Abstract
Cancer genome sequencing has enabled the rapid identification of the complete repertoire of coding sequence mutations within a patient’s tumor and facilitated their use as personalized immunogens. While a variety of techniques are available to assist in the selection of mutation-defined epitopes to be included within the tumor vaccine, the ability of the peptide to bind patient MHC is a key gateway to peptide presentation. With advances in the accuracy of predictive algorithms for MHC class I binding, choosing epitopes on the basis of predicted affinity provides a rapid and unbiased approach to epitope prioritization. We show herein the retrospective application of a prediction algorithm to a large set of bona fide T-cell defined mutated human tumor antigens that induced immune responses most of which were associated with tumor regression or long-term disease stability. The results support the application of this approach for epitope selection and reveal informative features of these naturally occurring epitopes to aid in epitope prioritization for use in tumor vaccines.
Keywords: somatic mutations, neoantigens, whole-genome sequencing, whole-exome sequencing, T cell, immunotherapy, vaccine, next-generation sequencing, T cell receptor
We and others (1–3) have suggested that the vast number of personal, tumor-specific mutations found in the genome of cancer patients provides a rich source of unique immunogens (“neoantigens”) for use in tumor vaccination strategies. These tumor neoantigens are attractive as vaccine targets since they are expected to bypass the immune dampening effects of central tolerance and because their expression is exquisitely tumor-specific. In order to bring this highly personalized treatment approach to cancer patients, one crucial challenge is the choice of which of the many possible personal mutated epitopes to incorporate in the vaccine.
Cancer genomes vary widely in the number of total and coding sequence mutations depending on the tumor type (4). The five most common tumor types in the United States (prostate, breast, lung, colon and melanoma) harbor an average of 25 to 500 non-synonymous coding sequence mutations. Vaccination approaches that utilize irradiated whole tumor cells or various forms of cell lysates (5–11) have attempted to capture all such neoantigens (in addition to native tumor-associated antigens). Although these strategies appear comprehensive and have resulted in clinical benefit in some cases, they do not favor any particular T cell immunogen. Hence, potentially highly effective immunogens may be drowned out within the vast sea of immunologically irrelevant antigens. Such complete antigen preparations are similar to the endogenous presentation of the tumor cell to the immune system and lack the “pharmacologic specificity” of a rationally designed vaccine.
A more selective but still comprehensive approach for neoantigens could be envisioned by utilizing every identified coding mutation as a separate immunogen. Although possibly feasible from the technical standpoint -- especially for tumors with a low mutation load -- the dilution of the potent immunogens is likely to reduce its effectiveness, and thus, a more discriminating approach to identify the most effective subset seems advisable.
Potential strategies to identify and prioritize mutated antigens
Multiple biochemical and biologic techniques are available that can help prioritize candidate mutated antigens for inclusion in tumor vaccines.
Mass Spectrometry
Great strides in the fields of mass spectrometry (MS) and associated computational algorithms have enabled the characterization of the MHC-displayed “ligandome” (12,13). This approach can be used to test if a mutated peptide (or a native tumor-associated peptide) is displayed by tumor cells. This information is important since the peptide-MHC complexes are the substrates recognized by T cell receptors (TCR). However, the approach is limited technically by insufficient amounts of tumor tissue and conceptually by the observation that few peptide-bound MHC targets are needed for an effective T cell response (14,15). As a result, many useful but less abundant targets on the cell may be bypassed in favor of those that are less potent but more highly represented.
Ex vivo T cell assays
Peripheral blood monocytes (PBMC) or tumor-infiltrating lymphocytes (TIL) can be tested in antigen-specific ex vivo assays to identify neoantigens that stimulate existing T-cell populations. This strategy would be expected to reveal the patient’s natural response to neoantigens (2,16). However, routine clinical application of ex vivo assays is costly and technically challenging given the number of neoantigen mutations (requiring rapid preparation of many stimulatory immunogens), the requirement of MHC-matched antigen presenting cells (APC) for some of these assays and the relative insensitivity of these techniques. Most importantly, using ex vivo assays as a filter for neoantigen selection limits the spectrum of T cell reactivity to existing T cell responses. In patients with clinically evident tumors, this would restrict the selected neoantigen repertoire to the existing and possibly ineffective T cell responses. It is currently unknown whether enhancement of an ongoing T cell response or generation of de novo responses is clinically relevant for an effective tumor vaccine. Other biological assays such as in vitro or in vivo immunization of a humanized mouse could also be considered but such assays are likewise technically challenging, costly and conceptually limited.
In silico prediction of peptide-MHC binding
Generation of an immune response to any mutated peptide sequence and recognition of tumor cells containing that peptide depend critically on the ability of the patient’s MHC molecules to effectively bind the mutated peptide and present it to a T cell. Advanced algorithms utilizing neural network-based learning approaches have been developed to capitalize on large amounts of data describing peptides that bind with different strengths to a wide variety of class I MHC molecules (17). These algorithms allow rapid in silico prediction of peptide-binding strength to patient-specific MHC alleles, and potentially enable a more rapid and less restrictive approach to filter the list of candidate neoantigens from sequencing data. Using results from the next-generation DNA sequencing, we have evaluated the binding for more than 100 different predicted peptides to understand the boundaries of the accuracy of prediction by these algorithms (18). In order to link this in silico analysis to potentially clinically and biologically relevant observations, we present here an analysis of 40 neoantigens previously identified as CD8+ T-cell targets in the literature.
The predicted binding characteristics of tumor neoepitopes recognized by T cells in patients with antitumor immunity
We have conducted an extensive search of the literature including recent reviews on neoantigens (1,2) from PUBMED, and the most comprehensive list of cancer vaccine antigens compiled by the Cancer Research Institute (19), identifying reports of spontaneous CD8+ T-cell responses in cancer patients in whom the target epitopes were discovered subsequently. To avoid bias of the results, reports of vaccinations with known epitopes or of selected searches for single T-cell epitopes (such as for an immune response to a known mutated oncogene) were not included. Multiple reports of spontaneous CD8+ T-cell epitopes were identified, and remarkably in each case following an unbiased search for the dominant T-cell epitope, the target epitope was a neoantigen. Two-thirds of the patients in these reports experienced significant partial or complete tumor regression or long-term stable disease, either spontaneously or following therapy.
As shown in Table 1, 31 of these 40 neoepitopes were identified in an unbiased manner based on cDNA expression cloning or MHC/peptide elution, while the remaining 9 were found based on genomic mutation and epitope-binding predictions. These neoantigens resulted from 35 missense mutations and 5 frame-shift mutations (that led to novel open reading frames, neoORFs) and are restricted by 11 different HLA alleles, representing both common and less common alleles as expected from sampling of the population at large. Approximately 80% of these are somatic mutations found exclusively in the tumors of individual patients. The remaining alterations are polymorphic loci within hematopoietically-restricted minor histocompatiblility antigens (miHAgs) identified following hematopoietic stem cell transplantation for blood malignancies. In almost every case, the mutated peptide was significantly (>100X) more potent than the cognate native peptide in the induction of T cell IFNγ production or cytotoxicity. These examples represent seven different cancer types (non-small cell lung cancer, melanoma, renal cell carcinoma, bladder cancer, B-cell acute lymphoblastic leukemia, multiple myeloma, chronic lymphocytic leukemia).
Table 1.
Biological features and predicted binding affinities of neoantigen-directed T-cell responses in humans
| Group | Gene Ref | Identification Approach* |
Favorable Clinical Response |
MUT>>>NAT T cell Response Detected (approach)§ |
HLA Allele |
Mutated Epitope (Native allele) |
Predicted Binding Affinity (IC50 nM)♦ |
|
|---|---|---|---|---|---|---|---|---|
| MUT | NAT | |||||||
| 1 | ECGF-1 32 (miHAg) | cDNA | Yes | Yes (γ) | B*07:02 | RPHAIRRPLAL(R) | 3 | 2 |
| ME-1 31 | cDNA | Yes | Yes (C) | A*02:01 | FLDEFMEGV(A) | 3 | 2 | |
| PLEKHM2 16 | WES | Yes | Yes (γ) | A*01:01 | LTDDRLFTCY(H) | 3 | 97 | |
| FNDC3B 18 | WES | Yes | (T) (≥10X) | A*02:01 | VVMSWAPPV(L) | 4 | 7 | |
| PRDX5 27 | cDNA | NR | Yes (C) | A*02:01 | LLLDDLLVSI(S) | 5 | 7 | |
| MATN2 16 | WES | Yes | Yes (γ) | A*11:01 | KTLTSVFQK(E) | 5 | 20 | |
| DDX21 33 | cDNA | Yes | Yes (C) | A*68:01 | EAFIQPITR(S) | 10 | 29 | |
| RBAF 29 | cDNA | Yes | Yes (C) | B*07:02 | RPHVPESAF(G) | 10 | 68 | |
| GAS7 16,34 | cDNA(and later WES) | Yes | Yes (C) | A*02:01 | SLADEAEVYL(H) | 12 | 39 | |
| ATR 35 | WES | Yes | (T) (≥ 10X) | A*03:01 | KLYEEPLLK(S) | 13 | 13 | |
| SIRT2 29 | cDNA | Yes | C (10X) | A*03:01 | KIFSEVTLK(P) | 14 | 16 | |
| EF2 36 | MS | NR | Yes (C) | A*68:02 | ETVSEQSNV(E) | 16 | 27 | |
| KIAA0223 (HA-1) 37 (miHAg) | MS | Yes(Severe GvHD) | NR | A*02:01 | VLHDDLLEA(R) | 17 | 140 | |
| GAPDH 34 | cDNA | Yes | Yes (C) | A*02:01 | GIVEGLITTV(M) | 21 | 27 | |
| BCL2A1 38 (miHAg)✱ | Linkage | NR | Yes (C) | A*24:02 | DYLQYVLQI(C) | 22 | 34 | |
| HSP 70 39 | cDNA | NR | Yes (C) | A*02:01 | SLFEGIDIYT(F) | 23 | 7 | |
| ACTININ 30 | cDNA | Yes | Yes (C) | A*02:01 | FIASNGVKLV(K) | 29 | 44 | |
| CDK12 16 | WES | Yes | Yes (γ) | A*11:01 | CILGKLFTK(E) | 33 | 42 | |
| KIAA1440 40 | cDNA | Yes | Yes (C) | A*01:01 | QTACEVLDY(T) | 33 | 78 | |
| HAUS3 16 | WES | Yes | Yes (γ) | A*02:01 | ILNAMIAKI(T) | 34 | 36 | |
| BCL2A1 38 (miHAg)* | Linkage | NR | Yes (C) | B*44:03 | KEFEDDIINW(G) | 36 | 27 | |
| PPP1R3B 16,28 | cDNA(and later WES) | Yes | Yes (γ) | A*01:01 | YTDFHCQYV(P) | 49 | 72 | |
| HB-1 41 (miHAg) | cDNA | NR | Yes (C) | B*44:03 | EEKRGSLHVW(Y) | 81 | 67 | |
| MUM-2 42 | cDNA | Yes | Yes (C) | B*44:02 | SELFRSGLDSY(R) | 184 | 182 | |
| KIAA0205 43 | cDNA | NR | Yes (C) | B*44:03 | AEPIDIQTW(N) | 258 | 288 | |
| GPNMB 29 | cDNA | Yes | Yes (C) | A*03:01 | TLDWLLQTPK(G) | 282 | 179 | |
| 2 | CSNK1A1 16 | WES | Yes | Yes (γ) | A*02:01 | GLFGDIYLAI(S) | 6 | 1312 |
| CLPP 44 | cDNA | Yes | Yes (C) | A*02:01 | ILDKVLVHL(P) | 32 | 7566 | |
| CTNNB1 45 | cDNA | Yes (?) | Yes (C) | A*24:02 | SYLDSGIHF(S) | 41 | 18746 | |
| SNRP116 29 | cDNA | Yes | Yes (C) | A*03:01 | KILDAVVAQK(E) | 48 | 14976 | |
| OS9 46 | cDNA | NR | Yes (C) | B*44:03 | KELEGILLL(P) | 60 | 1161 | |
| MYH2 47 | cDNA | Yes | Yes (C) | A*03:01 | KINKNPKYK(E) | 141 | 4960 | |
| 3 | MART-2 48 | cDNA | Yes (weak) | Yes (C) | A*01:01 | FLEGNEVGKTY(G) | 1115 | 4504 |
| NFYC 49 | cDNA | NR | Yes (C) | B*52:01 | AQQITKTEV(Q) | 7314 | 5701 | |
| CDK4 50 | cDNA | NR | Yes (C) | A*02:01 | ACDPHSGHFV(R) | 11192 | 25222 | |
| neoORF | pARF14-ORF3 51 | cDNA | Yes | Not Relevant | A*11:01 | AVCPWTWLR | 25 | Not Relevant |
| HMSD-ν 52 (miHAg) | cDNA | Yes | Not Relevant | B*44:03 | MEIFIEVFSHF | 36 | Not Relevant | |
| PANE-1 53 (miHAg) | MS | NR | Not Relevant | A*03:01 | RVWDLPGVLK | 44 | Not Relevant | |
| MUM1 54 | cDNA | Yes | Yes(C) | B*44:02 | (L)EEKLIVVLF▲ (S) | 434 (409) | Not Relevant | |
| P2X5 55 (miHAg) | Linkage | Yes | Not Relevant | B*07:02 | TPNQRQNVC | 1769 | Not Relevant | |
miHAg -minor Histocompatiblility Antigens
NR Not Reported
Bold - identification by binding predictions and either genomic sequence or genetic linkage; WES - whole-exome sequencing; cDNA - cDNA library expression cloning; MS - mass spectrometry.
C - cytotoxicity; γ -IFNγ-stimulation; T - CD107a stimulation.
Underlined values: comparable experimentally measured binding affinity of mutated and native peptides were reported
Two separate CTL clones specific for peptides restricted by distinct MHC alleles (A2402 and B4403) were identified. Each peptide spanned a distinct missense mutation in the BCL2A1 gene
Special case of a missense, partial neoORF
| Predicted Affinity Highlighting: | Green - Strong/moderate binder | IC50≤ 150 nM |
| Blue - Weak binder | IC50 between 150 and 500 nM | |
| Black - Non-binder | IC50 ≥ 500 nM |
Because these neoepitopes are associated with biological responses, they provide an ideal set of sequences for retrospective peptide affinity predictions to “reverse engineer” predictable characteristics of effective epitopes. For this analysis, we utilized the netMHCpanv2.4 algorithm (Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark, www.dtu.dk) (20). NetMHCpan is an artificial neural network trained algorithm with an extensive training data set (17), including 43 HLA-A and -B alleles, representing ~90% and ~60%, respectively, of the allelic population distribution, with more than 1000 members each in the training set. NetMHCpan was determined to be one of the most accurate predictive algorithms in a 2012 competition (21). We applied this algorithm to individually predict MHC binding for all possible tiled peptides containing the mutated or the corresponding unmutated residues of these observed spontaneously epitopes in order to determine:
whether the naturally recognized epitopes would have been predicted;
the predicted affinity of each mutated epitope;
the predicted affinity of each cognate native epitope (focusing on only the missense mutations).
Functional neoepitopes are correctly predicted by a class I MHC-peptide binding algorithm
Thirty-one of the epitopes shown in Table 1 were identified by ex vivo T-cell reactivity or mass spectrometry and did not utilize genomic sequence or binding prediction information as a component of their identification. For all but one of these 31, we found that the reported epitope was the peptide with the strongest predicted MHC-binding affinity among the tiled peptides containing the mutation. The only exception was a MUM1-derived 10mer containing an additional leucine at the N-terminus that had a slightly better predicted affinity (IC50 of 409 nM) than the observed 9mer (IC50 434 nM). We conclude that the MHC-peptide binding prediction algorithm netMHCpan consistently predicts the naturally recognized tumor neoepitope from all of the possible epitopes harboring a specific mutation.
Most functional neoepitopes have high to moderate predicted IC50
Twenty of 31 (65%) of the naturally recognized missense and neoORF epitopes had predicted IC50 < 50 nM (strong binders) and 3 of 31 (10%) had a predicted IC50 between 50 nM and 150 nM (moderate binders). Thus, 75% of the dominant T cell clones isolated from the naturally occurring T cell populations recognize an epitope with a strong or moderate predicted affinity (IC50 <150 nM) for the patient’s MHC allele. Since an unbiased functional assay (cytolysis, IFNγ production, or MS) was the critical test used to identify each of the stimulating peptides in these 31 examples, it is unlikely that there was an experimental bias towards the identification of epitopes with higher predicted affinity. Four of 31 naturally recognized peptides were predicted to be “weak” binding peptides (IC50 between 150 and 500 nM), indicating that a total of 27 of 31 (87%) of the naturally occurring epitopes would have been considered as binding peptides (IC50 < 500 nM affinity) using netMHCpan.
Conversely, only four of 31 naturally recognized peptides were predicted by netMHCpan to be non-binders (IC50>500nM); they may be false negatives from the prediction algorithm or may represent low affinity yet functional epitopes. Although these alternatives cannot be distinguished based on the available data, three observations are relevant. First, for the three epitopes arising from missense mutations (MART-2, NFYC, CDK4), cytolytic activity was preferentially induced by the mutated peptide and not by the native peptide at a range of peptide concentrations (1 – 10 nM) comparable to those observed with more strongly predicted binding peptides. Second, for the Arg → Cys CDK4 mutation, the highly oxidizable sulfur residue may contribute serendipitously to MHC binding as a “pseudo-” anchor residue that could not have been accounted for by the prediction algorithms. Finally, T cells recognizing the fourth epitope (the miHAg P2X5) represented as much as 1.6% of all circulating T cells following the therapeutic infusion of donor lymphocytes. Results from these 40 examples dataset suggest that there are limitations to the capability of predictive algorithms and that up to 15% of target T-cell epitopes may be missed by the prediction algorithms.
Most of the cognate native peptides are predicted to bind MHC equally to the mutated peptides
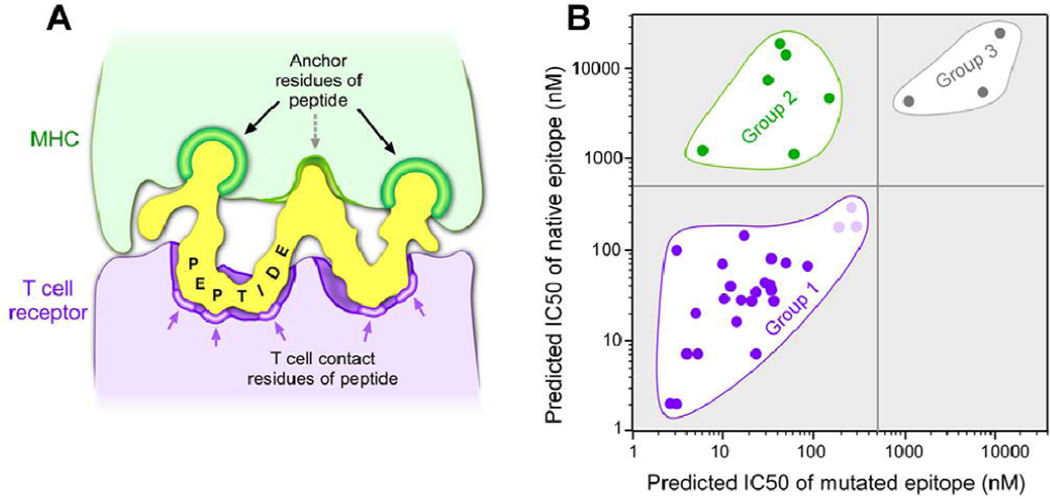
In addition to analyzing MHC binding to the mutated epitopes we also compared the predicted affinities of the cognate native epitopes corresponding to all 35 missense epitopes in Table 1 and identified 3 distinct classes. The predominant class (26 of 35 or 74%; Group 1) was composed of native/mutated pairs that were predicted to bind with comparable affinity (with 23 of 26 showing strong to moderate predicted binding [IC50 <150 nM] and the remaining 3 showing weak binding [IC50 between 150 and 500 nM]). Despite comparable predicted binding, in almost all cases the mutated peptide had been found to be significantly more potent in stimulating T cells than the native peptide. A smaller group (6 of 35 or 17%; Group 2) showed low predicted binding for the native epitope and strong binding for the mutated epitope, directly correlating with the differential T-cell responses to the mutated and native peptides. Finally, in a minority of cases both mutated and native peptides were predicted to be non-binding (3 of 35 or 9%; Group 3). We note that each Group in Table 1 comprises multiple HLA with no apparent bias in representation. While the existence of the Group 1 and Group 2 epitopes is not surprising, the predominance of the Group 1 epitopes (containing 74% of all missense epitopes and 4 times more abundant than Group 2) with comparable affinities for both the mutated and native peptides is unexpected.
DISCUSSION
The MHC-bound peptide can be considered as a double-sided “key”, which must fit both the MHC and the TCR “locks” in order to stimulate an immune response and for subsequent target-cell cytolysis (Figure 1A). Sequence-specific binding of peptides to the MHC molecule is highly dependent on the interactions of the peptide side chains at particular positions (“anchors”) along the length of the peptide with chemical moieties defined by the polymorphic residues that constitute the MHC binding pocket (22–24); hence, predictive calculations are sequence-dependent (25). Furthermore, analysis of these critical MHC-binding positions and residues over a wide range of MHC alleles shows that only a few positions of the peptide are anchor positions and only a few amino acids at the anchor positions of the peptide contribute to binding in a positive manner (26). On the other side of the “key”, TCR recognition of the peptide/MHC complex gains specificity from the ordered presentation of the other face of the peptide conferred by the anchoring residues.
Figure 1.
(A) The two faces of a bound peptide to the MHC and TCR molecules form a “double-sided key” that must be present in order to stimulate an antigen-specific immune response. Green--Anchor residues in the peptide that interact with MHC. Purple--Regions of the peptide that interact with the TCR surface. (B) A scatter plot of the predicted affinities of epitopes that stimulate detectable neoantigen T-cell responses, shown in Table 1. Group 1 epitopes demonstrate comparable predicted affinities of native and mutated peptides and were determined to have mutations in regions of the peptide critical for interactions with the TCR (dark purple – strong/moderate binders; light purple – weak binders). Group 2 epitopes (green) are mutated peptides with strong/moderate predicted affinity whose corresponding native peptides are not predicted to bind MHC, and were found to have mutations in the peptide residues critical for the interaction with MHC. Group 3 epitopes (grey) represent peptides where neither the native or mutated peptide are predicted to be HLA-binding peptides and may be either false negatives of the prediction algorithm or very low affinity functional epitopes.
For the majority of the missense mutations, both the native and the mutated peptides were predicted to be binding peptides (Group 1, Figure 1B). This observation is almost certainly a consequence of the mutations affecting the region of the peptide “key” that is involved in TCR recognition. In all but 2 of these 26 examples, the mutation was in a non-anchor position (as identified by the online tool provided at www.sypeithi.de ; Ref. # 26). In the two non-conforming examples (PLEKHM2 and KIAA1440), a second anchor residue was already present in the native peptide. Other investigators have also reported mutations with equivalent affinity predictions for the native and the mutated peptides pairs (16,27). Our broader analysis suggests that such mutant epitopes are a common phenomenon. Only a minority of the missense mutations was found in Group 2 characterized by non-binding of the native peptide. All of the Group 2 examples except for MYOSIN were mutations to preferred anchor residues at critical anchor positions.
Although the majority of the naturally occurring tumor epitopes were derived from the corresponding native peptides predicted to bind MHC, the vast majority (>98%) of the native human peptidome is not predicted to contain peptides that are binding epitopes of human MHC (our unpublished analysis using netMHCpan). Random mutational events that convert a non-binding peptide (the vastly predominant target) to a binding peptide (Group 2) are expected to be rare because they require mutation to one of only a few specific amino acids at a small number of anchor positions. For most MHC molecules, there are only one or two important anchor positions and usually only 2 or 3 amino acids at those positions promote binding. Conversely, non-anchor positions are 3–4 times more abundant than anchor positions, and most mutations to native binding epitopes in these non-anchor positions would maintain MHC binding (Group 1). This simple probabilistic explanation may be sufficient to account for the predominance of the observed Group 1 epitopes (derived from the vastly under-represented class of native peptides that are predicted to bind MHC). Alternatively, more complex explanations may be required. For example, aspects of central immune tolerance that are currently not well understood may cause the extant TCR repertoire to more effectively respond to peptides presenting a surface chemically distinct from any native peptide (Group 1) than to peptides which more efficiently present an otherwise native peptide surface (Group 2).
We did not observe weak native peptide binders that converted to strong/moderate mutated binders or strong/moderate native peptide binders that converted to weak mutated peptide binders. While this may reflect the relatively limited dataset we used, it could also be that such upgrading or downgrading of binding involves anchor residue changes that moderately increase/decrease binding affinity but retain similar chemical structure of the non-anchor residues available for TCR recognition. In these scenarios, because the native peptides could bind MHC, central immune tolerance may have effectively deleted cells with the reactive TCR, rendering both native and mutated peptides non-immunogenic.
From the perspective of efficacy, we propose that mutations resulting in either Group 1 or Group 2 binders should be considered as acceptable for use as immunogens as both types of mutated neoepitopes have been found in vivo in cancer patients with spontaneous tumor regressions and in long-term cancer survivors. Notably, in long-term survivors, T cells specific to mutated tumor epitopes from both Group 1 and Group 2 have been found to persist over many years (28,29).
From the perspective of safety, there have been no reports of immune-mediated toxicities (except for the expected occurrence of graft vs host disease (GVHD) as a result of responses against miHAgs) despite the observation that for most of the mutations, the cognate native peptide was predicted, and experimentally demonstrated in some cases (18,27,30,31), to bind MHC as well as the mutated peptide (Group 1). Importantly, in almost all cases the mutant peptide was shown to be more potent than the native peptide in stimulating T cell cytotoxicity or IFNγ production. The absence of autoimmune toxicity in these patients fits with the model that T cells reactive to the native epitope were eliminated by central immune tolerance and that T cells reactive to the mutated epitope do not cross-react to the native epitope as the mutation exclusively affects the TCR binding region.
In conclusion, in silico peptide-binding predictions provide a useful and rapidly deployable tool to capture the types of immunogens that are naturally observed in cancer patients, many of whom experienced tumor regression and sometimes long-term tumor control. Moreover, results from our retrospective prediction study reveal features of these epitopes to further guide inclusion as immunogens in vaccines. We have recently initiated a clinical study employing personalized neoantigen epitopes identified by whole-exome sequencing and prioritized by MHC-binding predictions in which we will carefully monitor the immune response to each mutation (NCT01970358).
Acknowledgements
We thank Sachet Shukla for helpful and insightful discussions. We acknowledge the generous support of the Blavatnik Family Foundation and the NIH (NHLBI:5 R01 HL103532-03; NCI:1R01CA155010-02) for our work on neoepitope-based vaccines.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Authors' Contributions
Conception and design: E.F. Fritsch, N. Hacohen, C.J. Wu
Development of methodology: E. F. Fritsch, N. Hacohen, C.J. Wu, M. Rajasagi
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): N. Hacohen, C.J. Wu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): E.F. Fritsch, N. Hacohen, C.J. Wu, M. Rajasagi,V.A. Brusic
Writing, review, and/or revision of the manuscript: E.F. Fritsch, C.J. Wu, N. Hacohen, M Rajasagi, P. A. Ott, V. Brusic
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): N. Hacohen, C.J. Wu
Study supervision: N. Hacohen, C.J. Wu
REFERENCES
- 1.Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu C. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol Res. 2013;1:11–15. doi: 10.1158/2326-6066.CIR-13-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. EMBO J. 2013;32:194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castle JC, Kreiter S, Diekmann J, Lower M, van der Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berd D, Murphy G, Maguire HC, Jr, Mastrangelo MJ. Immunization with haptenized, autologous tumor cells induces inflammation of human melanoma metastases. Cancer Res. 1991;51:2731–2734. [PubMed] [Google Scholar]
- 6.Soiffer R, Hodi FS, Haluska F, Jung K, Gillessen S, Singer S, et al. Vaccination with irradiated, autologous melanoma cells engineered to secrete granulocyte-macrophage colony-stimulating factor by adenoviral-mediated gene transfer augments antitumor immunity in patients with metastatic melanoma. J Clin Oncol. 2003;21:3343–3350. doi: 10.1200/JCO.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Randazzo M, Terness P, Opelz G, Kleist C. Active-specific immunotherapy of human cancers with the heat shock protein Gp96-revisited. Int J Cancer. 2012;130:2219–2231. doi: 10.1002/ijc.27332. [DOI] [PubMed] [Google Scholar]
- 8.Senzer NN, Kaufman HL, Amatruda T, Nemunaitis M, Reid T, Daniels S, et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J Clin Oncol. 2009;27:5763–5771. doi: 10.1200/JCO.2009.24.3675. [DOI] [PubMed] [Google Scholar]
- 9.Morton DL, Foshag LJ, Hoon DS, Nizze JA, Famatiga E, Wanek LA, et al. Prolongation of survival in metastatic melanoma after active specific immunotherapy with a new polyvalent melanoma vaccine. Ann Surg. 1992;216:463–482. doi: 10.1097/00000658-199210000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaishampayan U, Abrams J, Darrah D, Jones V, Mitchell MS. Active immunotherapy of metastatic melanoma with allogeneic melanoma lysates and interferon alpha. Clin Cancer Res. 2002;8:3696–3701. [PubMed] [Google Scholar]
- 11.Bystryn JC, Zeleniuch-Jacquotte A, Oratz R, Shapiro RL, Harris MN, Roses DF. Double-blind trial of a polyvalent, shed-antigen, melanoma vaccine. Clin Cancer Res. 2001;7:1882–1887. [PubMed] [Google Scholar]
- 12.Rammensee HG, Singh-Jasuja H. HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev Vaccines. 2013;12:1211–1217. doi: 10.1586/14760584.2013.836911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 14.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 15.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 16.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods. 2011;374:26–34. doi: 10.1016/j.jim.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajasagi M, Keskin D, Kim J, Zhang W, Shukla S, DeLuca D, et al. Systematic Identification of Personal Mutated Tumor-Specific Neoantigens in CLL. Abstract number 954 for the 54th ASH Annual Meeting and Exposition; December 8–11, 2012; ( https://ash.confex.com/ash/2012/webprogram/Paper51216.html) [Google Scholar]
- 19.van der Bruggen P, Strroobant V, Vigneron N, Van den Eynde BJ. Peptide database: T cell-defined tumor antigens resulting from mutations. http://cancerimmunity.org/peptide/mutations/
- 20.Center for Biological Sequence Analysis. Lyngby, Denmark: Technical University of Denmark; www.dtu.dk. [Google Scholar]
- 21.Zhang GL, Ansari HR, Bradley P, Cawley GC, Hertz H, Hu X, et al. Machine learning competition in immunology - Prediction of HLA class I binding peptides. J Immunol Methods. 2011;374:1–4. doi: 10.1016/j.jim.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Yanover C, Bradley P. Large-scale characterization of peptide-MHC binding landscapes with structural simulations. Proc Natl Acad Sci U S A. 2011;108:6981–6986. doi: 10.1073/pnas.1018165108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 24.Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 25.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. www.sypeithi.de. [DOI] [PubMed] [Google Scholar]
- 27.Sensi M, Nicolini G, Zanon M, Colombo C, Molla A, Bersani I, et al. Immunogenicity without immunoselection: a mutant but functional antioxidant enzyme retained in a human metastatic melanoma and targeted by CD8(+) T cells with a memory phenotype. Cancer Res. 2005;65:632–640. [PubMed] [Google Scholar]
- 28.Lu YC, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, et al. Mutated PPP1R3B Is Recognized by T Cells Used To Treat a Melanoma Patient Who Experienced a Durable Complete Tumor Regression. J Immunol. 2013;190:6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Echchakir H, Mami-Chouaib F, Vergnon I, Baurain JF, Karanikas V, Chousib S, Coulie PG. A point mutation in the alpha-actinin-4 gene generates an antigenic peptide recognized by autologous cytolytic T lymphocytes on a human lung carcinoma. Cancer Res. 2001;61:4078–4083. [PubMed] [Google Scholar]
- 31.Karanikas V, Colau D, Baurain JF, Chiari R, Thonnard J, Gutierrez-Roelens I, et al. High frequency of cytolytic T lymphocytes directed against a tumor-specific mutated antigen detectable with HLA tetramers in the blood of a lung carcinoma patient with long survival. Cancer Res. 2001;61:3718–3724. [PubMed] [Google Scholar]
- 32.Slager EH, Honders MW, van der Meijden ED, van Luxemburg-Heija SA, Kloosterboer FM, et al. Identification of the angiogenic endothelial-cell growth factor-1/thymidine phosphorylase as a potential target for immunotherapy of cancer. Blood. 2006;107:4954–4960. doi: 10.1182/blood-2005-09-3883. [DOI] [PubMed] [Google Scholar]
- 33.Baurain JF, Colau D, van Baren N, Landry C, Martelange V, Vikkula M, et al. High frequency of autologous anti-melanoma CTL directed against an antigen generated by a point mutation in a new helicase gene. J Immunol. 2000;164:6057–6066. doi: 10.4049/jimmunol.164.11.6057. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Dudley ME, Rosenberg SA, Robbins PF. Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy. J Immunother. 2005;28:53–62. doi: 10.1097/00002371-200501000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor Exome Analysis Reveals Neoantigen-Specific T-Cell Reactivity in an Ipilimumab-Responsive Melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogan KT, Eisinger DP, Cupp SB, 3rd, Lekstrom KJ, Deacon DD, Shabanowitz J, et al. The peptide recognized by HLA-A68.2-restricted, squamous cell carcinoma of the lung-specific cytotoxic T lymphocytes is derived from a mutated elongation factor 2 gene. Cancer Res. 1998;58:5144–5150. [PubMed] [Google Scholar]
- 37.den Haan JM, Meadows LM, Wang W, Pool J, Blokland E, Bishop TL, et al. The minor histocompatibility antigen HA-1: a diallelic gene with a single amino acid polymorphism. Science. 1998;279:1054–1057. doi: 10.1126/science.279.5353.1054. [DOI] [PubMed] [Google Scholar]
- 38.Akatsuka Y, Nishida T, Kondo E, Miyazaki M, Taji H, Lida H, et al. Identification of a polymorphic gene, BCL2A1, encoding two novel hematopoietic lineage-specific minor histocompatibility antigens. J Exp Med. 2003;197:1489–1500. doi: 10.1084/jem.20021925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaudin C, Kremer F, Angevin E, Scott V, Triebel F. A hsp70-2 mutation recognized by CTL on a human renal cell carcinoma. J Immunol. 1999;162:1730–1738. [PubMed] [Google Scholar]
- 40.Zhou X, Jun DY, Thomas AM, Huang X, Huang LQ, Mautner J, et al. Diverse CD8+ T-cell responses to renal cell carcinoma antigens in patients treated with an autologous granulocyte-macrophage colony-stimulating factor gene-transduced renal tumor cell vaccine. Cancer Res. 2005;65:1079–1088. [PubMed] [Google Scholar]
- 41.Dolstra H, Fredrix H, Maas F, Coulie PG, Brasseur F, Mensink E, et al. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J Exp Med. 1999;189:301–308. doi: 10.1084/jem.189.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiari R, Foury F, De Plaen E, Baurain JF, Thonnard J, Coulie PG. Two antigens recognized by autologous cytolytic T lymphocytes on a melanoma result from a single point mutation in an essential housekeeping gene. Cancer Res. 1999;59:5785–5792. [PubMed] [Google Scholar]
- 43.Gueguen M, Patard JJ, Gaugler B, Brasseur F, Renauld JC, Van Cangh PJ, et al. An antigen recognized by autologous CTLs on a human bladder carcinoma. J Immunol. 1998;160:6188–6194. [PubMed] [Google Scholar]
- 44.Corbiere V, Chapiro J, Stroobant V, Ma W, Lurguin C, Lethe B, et al. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer Res. 2011;71:1253–1262. doi: 10.1158/0008-5472.CAN-10-2693. [DOI] [PubMed] [Google Scholar]
- 45.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, Rosenberg SA. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigneron N, Ooms A, Morel S, Degiovanni G, Van Den Eynde BJ. Identification of a new peptide recognized by autologous cytolytic T lymphocytes on a human melanoma. Cancer Immun. 2002;2:9. [PubMed] [Google Scholar]
- 47.Zorn E, Hercend T. A natural cytotoxic T cell response in a spontaneously regressing human melanoma targets a neoantigen resulting from a somatic point mutation. Eur J Immunol. 1999;29:592–601. doi: 10.1002/(SICI)1521-4141(199902)29:02<592::AID-IMMU592>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 48.Kawakami Y, Wang X, Shofuda T, Sumimoto H, Tupesis J, Fitzgerald E, Rosenberg S. Isolation of a new melanoma antigen, MART-2, containing a mutated epitope recognized by autologous tumor-infiltrating T lymphocytes. J Immunol. 2001;166:2871–2877. doi: 10.4049/jimmunol.166.4.2871. [DOI] [PubMed] [Google Scholar]
- 49.Takenoyama M, Baurain JF, Yasuda M, So T, Sugaya M, Hanagiri T, et al. A point mutation in the NFYC gene generates an antigenic peptide recognized by autologous cytolytic T lymphocytes on a human squamous cell lung carcinoma. Int J Cancer. 2006;118:1992–1997. doi: 10.1002/ijc.21594. [DOI] [PubMed] [Google Scholar]
- 50.Wölfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 51.Huang J, El-Gamil M, Dudley ME, Li YF, Rosenberg SA, Robbins PF. T cells associated with tumor regression recognize frameshifted products of the CDKN2A tumor suppressor gene locus and a mutated HLA class I gene product. J Immunol. 2004;172:6057–6064. doi: 10.4049/jimmunol.172.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawase T, Akatsuka Y, Torikai H, Morishima S, Oka A, Tsujimura A, et al. Alternative splicing due to an intronic SNP in HMSD generates a novel minor histocompatibility antigen. Blood. 2007;110:1055–1063. doi: 10.1182/blood-2007-02-075911. [DOI] [PubMed] [Google Scholar]
- 53.Brickner AG, Evans AM, Mito JK, Xuereb SM, Feng X, Nishida T, et al. The PANE1 gene encodes a novel human minor histocompatibility antigen that is selectively expressed in B-lymphoid cells and B-CLL. Blood. 2006;107:3779–3786. doi: 10.1182/blood-2005-08-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coulie PG, Lehmann F, Lethe B, Herman J, Luiguin C, Andrawiss M, Boon T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci U S A. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Rijke B, van Horssen-Zoetbrood A, Beekman JM, Otterud B, Maas F, Woestenenk R, et al. A frameshift polymorphism in P2X5 elicits an allogeneic cytotoxic T lymphocyte response associated with remission of chronic myeloid leukemia. J Clin Invest. 2005;115:3506–3516. doi: 10.1172/JCI24832. [DOI] [PMC free article] [PubMed] [Google Scholar]