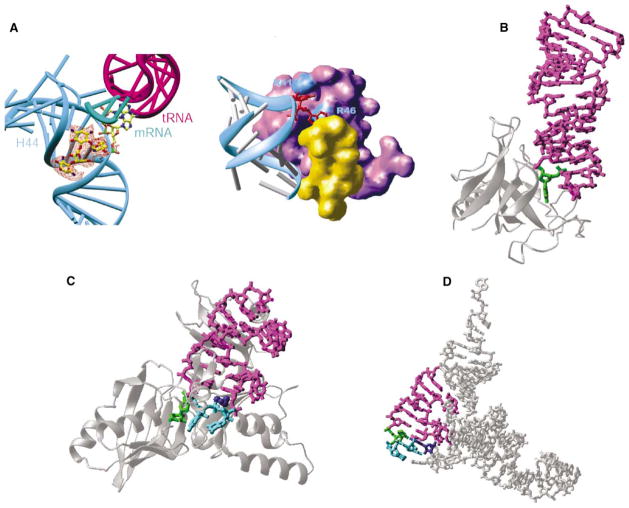
Figure. Examples of RNA Base Flipping.
(A) 30S ribosomal subunit: flipped-out A1492 and A1493 from helix 44 (in cyan) of 16S RNA by binding of paromomycin (left, seen in the orange difference density) and initiation factor IF1 (right, in purple). The protein S12 is in yellow.
(B) Restrictocin-RNA complex (Protein Data Bank code 1JBR). The RNA is presented as a magenta stick model, with the flipped nucleotide in green. The enzyme is represented as silver ribbons.
(C) TruB-tRNA complex (Protein Data Bank code 1K8W). The flipped nucleotides are in green (U55) and cyan (C56 and G57). The amino acid His43 in dark blue stacks with U54:A58 pair.
(D) Stick model of yeast tRNAPhe (Protein Data Bank code 1EHZ), with the stem loop in magenta, U55 in green, C56 and G57 in cyan, and G18 of D loop in dark blue.
There has also been earlier structural evidence that proteins use RNA base flipping. This includes structures for the fungal ribotoxin restrictocin (see Figure, panel B) [6] and the 30S ribosomal subunit interacting with initiation factor IF1 [7]. Binding of IF1 occludes the A site and, like paromomycin, flips out adenines 1492 and 1493 so that they are buried in IF1 pockets (see Figure, panel A, right) [7]. Enzymes that modify tRNA or rRNA within base-paired regions, either by methylation or by introducing more complex changes, might use this mechanism, and there has been indirect evidence (such as 2-aminopurine fluorescence) to support this view.
Pseudouridine synthases are a family of enzymes that catalyze the synthesis of pseudouridine, the C5-C1′ isomer of uridine, in a variety of RNAs including tRNA, rRNA, and snRNA. The mechanism employed by these synthases, originally proposed by Gu et al. [8], begins with nucleophilic attack by a conserved aspartate (Asp48 in TruB) on C6 of the substrate uridine, leading to cleavage of the N1-C1′ glycosidic bond between the base and the ribose and formation of a covalent bond between C6 and Asp. After 180° rotation of the uracil ring on the new C6-Asp bond, the base reattaches to the ribose via carbon-carbon bonding between C5 and C1′. Abstraction of the proton at C5 is followed by breakage of the covalent enzyme-tRNA bond to yield pseudouridine. Key evidence for this mechanism is that a suicide substrate, in which the C5 proton is replaced by the poor leaving group fluorine, allows trapping of the covalently bonded enzyme-tRNA reaction intermediate [8]. This is formally analogous to the trapping of a 5-fluorocytosine-cysteine covalent intermediate between HhaI DNA methyltransferase and its DNA substrate, which was used in the initial discovery of base flipping [1], and Hoang and Ferré-D’Amaré have put this approach to similarly productive use.
Now Hoang and Ferré-D’Amaré [9] present the structure of TruB tRNA pseudouridine synthase bound to a portion of its RNA substrate (containing the T stem-loop hairpin), using the 5-fluorouridine suicide substrate to trap the reaction intermediate (see Figure, panel C). The TruB structure shows base flipping as clearly and dramatically as did the original DNA methyltransferase structures. In the native tRNA, the target uridine 55 is buried by base pairing with a D loop nucleotide, G18 (dark blue in Figure, panel D) and stacking interactions with U54:A58. Without substantially distorting the overall tRNA structure, U55 (green in Figure, panels C and D), together with the adjacent nucleotides 56 and 57 (cyan in Figure, panels C and D), is swung completely out of the helix and occupies the TruB active site pocket. To fill the space left by this base flipping, TruB inserts an amino acid side chain (His43; dark blue in Figure, panel C) into the T loop to stack with the U54:A58 pair, taking the place of G18 from the D loop in intact tRNA.
The TruB-RNA complex also validates the proposed reaction mechanism. The ribose-base disconnection between N1-C1′ has occurred, rotation along the C6-Asp bond is apparent, and reattachment of 5-fluoroura-cil via C5-C1′ has taken place. Interestingly, the structure reveals no obvious candidate for the enzymic base required to abstract the proton at C5, though it seems reasonable to propose that a water molecule could diffuse into the active site to play this role. However, the structure also provides an unexpected observation—the Asp48 carboxylate is detached from C6 of the base despite the presence of fluorine at C5. Hoang and Ferré-D’Amaré [9] suggest that hydrolysis has occurred, generating 5-fluoro-6-hydroxy-pseudouracil, instead of breaking the C5-F bond. Furthermore, the base must have undergone further conformational change to yield the observed configuration: C6 is further away than C4 from the carboxylate of Asp48 (see Figure 5B of [9]).
RNA base flipping may also serve a chaperone function. A most interesting suggestion by Hoang and Ferré-D’Amaré, based on work from the Ofengand laboratory [10], is that RNAs with complex three-dimensional structures may be pulled apart and allowed to refold by base-flipping enzymes. For this to work, however, it would have to be shown that the base-flipping enzyme could recognize a misfolded substrate in the first place, and that the limited local unfolding (a few flipped nucleotides in a structure that is otherwise relatively undisturbed, in the case of TruB) could significantly contribute to correcting a misfolded structure. Nevertheless, this work dramatically confirms base flipping among enzymes that act on RNA, and raises the question of whether RNA base flipping may play roles yet to be discovered that are not played in the DNA world.
Selected Reading
- 1.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RJ, Cheng X. Base flipping. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 3.Rupert PB, Ferré-D’Amaré AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 4.Andersen AA, Collins RA. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 5.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Gerczei T, Glover L, Correll CC. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat Struct Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 7.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 8.Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluoro-uracil-tRNA. Proc Natl Acad Sci USA. 1999;96:14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang C, Ferré-D’Amaré AR. Cocrystal structure of a tRNA 55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 10.Gutgsell NS, Del Campo MD, Raychaudhuri S, Ofengand J. A second function for pseudouridine synthases: a point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA. 2001;7:990–998. doi: 10.1017/s1355838201000243. [DOI] [PMC free article] [PubMed] [Google Scholar]