Abstract
Histone variants have been proposed to act as determinants for post-translational modifications (PTM) with widespread regulatory functions. In this report, we identify a histone-modifying enzyme that selectively methylates the replication-dependent histone H3 variant H3.1. The crystal structure of the SET domain of the histone H3 lysine 27 (H3K27) methyltransferase ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) in complex with a H3.1 peptide shows that ATXR5 contains a bipartite catalytic domain that specifically “reads” alanine 31 of H3.1. Variation at position 31 between H3.1 and replication-independent H3.3 is conserved in plants and animals, and threonine 31 in H3.3 is responsible for inhibiting the activity of ATXR5 and its paralog ATXR6. Our results suggest a simple model for the mitotic inheritance of the heterochromatic mark H3K27me1 and the protection of H3.3-enriched genes against heterochromatization during DNA replication.
During the S phase of the cell cycle, patterns of histone PTMs must be re-established after passage of the replication fork to restore the correct epigenetic status to each region of the genome (1). Because many different chromatin states are encountered during replication, the deposition of histone PTMs on newly replicated chromatin must be precisely regulated.
The H3K27 methyltransferases ATXR5 and ATXR6 (ATXR5/6) are thought to maintain the heterochromatic mark H3K27me1 during DNA replication in plants (2). In atxr5 atxr6 double mutants, H3K27me1 levels are reduced, heterochromatin is decondensed, some repetitive sequences are transcribed and heterochromatic over-replication is observed (3–5). In both animals and plants, chromatin restoration after DNA replication also depends on the histone chaperone CAF-1 and involves deposition of the S-phase-expressed histone H3 variant H3.1 (6, 7). In contrast, histone H3.3 is inserted by other histone chaperones, mainly during transcription, and acts as a replacement histone (7–11). Canonical histone H3.1 and histone H3.3 are >96% identical in most eukaryotes (12) and differ only by four and five residues in flowering plants and mammals, respectively (Fig. 1A). H3.1 and H3.3 variants have been shown to contain different histone PTMs, but the mechanisms involved in H3 variant-specific marking are not known (12). It is possible that sequence variation between the variants could directly affect their PTMs (13, 14).
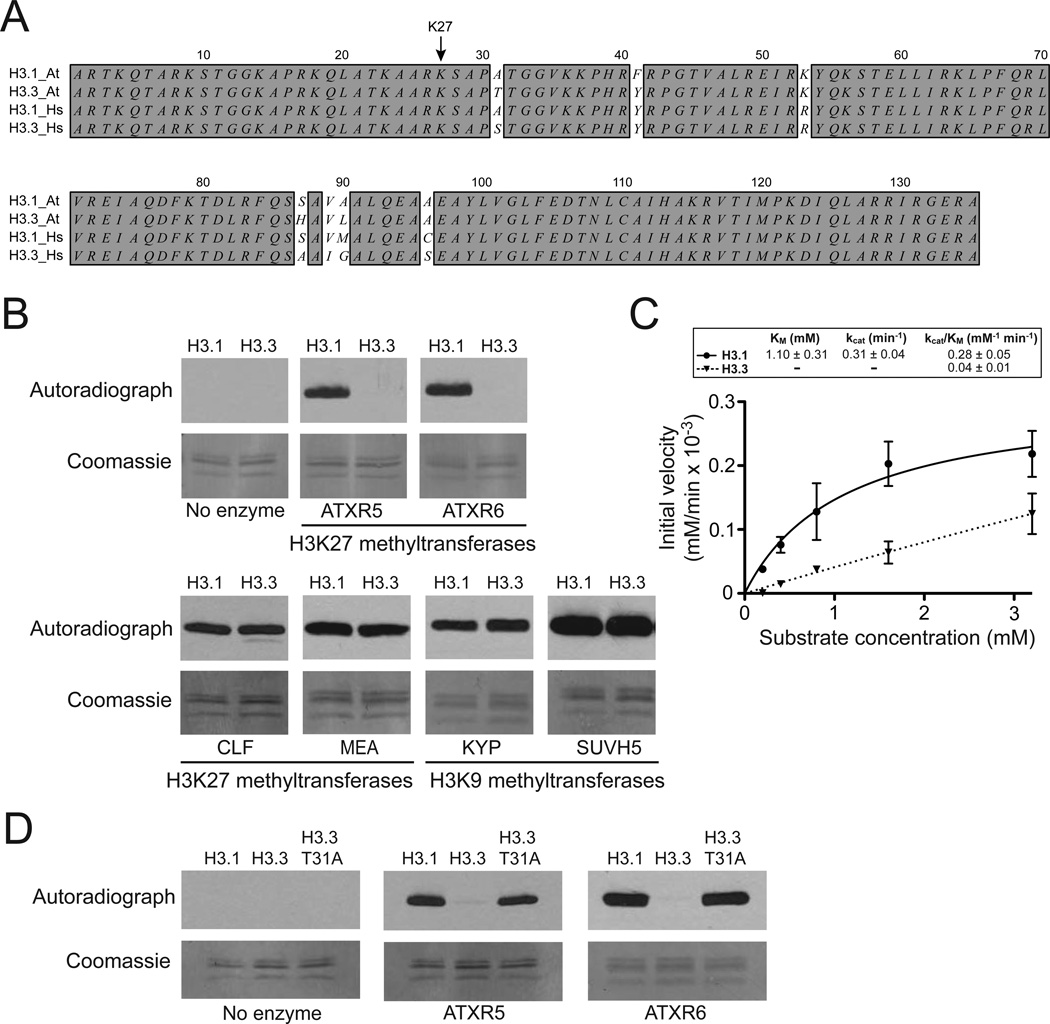
Figure 1. ATXR5 and ATXR6 selectively methylate the histone H3 variant H3.1.
A. Alignment of the canonical histone H3 variants H3.1 and H3.3 from A. thaliana (At) and human (Hs). Identities are dark-shaded. B. In vitro HKM assay using recombinant chromatin containing plant histone H3.1 or plant histone H3.3 as substrates and various histone methyltransferases from A. thaliana. C. Michaelis–Menten plot of initial velocity vs peptide substrate concentration. The kcat and Km values for the H3.1 and H3.3 peptides are shown as inlet. Errors bars represent the standard deviations of three independent experiments each performed in triplicates with three different batches of RcATXR5. D. In vitro HKM assay using recombinant chromatin containing plant histone H3.1, plant histone H3.3 or plant histone H3.3 T31A.
One of the conserved differences between H3.1 and H3.3 is at position 31, with alanine (H3.1) or serine/threonine (H3.3) (Fig. 1A). Because residue 31 of histone H3 is close to the modifiable and functionally important residue K27, we hypothesized that H3 variants could selectively regulate methylation at K27. To test this, we performed histone lysine methyltransferase (HKM) assays using methyltransferases from Arabidopsis thaliana and recombinant chromatin containing either plant histone H3.1 or plant histone H3.3. Our results show that the H3K27 methyltransferases ATXR5/6 have much higher activity on nucleosomes containing histone H3.1 than H3.3 (Fig 1B). Furthermore, steady-state kinetic analysis of ATXR5 confirms that the enzyme exhibits strong preference toward the H3.1 variant (Fig. 1C). This ability to favor H3.1 nucleosomes over H3.3 nucleosomes as substrates was not observed for two Polycomb Repressive Complex 2 (PRC2) complexes (MEDEA and CURLY LEAF), which also methylate K27, or the histone H3 lysine 9 (H3K9) methyltransferases KRYPTONITE (KYP)/SU(VAR)3–9 HOMOLOG 4 (SUVH4) and SUVH5 (Fig. 1B). We tested if alanine at position 31 (Ala-31) of H3.1 is required for H3K27 methylation by ATXR5/6. When using H3.3 nucleosomes with threonine 31 (Thr-31) replaced with alanine (T31A), we observed levels of H3K27 methylation similar to levels obtained when H3.1 nucleosomes are used (Fig. 1D). Taken together, these results demonstrate that ATXR5/6 selectively methylate the replication-dependent histone H3 variant H3.1 in vitro and Thr-31 in histone H3.3 is responsible for inhibiting the activity of ATXR5/6.
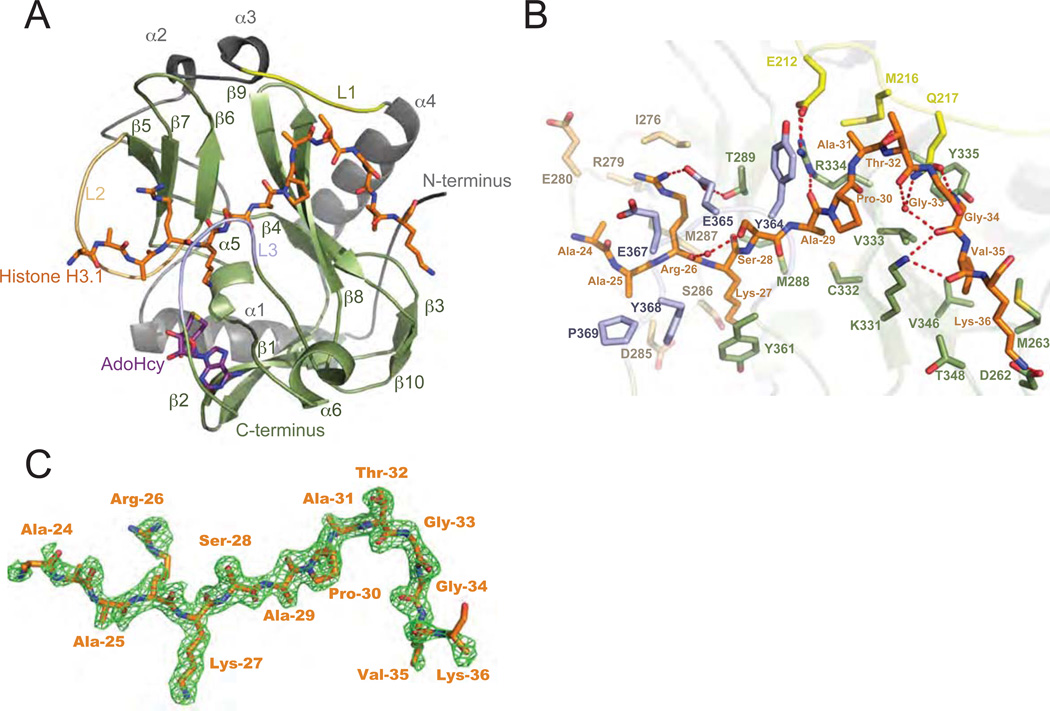
To gain a better understanding of how ATXR5/6 specifically methylate H3.1, we solved the crystal structure of an ATXR5-H3.1 complex. We focused on the C-terminal half of ATXR5, which contains the catalytic SET domain preceded by a conserved sequence (hereafter named nSET) of unknown function (fig. S1). The structure of the ATXR5 homologue from the plant Ricinus communis (RcATXR5 a.a. 158–374) in complex with a histone H3.1 peptide (a.a. 18–36, strictly conserved between A. thaliana and R. communis) and the product cofactor S-adenosylhomocysteine (AdoHcy) was solved at 2.1 Å resolution (fig. S2 and Table S1). Collectively, the structure shows that the SET domain comprises two short α-helices (α5–α6) and ten β-strands (β1–β10) all forming twisted anti-parallel β-sheets (Fig. 2A). The nSET domain folds as four consecutive α- helices (α1–α4) interspersed by loops that pack onto the SET domain. The histone H3.1 peptide binds in a tight binding cleft in an L-shaped conformation and engages in several hydrogen bonds and hydrophobic contacts (see supplementary discussion) with RcATXR5 (Figs. 2A and 2B). A simulated annealing Fo - Fc omit map revealed clear electron density for residues 24–36 of histone H3.1 (Fig. 2C). Comparative analysis of the ATXR5/H3.1 complex reveals structural divergence in the histone-binding mode (fig. S3 and supplementary discussion).
Figure 2. ATXR5/6 contain a bipartite catalytic domain composed of nSET and the SET domain.
A. Ribbons representation of the RcATXR5-H3.1-AdoHcy ternary complex in which nSET and the SET domain are highlighted in grey and green, respectively. Carbon atoms of histone H3.1 and product cofactor are colored in orange and magenta, respectively. L1-L3: loop 1-loop 3. β1–β10: beta strand 1-beta stand 10. α1–α6: alpha helix 1-alpha helix 6. B. Zoomed-view of the peptide binding cleft of RcATXR5. Three-letter code refers to histone H3.1 residues; one-letter code refers to RcATXR5 residues. Carbon atoms of residues found in the L1, L2 and L3 loops are rendered in yellow, beige and purple, respectively. The carbon atoms of other residues interacting with histone H3.1 are highlighted in green. Carbon atoms of histone H3.1 residues are colored in orange while oxygen and nitrogen atoms are highlighted in red and blue. Hydrogen bonds and water molecules are illustrated as red dashed lines and red spheres, respectively. C. Simulated annealing Fo – Fc omit map (green) contoured at 2σ. The histone H3.1 peptide is rendered as in panel A.
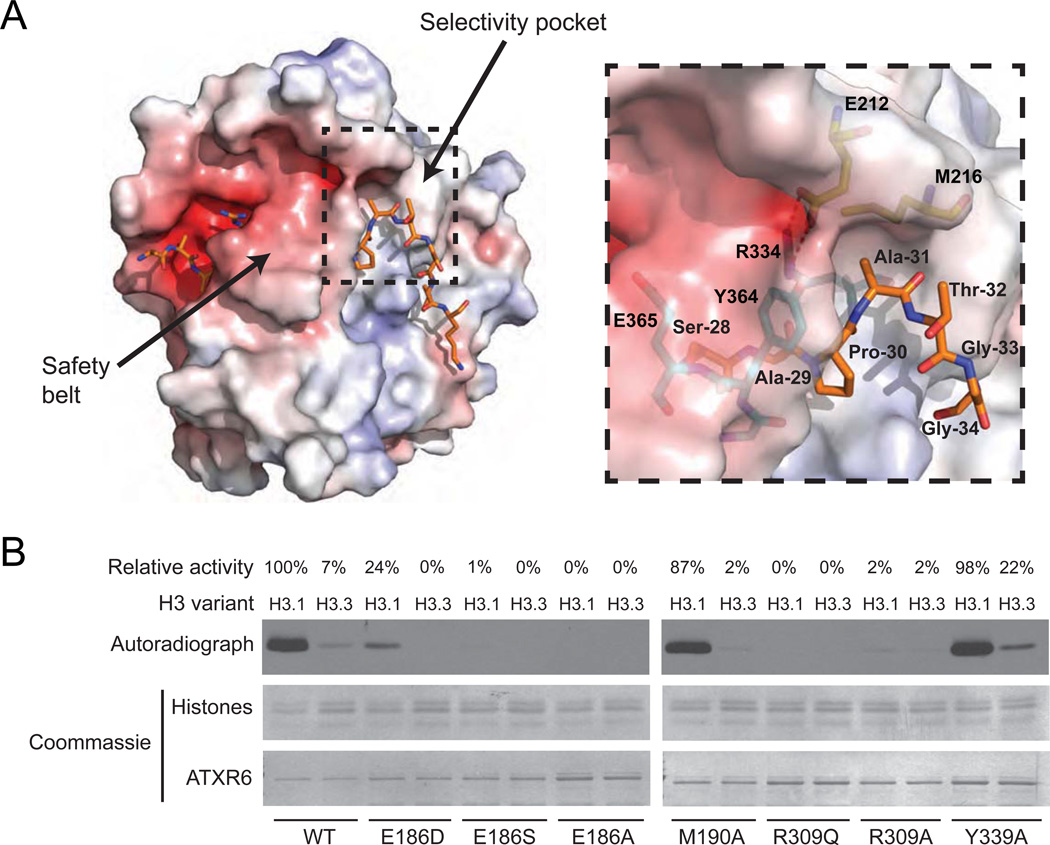
The ternary structure shows that both SET and nSET of RcATXR5 are involved in selective H3.1 binding. The residues E212 and M216 of the α3–α4 loop (L1) of nSET along with R334 in the SET domain form a shallow binding pocket (referred to as selectivity pocket) accommodating the small side chain of Ala-31 (Fig. 3A). Accordingly, E212, M216 and R334 are strictly conserved in ATXR5/6 homologues from mosses to flowering plants (fig. S1). The side chain of Ala-31 makes hydrophobic and van der Waals contacts with the side chains of M216 and R334 and the guanidium group of R334 engages in two short hydrogen bonds with the carboxylate group of E212, which likely rigidify the specificity pocket. Consistent with our HKM assays (Fig. 1D), we found that replacing Ala-31 with Thr-31 generates van der Waals clashes between Thr-31 Cγ methyl group and the side chain of R334 (fig. S4). In addition, amino acid substitutions at E212 and R334 (A. thaliana ATXR6 residues E186 and R309) drastically reduced methylation on nucleosomes (Fig. 3B), suggesting that residues forming the specificity pocket are important for conferring specificity and high affinity binding to histone H3.1.
Figure 3. The selectivity pocket and safety belt of ATXR5/6-type H3K27 methyltransferases are responsible for histone H3.1 preference over histone H3.3.
A. The structure of the ATXR5-H3.1-SAH complex in electrostatic potential surface representation with the selectivity pocket and safety belt highlighted. Positive and negative potentials are in blue and red, respectively. Inlet figure shows a zoomed view of the residues forming the surface of the selectivity pocket (three-letter code refers to histone H3.1 residues; one-letter code refers to RcATXR5). Hydrogen bonds are shown as dashed red lines. B. In vitro HKM assay using recombinant chromatin containing plant histone H3.1 or H3.3 as substrates and wild-type or point mutants of ATXR6 from A. thaliana. The enzymatic activity indicated for each reaction is relative to the activity of ATXR6 (WT) on H3.1 nucleosomes.
Another unique structural feature of ATXR5/6 likely contributes to the selective methylation of histone H3.1. A loop (L3) comprising residues G363, Y364, E365 and E367 folds back on top of H3.1 shielding the peptide from the solvent in a “safety belt” conformation (Figs. 2A, 2B and 3A). Y364 makes hydrophobic contact with Pro-30 while the carboxylate group of E365 engages in hydrogen bonds with the guanidium group of Arg-26 and the hydroxyl group of T289. Together, these residues help bind the peptide tightly to the histone H3.1 binding cleft. The role of the L3 loop is likely twofold: 1) locking the peptide in a conformation that forces the side chain of Ala-31 into the specificity pocket and 2) packing the structurally constrained residue Pro-30 onto the peptide-binding pocket of RcATXR5. This hypothesis is supported by our HKM assays showing that substitution of Y364 (A. thaliana ATXR6 Y339) by an alanine residue reduces the specificity of the enzymes for H3.1 by 3–fold (Fig. 3B).
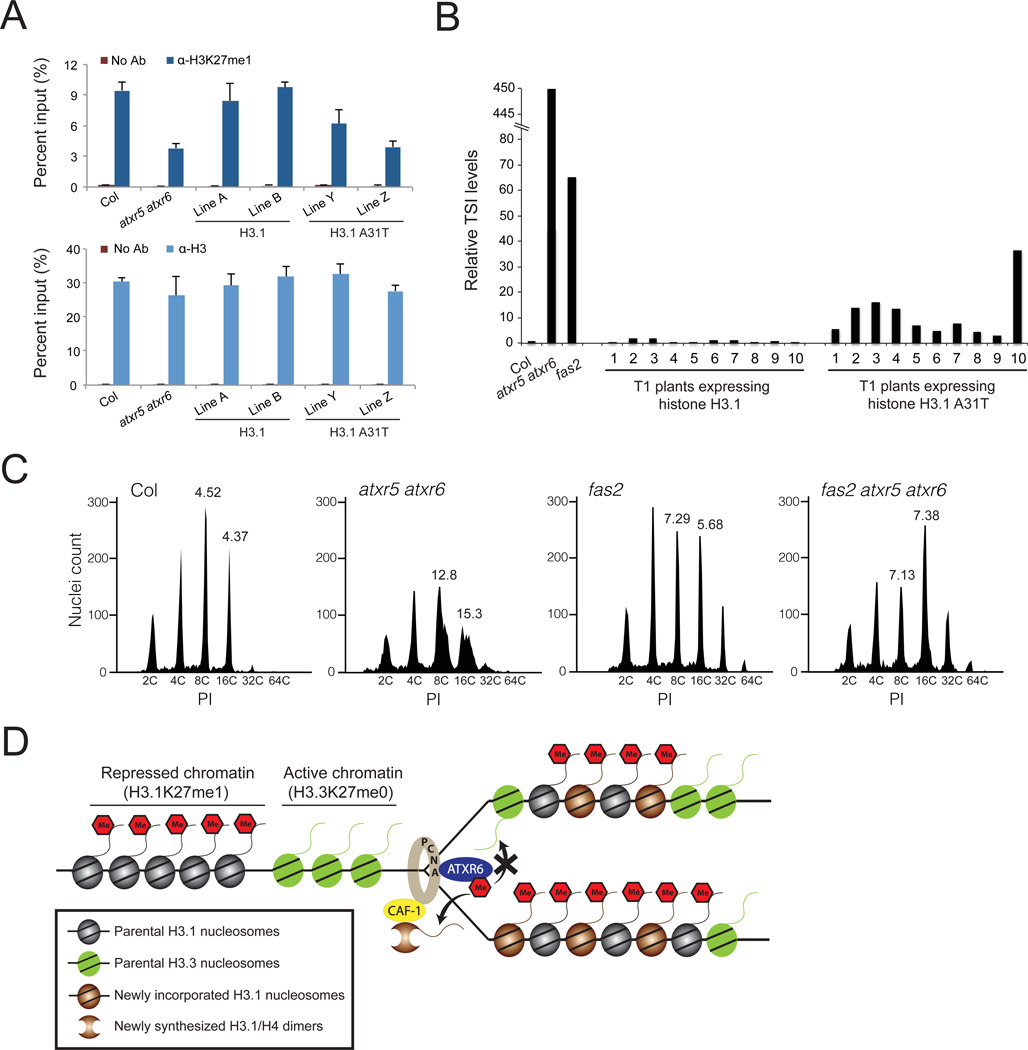
In Arabidopsis, H3K27me1 is enriched on H3.1 (fig. S5) (15, 16), and more than 80% of H3.1 was found to be methylated at K27 by mass spectrometry (17). To validate that Thr- 31 in histone H3.3 directly interferes with the activity of ATXR5/6 in vivo, we generated transgenic Arabidopsis plants expressing the tandem histone H3.1 genes HTR9 and HTR13 as wild-type proteins (H3.1) or with an alanine-to-threonine replacement at position 31 (H3.1 A31T). The transgenes were expressed in H3.1 quadruple mutants (A. thaliana contains five H3.1 genes). We quantified by chromatin immunoprecipitation (ChIP) the levels of H3K27me1 at genomic regions enriched in H3.1 (fig. S6) and that have been shown to be dependent on ATXR5/6 for H3K27me1 (4), as plant PRC2 complexes also have the ability to monomethylate H3K27 (fig. S7). When we measured H3K27me1 levels in the two sets of transgenic plants, we observed lower levels of the epigenetic mark in plants expressing H3.1 A31T compared to wild-type (Col), but not in plants expressing H3.1 (Figs. 4A and S8). As in atxr5 atxr6 double mutants, silencing of Athila ORF1 (also known as transcriptionally silent information (TSI)) was lost in transgenic plants expressing H3.1 A31T (Fig. 4B).
Figure 4. Thr-31 of H3.3 inhibits the activity of ATXR5/6 in vivo.
A. ChIP assays for H3K27me1 (upper panel) and H3 (lower panel) at TSI in transgenic T4 (homozygous) lines expressing H3.1 or H3.1 A31T. The average and standard deviation of three independent experiments is presented. ChIP for H3 serves as a control for nucleosome density. No Ab: no antibody control. B. RT-qPCR expression analysis of the repetitive element TSI in Col, atxr5 atxr6, fas2 and independent transgenic lines (first generation) expressing either wild-type plant H3.1 or the mutant H3.1 A31T. C. Flow cytometry profiles of Col, atxr5 atxr6, fas2 and fas2 atxr5 atxr6 leaf nuclei. The numbers below the peaks indicate the endoreduplication (ploidy) levels of the nuclei. The numbers above the 8C and 16C peaks correspond to the robust CV values (PI units that enclose the central 68% of nuclei) for those peaks. High robust CV values at 8C and 16C peaks characterize heterochromatic over-replication (4). PI: propidium iodide. D. Model for the role of ATXR5/6 during DNA replication in plants.
Reactivation of TSI and other heterochromatic defects has also been observed when the histone chaperone CAF-1 is mutated in Arabidopsis (Fig. 4B) (18–22). Depletion of CAF-1 in mammalian cell lines leads to H3.1 replacement with H3.3 (23). Consistently, Arabidopsis CAF-1 mutants show higher expression of H3.3 genes (18). Based on our finding that ATXR5/6 specifically methylate H3.1, we hypothesized that the heterochromatic defects of CAF-1 mutants in Arabidopsis could be due, at least in part, to depletion of K27me1 when histone H3.3 replaces histone H3.1. Our results show that H3K27me1 levels are indeed lower in fasciata2 (fas2: subunit of CAF-1) mutants compared to Col, and this is not caused by defects in nucleosome density or antibody preference for H3.1K27me1 over H3.3K27me1 (figs. S9 and S10).
One of the phenotypes associated with reduced levels of H3K27me1 in atxr5 atxr6 double mutants is over-replication of heterochromatic DNA (4). Lower levels of H3K27me1 in fas2 mutants or our transgenic lines expressing H3.1 A31T is not accompanied by a similar defect in heterochromatic DNA replication (Figs. 4C, S11 and S12) (19, 24–26). This suggests a model in which unmethylated histone H3 having alanine at position 31 (i.e. H3.1K27me0) allows for heterochromatic over-replication to occur. One prediction from this model is that heterochromatic over-replication should be suppressed in a fas2 mutant background, as H3.1K27me0 would now be replaced by H3.3K27me0. As predicted, the phenotype is strongly suppressed in atxr5 atxr6 fas2 triple mutants (Fig. 4C). This model also provides an explanation for the partial suppression of the heterochromatic over-replication phenotype of atxr5 atxr6 by mutations affecting DNA methylation (5). H3.3 is known to replace H3.1 at transcribed genes in plants and animals (7–9, 11, 15, 16). Because loss of DNA methylation leads to the transcriptional activation of normally silent loci (27), DNA methylation mutants (similar to fas2 mutants) would replace H3.1 with H3.3 in heterochromatin. Accordingly, suppression of over-replication is strongest in the DNA methylation mutants that have the greatest effect on transcriptional reactivation (5). Taken together, our results suggest a model in which H3.1K27me0 is the stimulus for heterochromatic over-replication.
Overall, this study demonstrates how histone variants can determine epigenetic states through direct modulation of chromatin-modifying enzyme activity. Further, the ability of ATXR5/6 to discriminate between the H3 variants H3.3 and H3.1 provides a mechanism for the mitotic inheritance and genome-wide distribution of H3K27me1 in plants. According to this model, ATXR5/6 are recruited to the replication fork during S phase through their interaction with PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) (2), where they specifically monomethylate K27 at newly incorporated, CAF-1-dependent histone H3.1 to rapidly restore pre-replication levels of this epigenetic mark (Fig. 4D). This model does not rule out the possibility that some H3.1 might escape DNA replication-coupled K27 monomethylation (fig. S5). The inability of ATXR5/6 to methylate H3.3 may contribute to the protection of transcriptionally-active, H3.3-enriched regions against H3K27me1 and repression during DNA replication.
Materials and Methods
Plant materials
Plants were grown under cool-white fluorescent light under long-day (16h light, 8h dark) conditions. The double mutant atxr5 atxr6 has been described previously (3) and fas2–4 mutants (SALK_033228) were obtained from the Arabidopsis Biological Resource Center (ABRC). The H3.1 quadruple mutant was made using the following four single mutants all obtained from the ABRC: htr1 (At5g65360; SALK_069666), htr2 (At1g09200; SALK_022688C), htr3 (At3g27360; SALK_078768) and htr9 (At5g10400; SALK_148171). Transgenic plants expressing WT H3.1 (HTR9-HTR13) and H3.1 A31T (HTR9 A31T-HTR13 A31T) were made by transforming the H3.1 quadruple mutant.
Constructs
Cloning of catalytic fragments of ATXR5 (a.a. 57–379), ATXR6 (a.a. 25–349) and KYP (a.a. 291–624) for heterologous protein expression and in vitro methyltransferase assays has been described previously (3, 28). To synthesize the plant PRC2 complexes CURLY LEAF and MEDEA, we used multigene baculoviral vectors that come with the MultiBac system for insect cell-mediated protein expression (29). Full-length CDS of the plant PRC2 subunits FIE, MSI1, MEA, FIS2 (Landsberg erecta), CLF and EMF2 were cloned using SLIC (30) into the following vectors: pFL (FIE), pUCDM (MSI1), pSPL (CLF: p10 site; MEA: p10 site; EMF2: ph site; FIS2: ph site). A 1× FLAG tag was added at the N-terminus of the FIE CDS for affinity purification of the PRC2 complexes. Progenitor constructs were combined using in vitro Cre-fusion to create multigene transfer vectors. The tandem histone H3.1 genes HTR9 and HTR13, including 1068 bp upstream of the start codon of HTR9 and 368 bp downstream of the stop codon of HTR13, were cloned as a single unit in pENTR/D-TOPO (Life Technologies) and then sub-cloned using Gateway Technology into the plant binary vectors pMDC107 (31). For histone H3 expression, Arabidopsis H3.1 (At1g09200) and H3.3 (At5g10980) genes were codon-optimized for expression in E. coli and cloned into pET3. Site-directed mutagenesis of different clones was performed using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies).
Protein expression and generation of recombinant chromatin
Catalytic fragments of ATXR5, ATXR6 and KYP were synthesized in E. coli and purified as described previously (3, 28). The FLAG tagged PRC2 complexes CLF and MEA were expressed in SF9 insect cells using the MultiBac system (29). To purify the complexes, the SF9 cells were resuspended in lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1mM PMSF and 0.1% Triton X-100), and sonication 10 × 20 seconds on ice. The cell lysate was centrifuged at 20 000 × g for 40 minutes at 4°C and the complexes were purified with anti-FLAG M2 Affinity Gel according to the manufacturer’s protocol (Life Technologies). The FLAG fusion complexes were eluted from the columns by competition with 100 µg/ml FLAG peptide (Life Technologies) in TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.4). A complete description of the protocol to make the recombinant chromatin used in our assay has been published previously (32)
RcATXR5 protein expression, purification and crystallization
The cDNA encoding residues 158–374 of Ricinus communis ATXR5 (RcATXR5) was purchased from GenScript with codons optimized for overexpression in E. coli and was subcloned in the parallel vector pGST2. RcATXR5 was overexpressed in fusion with a TEV cleavable glutathione sulfotransferase in Rosetta cells (EMD Millipore) with 0.1mM of IPTG during 16 hours at 18°C. Cells were centrifuged and the pellet was harvested in cold PBS buffer (pH 7.3). Cells were lysed by sonication and clarified by centrifugation. The supernatant was applied onto glutathione sepharose beads for 1 hour at 4°C and washed extensively with PBS buffer. GST-ATXR5 was TEV cleaved on beads for 16 hours at 4°C in PBS buffer. The supernatant was subsequently concentrated and purified by size exclusion chromatography on a Superdex 200 (GE Healthcare) preequilibrated in 20mM Tris pH 7.5, 250mM NaCl and 5mM beta-mercaptoethanol. Fractions corresponding to the monomeric species of RcATXR5 were pooled and concentrated to 30 mg ml−1. For crystallization trials, the concentrated protein was immediately mixed with equimolar ratio of S-adenosylhomocysteine (AdoHcy) and the histone H3.1 peptide (a.a. 18–36) and incubated on ice for one hour. Several crystals were obtained with Ammonium sulfate as precipitant; however, these crystals corresponded to the apo-form of RcATXR5. Crystals of the RcATXR5/H3.1/SAH ternary complex were grown at 18°C in 50% polypropylene glycol 400, 100mM Na-HEPES pH 6.0 and 5% DMSO. Crystals were harvested, soaked in the mother liquor and flash frozen in liquid nitrogen.
RcATXR5 structure determination
A single-wavelength anomalous dispersion (SAD) data set was collected at the 17-ID-D beamline of LS-CAT (Advanced Photon Source; Argonne National Laboratories). The reflections were recorded on a single crystal of a selenomethionyl-substituted RcATXR5 protein at 0.978 Å using a MarMosaic300 CCD detector (Rayonix). The data set was subsequently processed and scaled using HKL2000. Using Solve/Resolve from Phenix (33), eight selenium atoms were located in the asymmetric unit and a partial model (~75%) was automatically built. For the crystal structure of the RcATXR5/H3.1/SAH ternary complex, a full data set was collected on a MicroMax 007-HF equipped with Rigaku IV++ image plate detector. The reflections were indexed and scaled using d*Trek (34) and a molecular replacement solution was found using Phaser (33) and RcATXR5 partial structure as a search model. Two ATXR5 molecules were located in the asymmetric unit and were refined using Buster. The model was completed using iterative rounds of refinement and model building using Buster and Coot, respectively (35, 36).
Histone lysine methyltransferase assays
The procedure used in the histone lysine methyltransferase (HKM) assays has been previously described (32), with some modifications. Briefly, we used between 0.5–1 µg of ATXR5, ATXR6, or KYP and 1.5 µg of MEA or CLF (PRC2) complexes with 1 µg of recombinant chromatin substrates. The histone methyltransferase buffer contained 50mM Tris pH 8.0, 2.5mM MgCl2 and 4mM DTT, and enzymes and chromatin substrates were incubated for 1.5 h at 23°C. H3K27me1 primary antibody (cat. #07-448, Millipore) at 1:1000 dilution and peroxidase conjugated anti-rabbit secondary antibody (cat. #111-035-03, Jackson Immunoresearch Lab Inc.) at 1:10 000 dilution were used for detection by Western Blot of methylated substrates resulting from non-radioactive HKM assays. The Pierce ECL Western Blotting Substrate (Pierce Biotechnology, cat# 32106) was used for detection. Quantification of HKM assays using point mutants of ATXR6 was done using the software ImageJ and the following protocol (http://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-imagej/).
To define the minimal structural determinants recognized by RcATXR5, enzymatic assays were performed as previously described (37, 38). Briefly, 600nM of enzyme was incubated with 1mM of peptide in the HMT buffer during 1 h at 30°C. Reactions were stopped and activity was quantified by liquid scintillation counts as previously described (37, 38). The kinetic parameters for the methylation of H3.1 (KQLATKAARKSAPATGGVKY) and H3.3 (KQLATKAARKSAPTTGGVKY) by RcATXR5 were measured using purified recombinant RcATXR5 and synthetic H3 peptides (GenScript). A tyrosine residue was added at the C-terminus of the peptides for UV quantification. AdoMet was obtained from Sigma and radiolabeled [3H-methyl]-AdoMet from Perkin Elmer (16.5Ci/mmol). ATXR5 assays were performed at 30°C for 90 min in a buffer containing 50mM Tris pH 8.5, 20mM KCl, 10mM MgCl2, 10mM β-mercaptoethanol and 10% glycerol. AdoMet was provided at a ratio of 2.5µM:7.5µM labeled and un-labeled respectively. Reactions performed with the H3.1 peptide were initiated by the addition of 1µM of RcATXR5. Peptides were added at a concentration range of 0.1–3.2mM to a final volume of 30µL. Reactions were stopped and the activity was quantified as described above. Initial velocities were plotted versus substrate concentration and data were fitted to the Michaelis-Menten equation using GraphPad Prism version 5.00 for MAC, GraphPad Software, San Diego California USA, (www.graphpad.com).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (39). 2.5µl of H3K27me1 antibody (Life Technologies: 49–1012) or 2.5 µl of Histone H3 antibody (Abcam: ab1791) was used per immunoprecipitation.
Real-time qPCR
qPCR was performed on a CFX96 Real-Time System (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's protocol. The primers used for RTqPCR and ChIP-qPCR were described previously (3–5), except the primers (FWD: AGAGGGTACGTCGCATCCAA and REV: TCTGCCTCATGTCCCTCACA) for EF1A which served as our internal control in the experiments. Leaves from four week old plants were used as starting materials for the assays.
Flow cytometry
Flow cytometry was performed as described (4), with slight modifications. Leaves six and seven of 4-weeks-old plants were used for nuclei extraction. Quantification (nuclei counts and robust CV values) was performed using Flowjo 10.0.6 (Tree Star).
Validation of H3K27me1 antibodies for H3.1 and H3.3
Analysis by Western blot of binding affinity for H3.1 and H3.3 was performed using the following antibodies at the indicated concentrations: Active Motif-39377 (1:1000), Active Motif-61015 (1:500), Active Motif-39889 (1:500), Life Technologies-49-1012 (1:500), Millipore-07-448 (1:1000) and Millipore-CS200593 (1:1000). 1 µg of plant H3.1 and H3.3 peptides (a.a 18–41: United Biosystems) were used in the assay. Quantification of binding affinity was performed using the software ImageJ.
Analysis of H3.1 and H3.3 levels at heterochromatic loci
H3.1 and H3.3 ChIP-seq data (with their respective controls) were downloaded from GEO (15). Reads were uniquely mapped to Arabidopsis genome TAIR10, with duplicate reads removed. Read coverage was normalized as reads per million (RPM). Log2(ChIP/Input) was plotted at single base pair resolution for AT1G37110 and AT4G03745 and smoothed only for presentation purposes. Regions of H3.1 and H3.3 enrichment were identified genome-wide with SICER using input DNA as a control with the following parameters: W=200, G=400, FDR=0.001.
Supplementary Material
Acknowledgments
We thank Joseph Calarco for critically reading the manuscript. Justin Goodrich (MEA and CLF cDNA clones), Claudia Kohler (FIS2 cDNA clone), Steve E. Jacobsen and Dinshaw J. Patel (SUVH4 cDNA clone and the SUVH5 protein) kindly provided materials and Thomas Schalch helped with the MultiBac system. This work was supported by the Howard Hughes Medical Institute-Gordon and Betty Moore Foundation and by grants from the National Science Foundation (DBI-1025830) and the National Institutes of Health (5R01GM067014) to R.A.M. J.F.C. is supported by a grant from the Canadian Institute of Health Research (BMA-355900) and the Natural Sciences and Engineering Research Council of Canada (Discovery Grant 191666) and acknowledges an Ontario Early Research Award and a Canada Research Chair in Structural Biology and Epigenetics. Y.J. was supported by a Louis-Berlinguet postdoctoral fellowship (Fonds Québécois de la Recherche en Santé/Génome Québec). P.V. is supported by fellowships from the Deutsche Akademie der Naturforscher Leopoldina (LPDS 2009-5) and the Empire State Training Program in Stem Cell Research (NYSTEM, contract # C026880). Work in the Reinberg laboratory is supported by grants from the National Institute of Health (GM064844 and R37GM037120) and the Howard Hughes Medical Institute. S.D.M. is supported by a grant from the National Institutes of Health (R01GM075060). The Protein Data Bank (PDB) accession number for the RcATXR5-H3.1-AdoHcy ternary structure is 4O30.pdb
Footnotes
Materials/Methods, Supplementary Text, Tables, Figures, and References
Materials and Methods
Supplementary discussion
Figs. S1 to S12
Tables S1
References 28 to 40
References and Notes
- 1.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012 Mar;13:153. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 2.Raynaud C, et al. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006 Aug;47:395. doi: 10.1111/j.1365-313X.2006.02799.x. [DOI] [PubMed] [Google Scholar]
- 3.Jacob Y, et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol. 2009 Jul;16:763. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacob Y, et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010 Aug 19;466:987. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stroud H, et al. DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis. PLoS Genet. 2012 Jul;8:e1002808. doi: 10.1371/journal.pgen.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989 Jul 14;58:15. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 7.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004 Jan 9;116:51. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005 Apr 1;19:804. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010 Mar 5;140:678. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010 Jun 15;24:1253. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002 Jun;9:1191. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 12.Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 2007 Sep;32:425. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 14.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005 Oct;37:1090. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 15.Stroud H, et al. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2012 Apr 3;109:5370. doi: 10.1073/pnas.1203145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wollmann H, et al. Dynamic deposition of histone variant h3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 2012 May;8:e1002658. doi: 10.1371/journal.pgen.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson L, et al. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schonrock N, Exner V, Probst A, Gruissem W, Hennig L. Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J Biol Chem. 2006 Apr 7;281:9560. doi: 10.1074/jbc.M513426200. [DOI] [PubMed] [Google Scholar]
- 19.Kirik A, Pecinka A, Wendeler E, Reiss B. The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell. 2006 Oct;18:2431. doi: 10.1105/tpc.106.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003 Jun;130:2555. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- 21.Ono T, et al. Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells. 2006 Feb;11:153. doi: 10.1111/j.1365-2443.2006.00928.x. [DOI] [PubMed] [Google Scholar]
- 22.Takeda S, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004 Apr 1;18:782. doi: 10.1101/gad.295404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011 Dec 23;44:928. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Endo M, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. Embo J. 2006 Nov 29;25:5579. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Exner V, Taranto P, Schonrock N, Gruissem W, Hennig L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development. 2006 Nov;133:4163. doi: 10.1242/dev.02599. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007 May;144:105. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigal M, Mathieu O. A "mille-feuille" of silencing: epigenetic control of transposable elements. Biochim Biophys Acta. 2011 Aug;1809:452. doi: 10.1016/j.bbagrm.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007 Feb 20;17:379. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004 Dec;22:1583. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 30.Li MZ, Elledge SJ. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol. 2012;852:51. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- 31.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003 Oct;133:462. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012 Sep 28;151:181. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 34.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999 Oct;55:1718. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 35.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60:2210. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010 Apr;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avdic V, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011 Jan 12;19:101. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 38.Avdic V, et al. Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic. FASEB J. 2011 Mar;25:960. doi: 10.1096/fj.10-171959. [DOI] [PubMed] [Google Scholar]
- 39.Villar CB, Kohler C. Plant chromatin immunoprecipitation. Methods Mol Biol. 2010;655:401. doi: 10.1007/978-1-60761-765-5_27. [DOI] [PubMed] [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.