Summary
Microbial exposure following birth profoundly impacts mammalian immune system development. Microbiota alterations are associated with increased incidence of allergic and autoimmune disorders with elevated serum IgE as a hallmark. The previously reported abnormally high serum IgE levels in germ-free mice suggests that immunoregulatory signals from microbiota are required to control basal IgE levels. We report that germ-free mice and those with low-diversity microbiota develop elevated serum IgE levels in early life. B cells in neonatal germ-free mice undergo isotype switching to IgE at mucosal sites in a CD4 T-cell- and IL-4-dependent manner. A critical level of microbial diversity following birth is required in order to inhibit IgE induction. Elevated IgE levels in germ-free mice lead to increased mast-cell-surface-bound IgE and exaggerated oral-induced systemic anaphylaxis. Thus, appropriate intestinal microbial stimuli during early life are critical for inducing an immunoregulatory network that protects from induction of IgE at mucosal sites.
Graphical Abstract
Highlights
-
•
Germ-free and mice with low-diversity microbiota develop high serum IgE levels
-
•
B cells in germ-free mice undergo IgE class switch recombination at mucosal sites
-
•
A diverse microbiota early in life is required to inhibit IgE induction
-
•
Hyper IgE in germ-free mice leads to exaggerated oral-induced systemic anaphylaxis
Introduction
After birth, body surfaces transit from complete sterility to the densest microbial ecosystem known on Earth. The lower intestine alone is home to nearly 100 trillion microbes, forming a highly complex microbiota with over 1,000 operational taxonomic units (Hooper et al., 2012). Each individual harbors an idiosyncratic microbial consortia (Turnbaugh et al., 2010) that is thought to be shaped by host immunity, environment, and diet (De Filippo et al., 2010; Elinav et al., 2011; Faith et al., 2011; Garrett et al., 2007; Muegge et al., 2011). A sophisticated microbial-host crosstalk shapes immune adaptation and bacterial communities, which underlies mutualism. Intestinal microbes shape not only the neighboring intestinal epithelial cells (Cash et al., 2006; Chassin et al., 2010) but also the majority of sterile body compartments at mucosal and systemic sites (Cahenzli et al., 2013; Ganal et al., 2012; Hooper et al., 2012). The pervasive effect of commensal microbes is reflected by its contribution to health and disease, including inflammatory bowel disease, obesity, malnutrition, autoimmunity, and allergic asthma (Herbst et al., 2011; Markle et al., 2013; Olszak et al., 2012; Smith et al., 2013; Turnbaugh et al., 2009).
Commensal microbial communities are thought to be a causal link between westernization and increasing immune disorders (Noverr and Huffnagle, 2005; Okada et al., 2010). Over the past few decades, westernized countries have experienced drastic changes in dietary habits and sanitation, including water decontamination, food pasteurization, sterilization, and uninterrupted cold chain delivery, vaccination, and widespread antibiotic use. All these factors have contributed to decreased infectious diseases (Bach, 2002). Over this period of improved hygiene, autoimmune and atopic allergic disorders have increased in incidence; an epidemiologic effect postulated to result from decreased acute infections during early childhood (Holt, 1998; Martinez and Holt, 1999; Strachan, 1989) or from a shift in commensal microbial communities (Braun-Fahrländer et al., 1999; Noverr and Huffnagle, 2005; Okada et al., 2010). Certainly, the western lifestyle shapes the composition of commensal bacterial consortia and their development over time (De Filippo et al., 2010; Koenig et al., 2011; Palmer et al., 2007; Walter and Ley, 2011). Although some mechanistic links between microbiota-induced immune regulation have been experimentally elucidated (Markle et al., 2013; Olszak et al., 2012), most models depend on particular microbes triggering disease (Elinav et al., 2011; Garrett et al., 2007).
IgE antibodies play a central role in atopic allergic disease and immunity to parasites (Allen and Maizels, 2011; Gould and Sutton, 2008; Paul and Zhu, 2010). Healthy individuals maintain serum IgE concentrations at basal levels (<0.0001% of serum immunoglobulins) (Sutton and Gould, 1993) because of an immunoregulatory network that regulates isotype switch to IgE. Multiple immunodeficiencies, including Wiskott-Aldrich syndrome, Omenn syndrome, or immunodysregulation polyendocrinopathy enteropathy X-linked syndrome, are correlated with elevated serum IgE levels despite the absence of allergic reaction or parasite infection (Kotlarz et al., 2013; Liston et al., 2008; Ozcan et al., 2008). Mouse models of immunodeficiencies also phenocopy the high IgE levels (Antón et al., 2002; Fontenot et al., 2003; Giblin et al., 2009), and we have previously reported that CD4−/−, major histocompatibility complex class II (MHC II)−/−, and athymic nude mice display high IgE (McCoy et al., 2006). It has been postulated that elevated serum IgE in the absence of atopic allergic disease or parasitic infection is a biomarker for immunodeficiencies (Liston et al., 2008).
Abnormally high serum IgE levels in germ-free mice have been reported previously (Herbst et al., 2011; Hill et al., 2012; McCoy et al., 2006). This suggests that immunoregulatory signals stemming from the microbiota are required in order to maintain IgE levels at basal levels even in genetically immunocompetent mice. We hypothesized that the proper induction of immune regulation requires adequate microbial exposure during early life. Here, we show that only exposure to a diverse microbial community during early life is able to induce functional immune regulation that maintains IgE at basal levels and decreases disease severity in a model of antigen-induced oral anaphylaxis.
Results
Absence of Microbial Colonization Leads to Elevated Serum IgE Levels
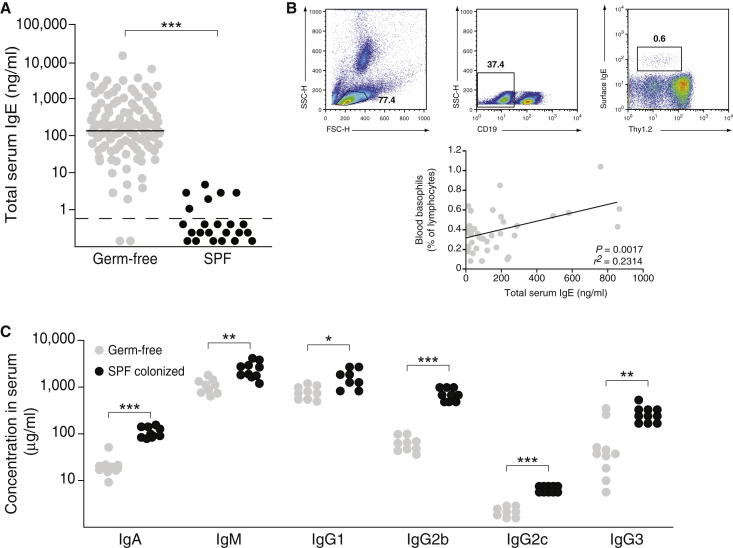
To investigate the role of the microbiota in the inhibition of IgE production, we first confirmed the presence of high IgE levels in a large cohort of germ-free C57BL/6 mice (Figure 1A). As expected, C57BL/6 mice born and raised with a specific pathogen-free (SPF) microbiota maintained IgE levels below the limit of detection (Figure 1A). High IgE levels were also observed in germ-free BALB/c mice (mean 292 ng/ml; range 15–2,393 ng/ml; n = 19), but not in germ-free Swiss Webster (n = 33) or NMRI (n = 21) mice (<0.8 ng/ml), possibly because of compensatory mechanisms frequently associated with outbred strains. As reported previously (Hill et al., 2012), we confirmed that elevated IgE levels correlated with an increased frequency of FSCloSSCloCD19−Thy1.2dull blood basophils (Lantz et al., 1997; Voehringer et al., 2004) with surface-bound IgE (Figure 1B). High IgE levels did not reflect a general hyperimmunoglobulinemia, given that all isotypes except IgE (IgA, IgM, IgG1, IgG2b, IgG2c, and IgG3) were reduced in germ-free and not SPF mice (Figure 1C), supporting the view of an immature immune system in germ-free mice (Macpherson and Harris, 2004). From these initial observations, we hypothesized that a diversified microbiota is required in order to maintain serum IgE levels at baseline.
Figure 1.
Absence of Microbial Colonization Leads to Elevated Serum IgE Levels
(A) Total serum IgE levels were measured in >4-week-old germ-free (n = 102; gray circles) and SPF (n = 24; black circles) mice. Each point represents an individual mouse, and the horizontal dotted line marks the lower limit of detection of the IgE ELISA assay (0.8 ng/ml).
(B) The gating strategy for identifying IgE+ basophils is shown for one representative germ-free mouse (top). Linear regression analysis of the frequency of FSCloSSCloCD19−Thy1.2dullIgE+ blood basophils and total serum IgE levels (n = 40; bottom). r2 = 0.2314; p = 0.0017.
(C) Total serum IgA, IgM, IgG1, IgG2b, IgG2c, and IgG3 were measured in germ-free (n = 8–10; gray circles) and SPF (n = 8–10; black circles) mice. Each dot represents an individual mouse.
∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05 by an unpaired Student’s t test.
Elevated Serum IgE Are Inherent to the Germ-free Status and Appear Early in Life
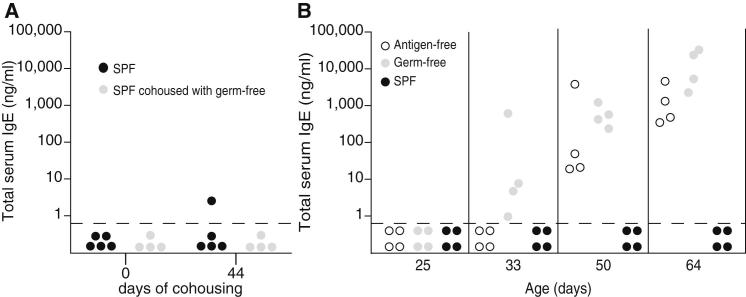
Although culture-dependent and -independent quality control methods were employed rigorously in order to confirm the absence of microbes, helminths, or viruses, we still faced the remote possibility that an undetected organism might trigger IgE induction. Therefore, we either cohoused SPF mice with undetectable serum IgE levels with germ-free mice (with high IgE) or left them with their SPF littermates. Serum IgE levels remained below the limit of detection in both groups of SPF mice, confirming that hygiene-mediated IgE was not due to the presence of a horizontally transferrable causing agent (Figure 2A). Next, we followed the kinetics of serum IgE levels longitudinally. IgE levels in SPF mice remained below the limit of detection at all ages (Figure 2B). In contrast, IgE levels in germ-free mice became readily detectable at around day 30 of age and increased onward (Figure 2B). Because the appearance of serum IgE coincided with the transition from lactation to solid food, we addressed the possibility of whether allergens in the sterilized chow might provoke increased serum IgE levels. SPF mice fed the same autoclaved chow did not develop increased IgE, and germ-free mice fed and raised on an antigen-free elemental diet also developed high IgE levels (Figure 2B). Therefore, and in agreement with the polyclonal and germline-encoded germ-free IgE (McCoy et al., 2006), a heightened allergic reaction to dietary antigens in the absence of intestinal microbes was not the cause of elevated IgE. Collectively, these observations indicated that, in the absence of commensal microbes, the immune regulatory pathways that normally maintain IgE at baseline levels were disrupted. Hyper-IgE is generated in genetically immunocompetent germ- and antigen-free mice, indicating that class-switch recombination to IgE reflects immune dysregulation in the absence of commensal microbiota.
Figure 2.
IgE Levels Become Elevated Early in Life and Are Not Triggered by Food Antigens
(A) Total serum IgE levels were determined in SPF mice (n = 4) cohoused with germ-free mice with high IgE levels (n = 4; gray circles) or left with SPF littermates (n = 5; black circles). IgE levels were measured before (day 0) and after cohousing (day 44).
(B) Serum IgE was determined longitudinally from 25–64 days of age in germ-free mice fed a conventional autoclaved chow diet (n = 4; gray circles) or an antigen-free elemental diet (n = 4; white circles) or in SPF mice (n = 4; black circles).
(A and B) Each point represents an individual mouse, and the horizontal dotted line marks the lower limit of detection of the IgE ELISA assay (0.8 ng/ml). Data for germ-free and SPF animals is representative of at least three independent experiments, and data for antigen-free mice are from one experiment.
B Cells Undergo Isotype Switch to IgE at Mucosal Sites
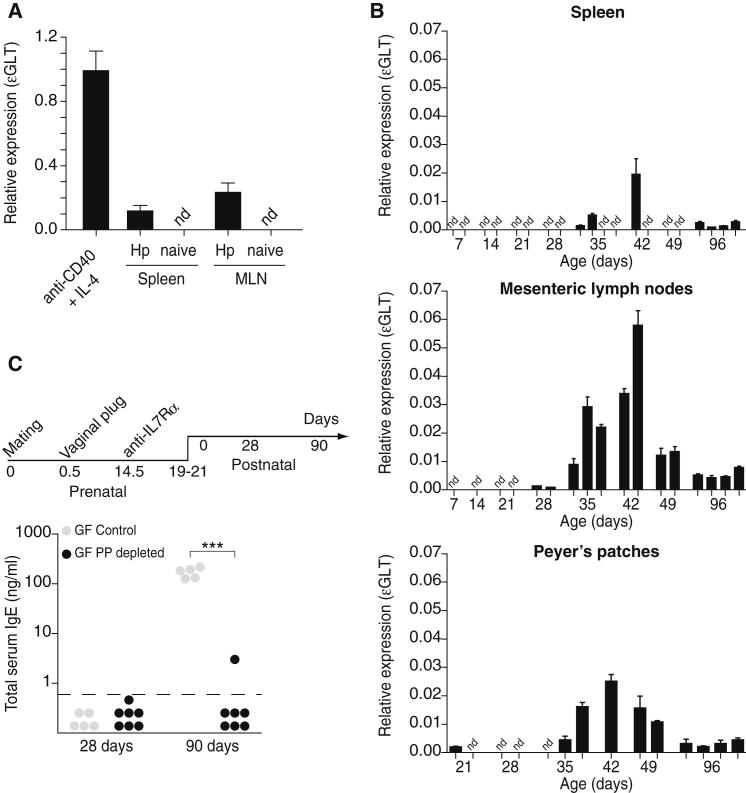
Next, we determined which lymphoid tissues foster IgE isotype switch in the absence of microbial exposure. Isotype switch is a DNA deletion-recombination process that exchanges the constant region exon of the immunoglobulin heavy chain with one of several sets of downstream exons, Cε in the case of isotype switch to IgE. Prior to productive class-switch recombination to Cε, transient transcription is initiated from an intronic (I) promoter (Iε), resulting in the generation of sterile ε germ-line transcripts (εGLTs) that do not encode for protein (Bacharier and Geha, 2000; Geha et al., 2003). Therefore, the expression of εGLTs is a hallmark of active isotype switch to IgE. Sterile εGLTs were readily detected via quantitative PCR (qPCR) in splenic B cells stimulated with anti-CD40 and IL-4 and B cells from the spleen and mesenteric lymph nodes (MLNs) of mice infected with the helminth Heligmosomoides polygyrus but not in splenic B cells isolated from naive SPF mice (Figure 3A). Then, we assessed the expression of εGLTs in germ-free mice during neonatal and adult life. Whereas low levels of εGLTs were found in the spleen of a few of the germ-free animals, εGLTs were readily detectable in the Peyer’s patches (PPs) and MLNs of all germ-free mice starting at day 35, peaking at day 42, and remaining detectable throughout adulthood (Figure 3B). The expression of εGLTs was not detected in the bone marrow, peripheral lymph nodes, or peritoneal cavity, the latter suggesting that peritoneal B-1 cells did not contribute significantly to high IgE levels. Therefore, in the absence of microbial exposure, IgE isotype switch is fostered at mucosal sites, where microbially triggered immune adaptations, such as IgA isotype switch, normally take place.
Figure 3.
Isotype Switch to IgE Is Initiated in Mucosal Lymphoid Tissues Early in Life
(A) Normalized expression (to gapdh) of εGLTs from B cells activated in vitro with IL-4 and anti-CD40 (anti-CD40 + IL-4) and ex vivo spleen and MLNs of naive or Heligmosomoides polygyrus (Hp)-infected SPF mice. Each bar represents the mean ± SD of three technical repeats. The MLN and spleen samples were pooled from 3–4 mice.
(B) Normalized expression (to gapdh) of εGLTs in spleen, MLNs, and PPs of germ-free mice at the indicated ages. Each bar represents an individual biological sample (one to two pooled mice; n = 1–4 biological samples per time point) ± SD of technical triplicates. nd, not detected. εGLTs was not detected in bone marrow, peripheral lymph nodes, and peritoneal cavity.
(C) PP ontogeny was inhibited in utero by intravenous administration of anti-iL-7Rα mAb in 14.5-day pregnant germ-free dams as depicted (top). Total serum IgE levels were measured in the progeny of untreated (n = 5–6; gray circles) and anti-iL-7Rα-treated (n = 7; black circles) dams. Each point represents an individual mouse, and the horizontal dotted line delineates the lower limit of detection of the IgE ELISA assay (0.8 ng/ml). Data are pooled from two independent experiments.
Given that the efferent lymphatics within the PPs drain into the MLNs (Macpherson and Smith, 2006), we tested whether IgE induction could be abrogated in mice lacking PPs. After the inhibition of PP ontogeny via IL-7R signaling blockade at embryonic stage E14.5 (Yoshida et al., 1999), germ-free IgE induction was fully abrogated (Figure 3C). These results suggest that PPs foster initial IgE isotype switch of B cells, which then rapidly egress to the MLNs.
Elevated IgE Levels in Germ-free Mice Are Dependent on CD4+ T Cells
Although classic IgE isotype switch requires cognate CD4+ T cell help coupled with IL-4 signals (Finkelman et al., 1988), numerous in vitro and in vivo experiments demonstrate alternative pathways for IgE isotype switch that can bypass some of these requirements. We have previously shown that cognate T cell help can be circumvented in immunodeficient CD4−/−, MHC II−/−, and athymic nude SPF mice (McCoy et al., 2006). However, this study addressed IgE generation in genetically immunodeficient hosts harboring a diverse microbiota with the possibility that dysbiosis, as a consequence of immunodeficiency, was the cause for the increased IgE levels.
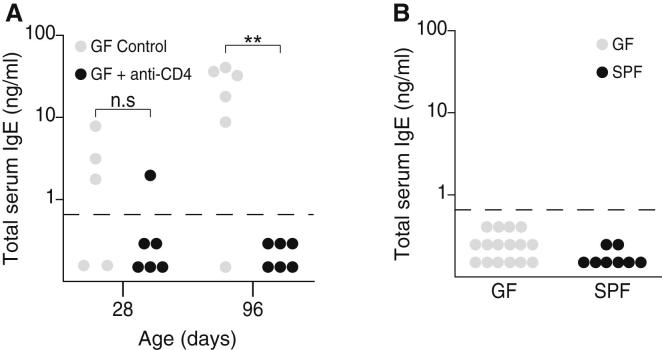
We sought to determine whether germ-free mice require CD4+ T cells in order to develop hyper-IgE. The administration of a CD4+ T cell-depleting antibody to germ-free mice starting at 28 days of age totally abrogated the induction of IgE (Figure 4A). IgE levels were also maintained below the limit of detection in germ-free mice genetically deficient in T cells (TCRβδ−/−; Figure 4B). These results indicate that IgE generated in the absence of commensal microbes is dependent on the presence of CD4+ T cells.
Figure 4.
IgE Induction in Germ-free Mice Is Dependent on CD4+ T Cells
(A) Germ-free mice were left untreated (n = 5; gray circles) or were depleted of CD4+ cells by intraperitoneal injection of anti-CD4 mAb (n = 6; black circles) two times per week starting on the day of weaning until the end of the experiment. IgE levels were measured at the start (28-days-old) and the end (96-days-old) of the experiment. Representative data from two independent experiments is shown.
(B) Total serum IgE levels were measured in germ-free (n = 16; gray circles) and SPF (n = 8; black circles) TCRβδ−/− mice.
(A and B) Each point represents an individual mouse, and the horizontal dotted line marks the lower limit of detection of the IgE ELISA assay (0.8 ng/ml). ∗∗p ≤ 0.01; n.s, not significant. p > 0.05 with an unpaired Student’s t test in (A).
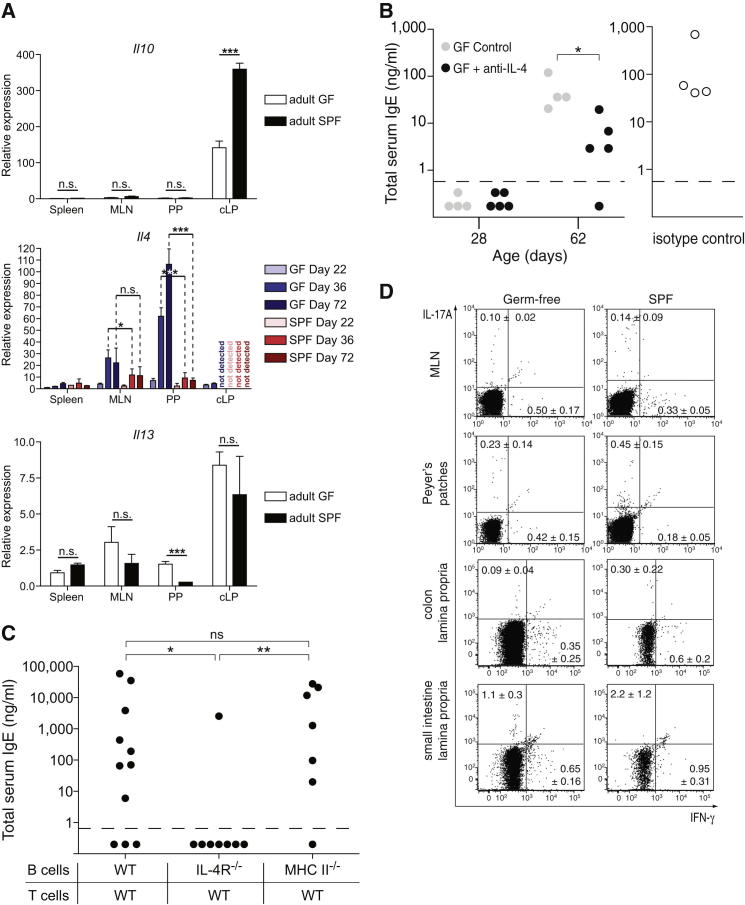
Elevated IgE Levels in Germ-free Mice Is Dependent on IL-4 but Not TSLP or MHC II Expression on B Cells
CD4+ T cells potentially provide help through cytokine production and/or MHC II cognate interactions. We assessed the cytokine expression profile of CD4+ T cells from the spleen, MLNs, PPs, and colon lamina propria (cLP) in germ-free and SPF mice. As expected, Il10 expression was higher in the cLP of SPF than that of germ-free mice (Atarashi et al., 2011; Geuking et al., 2011). Il4 expression in germ-free, but not SPF, mice was upregulated from day 36 of age onward, specifically in the MLNs and PPs. Thus, the expression of Il4 and εGLTs shared identical kinetics and occurred within identical immune geography, suggesting that the prototypic IgE-inducing cytokine IL-4 is also involved in germ-free IgE. Il13 expression was similar between germ-free and SPF mice with the exception of PPs, where expression was higher in germ-free than SPF mice (Figure 5A). Administration of an IL-4-neutralizing antibody to germ-free mice starting at 28 days of age led to a significant decrease in the concentration of serum IgE (Figure 5B). Failure to completely abrogate the development of detectable IgE may be explained by incomplete in vivo neutralization of IL-4 or compensation by other cytokines, such as IL-13. To further investigate this pathway, and because performing bone marrow chimeras remains a challenging task under germ-free conditions, we opted for a mixed adoptive transfer of FACS-sorted naive B and CD4+ T cells into germ-free Rag-1−/− mice. Homeostatic proliferation of T cells transferred into lymphopenic recipients is driven by both microbial and self-antigens. Rapid homeostatic proliferation is microbially driven, but, given that slow homeostatic proliferation in lymphopenic hosts is mediated exclusively by low-affinity self-antigens, cells transferred into germ-free recipients proliferate relatively slowly (Kieper et al., 2005). Cotransfer of wild-type (WT) B and T cells isolated from SPF mice into germ-free Rag-1−/− mice led to high IgE in the majority of reconstituted mice (Figure 5C), indicating that it is the environment within the germ-free host rather than intrinsic defects in germ-free T and B cells that is supportive of hygiene-induced IgE. Cotransfer of IL-4Rα-deficient B and WT T cells failed to induce IgE in seven out of eight mice (Figure 5C), suggesting that IL-4R-mediated signaling on B cells was crucial for IgE isotype switch under germ-free conditions. In contrast, cotransfer of MHC II-deficient B and WT T cells led to the induction of IgE in germ-free Rag-1−/− mice (Figure 5C), indicating that cognate interactions between B and T cells mediated by MHC II and antigenic peptide and T cell receptor (TCR) interactions was not required for IgE isotype switch under germ-free conditions. Flow cytometric analysis confirmed the absence of MHC II expression on the B cells isolated from Rag-1−/− recipients that received WT T and MHC II-deficient B cells, confirming there was no cross-contamination of MHC II-expressing B cells from the WT T cell preparation (Figure S1A available online). These results suggest that CD4+ T cell dependency may be limited to the production of cytokines, such as IL-4. Given that MLNs and PP CD4+ T cells of germ-free mice express higher levels of IL-4 and IL-13 in comparison to SPF mice (Figure 5A) and that the proportion of MLNs and colon CD4+IFN-γ+ T cells was lower under germ-free than SPF conditions (Figure 5D), it is plausible that a default Th2 pathway could be favored in the absence of microbes (Zhu et al., 2012).
Figure 5.
IgE Induction in Germ-free Mice Is Dependent on IL-4
(A) Normalized expression (to gapdh) of Il10, Il13, and Il4 from CD4+-enriched cells isolated from spleen, MLNs, PPs, and cLPs from the indicated groups of GF and SPF mice. Each bar represents the mean ± SD of 3–4 mice, except for samples from day 22 and cLPs, where the bar represents the mean ± SD of triplicate measurements of samples pooled from 3-4 mice.
(B) Germ-free mice were left untreated (n = 4; gray circles) or administered an IL-4-neutralizing monoclonal antibody (n = 5; black circles) or an isotype control (n = 4; hollow circles) by intraperitoneal injection two times per week from the time of weaning until the end of the experiment. IgE levels were measured at the start (28 days old) and/or the end (62 days old) of the experiment. Representative data from two independent experiments are shown.
(C) Lymphopenic germ-free Rag1−/− mice were adoptively transferred with purified wild-type CD4+CD3+CD25−CD45RBhigh naive T cells in combination with IgM+CD19+B220+CD3− naive B cells from wild-type C57BL/6 (n = 11), IL-4Rα−/− (n = 8), or MHC II (I-Ab)−/− (n = 7) donors, as indicated. Total serum IgE levels were measured 86 days posttransfer. Pooled data from two to three independent experiments per group are shown.
(B and C) Each point represents an individual mouse and the horizontal dotted line delineates the lower limit of detection of the IgE ELISA assay (0.8 ng/ml).
(D) Representative flow cytometry plots after intracellular staining for IFN-γ and IL-17A of MLNs, PPs, cLPs, and SI CD4+ T cells from germ-free and SPF mice after 4 hr of phorbol myristate acetate and ionomycin stimulation in the presence of Brefeldin A. The proportion of cytokine-producing CD4+ T cells is indicated in each quadrant ± SD.
∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.5; n.s, not significant. p > 0.05 with an unpaired Student’s t test in (A–C). See also Figure S1 and Table S1.
Thymic stromal lymphopoietin (TSLP) has been shown to be crucial for the initiation of Th2 responses and IgE isotype switch in some helminth infections and atopic allergic diseases (Paul and Zhu, 2010). Therefore, we investigated the role of TSLP in hygiene-mediated IgE in germ-free mice. The administration of a TSLP-neutralizing antibody to germ-free mice starting at 28 days of age failed to prevent the induction of IgE (Figure S1B). IgE levels were also not abrogated in germ-free TSLP receptor-deficient mice (Figure S1C). These data indicate that TSLP and signaling through its receptor are not required for IgE induction in the absence of commensal microbes.
Altogether, these observations suggest that excessive levels of IgE in germ-free mice are dependent on CD4+ T cells, IL-4, and possibly IL-13. Furthermore, this implies that similar immune mediators drive high IgE levels in germ-free C57BL/6 mice and immunodeficient SPF C57BL/6 mice (McCoy et al., 2006) and supports the notion that high IgE levels in the absence of microbes reflect immune dysregulation caused by the lack of microbial education of the immune system.
Neonatal Colonization with a Diversified Microbiota Is Required in Order to Inhibit the Induction of IgE
We have previously shown that intestinal microbial colonization with a limited diversity microbiota leads to the induction of intestinal regulatory T (Treg) cells and IL-10, which are required in order to limit the induction of proinflammatory Th1 and Th17 cells (Geuking et al., 2011). In an attempt to rescue the immune dysregulation and inhibit IgE while preserving the germ-free status, we tested whether adoptive transfer of microbially-instructed Treg cells or the administration of recombinant IL-10 could inhibit the induction of IgE in germ-free mice. Total CD4+, CD4+CD25− naive, or CD4+CD25+ Treg cells were isolated from the spleen or cLP of SPF donor mice and adoptively transferred into 28-day-old germ-free recipients. Although this protocol successfully limits Th1 and Th17 subset induction upon the colonization of Treg-cell-deficient mice (Geuking et al., 2011), it was not sufficient to limit IgE induction under germ-free conditions (Figure S2A). Similarly, the administration of recombinant IL-10 also failed to modulate IgE titers even when repetitively administered for the duration of the experiment (Figure S2B).
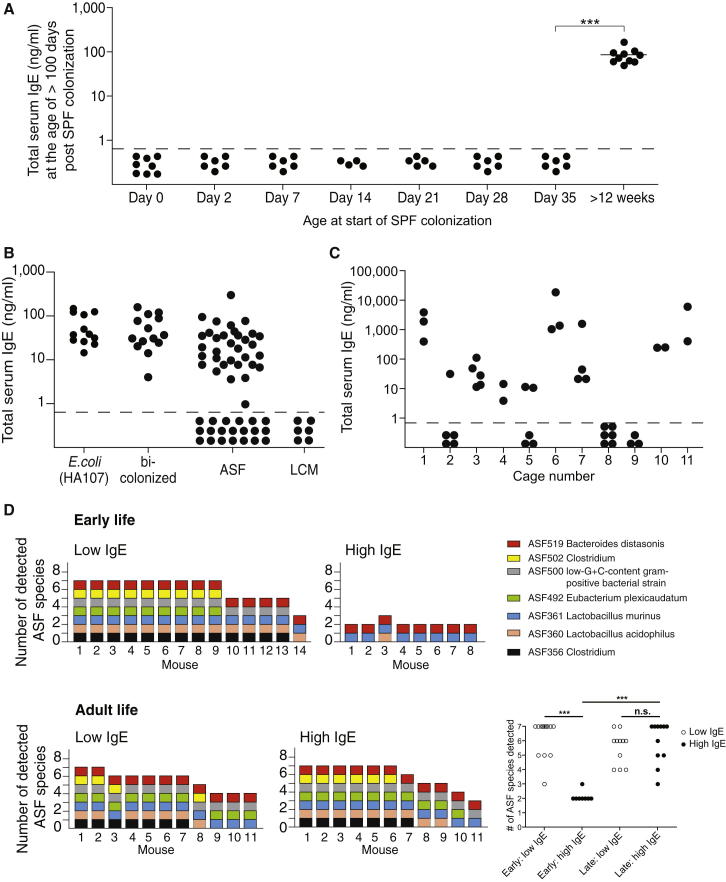
Our interpretation that IgE induction is a default pathway of mucosal immunity in the absence of commensal intestinal microbes predicts that IgE levels should be normalized in mice that are born germ free and later colonized by commensal microbes; the interesting question was whether there was a time window in which this could take place. Cohousing of adult germ-free mice (with high IgE) with SPF mice (with undetectable IgE and a complex intestinal microbiota) did not reduce serum IgE levels over an 8-week period (Figure 6A). In contrast, exposure to an SPF flora at the time of birth completely inhibited the induction of IgE (Figure 6A; day 0). To further refine the window of time whereby SPF colonization provides protection, germ-free pups were colonized at days 2, 7, 14, 21, 28, and 35 of age. In contrast to colonization during adulthood, the colonization of pups until 1 week postweaning (when serum IgE levels were still low) was able to fully protect them from the induction of elevated IgE during adulthood (Figure 6A), indicating that microbial signals shape IgE induction pathways early in life during a critical window of early-life development.
Figure 6.
Increased Intestinal Bacterial Diversity in Neonates Abrogates Hyper-IgE
(A) Germ-free C57BL/6 mice were colonized with an SPF flora by cohousing with an SPF sentinel at the indicated age after birth (n = 4–10 mice per group). Total serum IgE levels were measured >100 days after SPF colonization.
(B) Total serum IgE levels were measured in adult germ-free C57BL/6 mice that were orally administered three to six gavages of 2 × 109−1010 CFU of the auxotrophic E.coli strain HA107 before weaning (n = 12), bicolonized from birth with ASF519 (P. distasonis) and ASF361 (L. murinus) (n = 14), ASF colonized from birth (n = 52), or colonized from birth with a low-complexity microbiota (LCM) consisting of approximately 40 species (n = 6).
(C) Total serum IgE levels were measured in adult (>6 weeks old) ASF mice raised in a flexible film isolator within individual cages as indicated (n = 2–6 per cage).
(D) DNA was isolated from fecal pellets or cecal contents of ASF mice during early (30–40 days old) or adult (77–295 days old) stages, and individual ASF species were detected by species-specific qPCR. Each bar represents the diversity (number of species detected) within an individual mouse, and specific ASF species present are shown by colored squares as indicated. ASF457 (Mucispirillum schaedleri) was not detected in any of the samples.
(A–C) Each point represents an individual mouse, and the horizontal dotted line delineates the lower limit of detection of the IgE ELISA assay (0.8 ng/ml).
∗∗∗p ≤ 0.001; n.s, not significant. (p > 0.05) with an unpaired Student’s t test. See also Figure S2 and Table S2.
SPF mice contain a highly diverse and undefined microbiota ranging from symbionts to potentially opportunistic pathogens (Round and Mazmanian, 2009), making it difficult to use SPF mice to determine the minimal microbial diversity or composition required to inhibit IgE. To investigate the role of the level of diversity early in life in preventing the induction of high IgE later in life, we made use of four additional defined model microbiotas (Figure 6B). First, we exposed germ-free C57BL/6 pups three to six times between the ages of 2 and 4 weeks to the single bacterium E. coli HA107 by oral gavage. We used the auxotrophic HA107 strain because we have previously described that this protocol induces IgA responses (Hapfelmeier et al., 2010). Despite exposing the pups to multiple high doses (2 × 109−1010 CFU per gavage) before weaning, the mice developed hyper-IgE at adult age (Figure 6B). Next, we generated mice bicolonized from birth with the two altered Schaedler flora (ASF) species (Dewhirst et al., 1999), Parabacteroides distasonis ASF519 and Lactobacillus murinus ASF361. All bicolonized mice developed hyper-IgE (Figure 6B). Then, we performed the same experiment using the full ASF, a mutualistic benign microflora composed of eight commensal bacterial species (Dewhirst et al., 1999). We found that, even when housed within the same flexible film isolator, ASF mice clustered into two groups with some animals displaying hyper-IgE, whereas others were protected with undetectable IgE (Figure 6B). In order to investigate whether increasing the diversity further will protect from hyper-IgE, we also tested C57BL/6 mice colonized from birth with a low complexity microbiota (LCM) consisting of approximately 40 phylotypes (Stecher et al., 2010; Endt et al., 2010). At this level of diversity, all mice tested were protected and had undetectable levels of IgE in the serum. This indicates that increasing diversity is an important factor in providing protection from the induction of hyper-IgE. However, we cannot rule out the impact of individual microbial species in mediating protection.
In ASF-colonized mice, the presence of mice with both high and low IgE levels was intriguing and cage specific (Figure 6C), and we hypothesized that early-life colonization dynamics were responsible for this segregation.
To test this, the compositions of the ASF early (30–40 days of age) and late (77–295 days of age) in life were analyzed with qPCR and bacterial-specific primer pairs. Bacterial diversity during early life ranged from two to seven bacterial species, and the observed diversity inversely correlated with IgE levels; association with two to three detectable bacterial species (P. distasonis and L. murinus) correlated with high IgE, whereas association with a greater detectable bacterial diversity (four to seven bacterial species) correlated with low IgE levels (Figure 6D). In contrast, bacterial diversity in adult life ranged from three to seven bacterial species and was not predictive of IgE levels (Figure 6D). In our ASF colony, P. distasonis ASF519 (>90%) and L. murinus ASF361 (4%) are the two most abundant species present (high-throughput 16S amplicon sequencing) (data not shown). This suggests that increasing early-life diversity by adding less abundant additional species may already be sufficient to mediate protection from hyper-IgE.
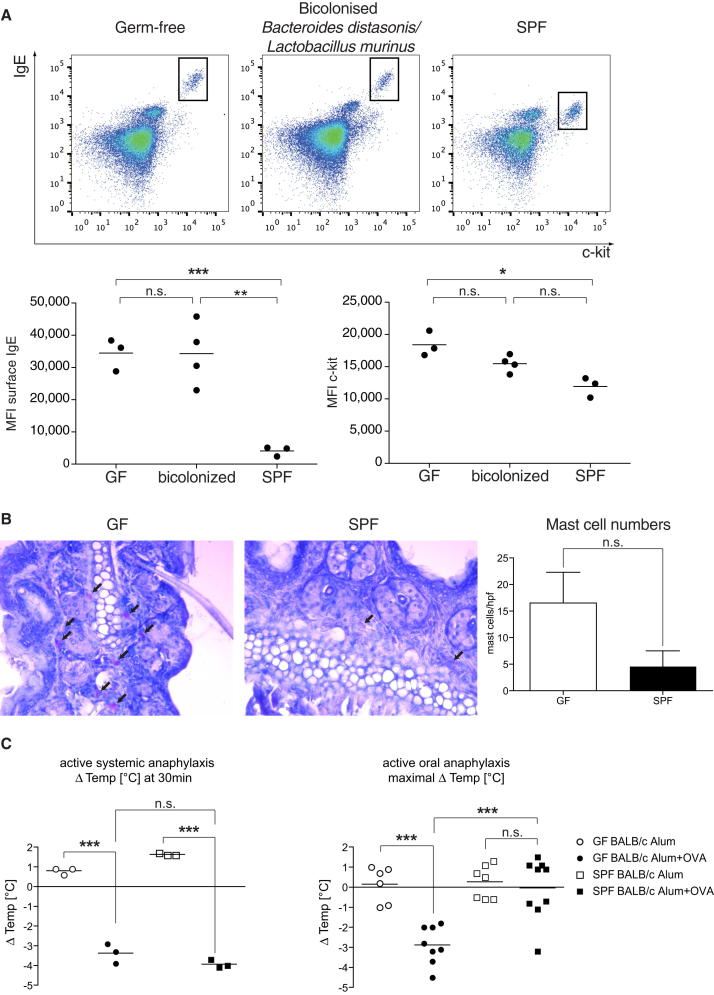
Elevated IgE Levels in Germ-free Mice Lead to Increased Amounts of Surface-Bound IgE on Mast Cells and Exaggerated Oral-Induced Systemic Anaphylaxis
Given that IgE is known to regulate mast cell homeostasis (Kalesnikoff et al., 2001; Kitaura et al., 2003), we investigated whether the elevated levels of IgE in germ-free mice could alter mast-cell-mediated pathologies. We found that the amount of surface-bound IgE was significantly increased in peritoneal mast cells from germ-free and bicolonized mice in comparison to SPF mice (Figure 7A). Interestingly, cell-surface levels of CD117 (c-kit) were also increased on mast cells from germ-free and bicolonized mice in comparison to SPF mice, which may hint to a more immature phenotype (Boyce, 2004). Next, we evaluated whether tissue mast cell numbers were increased because of the elevated IgE levels in germ-free mice. The number of cutaneous mast cells, as identified by chloroacetate esterase (CAE) staining in the ear tissue of germ-free mice, was slightly, although not significantly, elevated under steady-state germ-free conditions in comparison to SPF conditions (Figure 7B). In addition, the reduced number of mast cells in the ear skin of SPF animals showed weaker CAE staining (Figure 7B). We investigated whether elevated IgE levels in germ-free mice have an impact on the severity of active antigen-induced anaphylaxis. It has been reported that gastrointestinal, cutaneous, and cardiovascular symptoms in oral antigen-induced anaphylaxis are dependent on IgE and mast cells (Ahrens et al., 2012), whereas, in systemic antigen-induced anaphylaxis, these adverse symptoms are mediated by both IgE and IgG pathways (Osterfeld et al., 2010). Germ-free and SPF BALB/c mice were systemically primed with OVA and aluminum potassium sulfate (AIK(SO4)2-12H2O) (Alum) or Alum alone. Mice were challenged 2 weeks later by intravenous injection of ovalbumin (OVA; active systemic anaphylaxis) or by oral gavage of OVA (active oral anaphylaxis). In response to systemic antigen challenge, germ-free mice developed hypothermia to the same level as SPF animals (Figure 7C). In contrast, germ-free mice showed significantly increased susceptibility to oral antigen-induced anaphylaxis (Figure 7C). Bicolonized C57BL/6 mice also displayed increased susceptibility to active oral anaphylaxis in comparison to SPF C57BL/6 mice (Figure S3). These data suggest that increased serum IgE levels in germ-free mice impact on mast cell homeostasis, which, in turn, leads to greatly increased sensitivity to oral-induced anaphylaxis.
Figure 7.
Germ-free Mice Display Increased Antigen-Induced Oral Anaphylaxis
(A) Representative flow cytometry plots from peritoneal mast cells identified as c-kit+IgE+ and mean fluorescence intensity (MFI) of mast cell surface IgE and c-kit staining in germ-free, bicolonized, and SPF C57BL/6 mice. Data shown are representative of four independent experiments.
(B) Representative histological sections (20× magnification) from ears stained for chloroacetate esterase activity indicative of mast cells (arrows) and summary of mast cell quantification per high-power field (hpf; 20×) of germ-free and SPF mice. Data are indicated by mean ± SEM of 7–10 HPF per mouse and three to four mice per group.
(C) Germ-free (circles) and SPF (squares) BALB/c mice were subcutaneously sensitized with Alum alone (open symbols) or OVA + Alum (closed symbols) and, 2 weeks later, challenged with 100 μg OVA intravenously (n = 3 per group; active systemic anaphylaxis, left) or with 50 mg OVA orally (n = 6–9 per group; active oral anaphylaxis, right). Rectal temperature was measured every 5–10 min for 60 min, and the maximal Δtemperature [°C] is plotted. Data for the active oral anaphylaxis is pooled from two independent experiments.
(A and C) Each point represents an individual mouse, and the horizontal dotted line indicates the mean.
∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.5; n.s, not significant. p > 0.05 with an unpaired Student’s t test. See also Figure S3.
Discussion
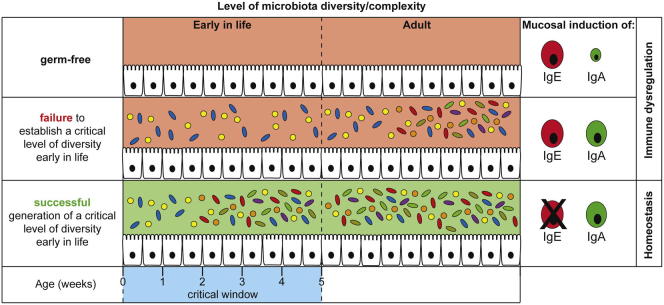
The hygiene hypothesis postulated that the quality and/or quantity of microbial exposure early in life might have an important impact on how the immune system behaves later in life (Strachan, 1989). Although the original hygiene hypothesis did not suggest a role for the intestinal commensal bacteria, in the past years, it has become clear that our intestinal inhabitants have a powerful influence on immune maturation at both mucosal and systemic sites. Systematic characterization of intestinal microbial communities in human babies has indicated that incidental environmental exposure plays a major role in determining the microbial community in each baby (Koenig et al., 2011; Palmer et al., 2007). Along with westernization, major changes in the environment have occurred that are likely to heavily influence the composition of the intestinal microbiota. Recent studies have also shown that children living in developing countries harbor greater intestinal bacterial diversity than children in developed countries (De Filippo et al., 2010; Lin et al., 2013). Decreased bacterial diversity and/or altered composition of the intestinal microbiota early in life are likely to contribute to the increased susceptibility to immune-mediated diseases in westernized countries. Using gnotobiotic mouse models, we provide experimental evidence that limited microbial association below a certain threshold of complexity or devoid of putative bacterial species or bacterial consortia with immunoregulatory properties during early life has profound negative and lasting effects on the induction of immune regulation, even when microbial diversity is increased above this threshold later in life. These data demonstrate the presence of a critical time window early in life during which appropriate microbial exposure has to occur in order to induce functional, life-long immune regulation that maintains serum IgE levels at baseline.
In addition to allergic disorders and certain helminth infections, elevated IgE has been suggested to be a biomarker for primary immunodeficiencies (Liston et al., 2008). Data from numerous mouse models of genetic immunodeficiency suggest that reduced functional T cell populations may lead to immune dysregulation and elevated IgE levels as a consequence. There are most likely many pathways involved in the immune regulatory network that function to maintain IgE at low levels. The development of hyper-IgE in genetically immunocompetent germ-free mice indicates that exposure to commensal bacteria is a key factor in the development of a functional immune regulatory network. Germ-free mice also show elevated invariant natural killer T cells at mucosal sites (Olszak et al., 2012), which trigger heightened pathology in models of inflammatory bowel disease and allergic asthma. In this study, we show that elevated serum IgE titers leads to increased levels of surface-bound IgE on mast cells, and germ-free mice displayed exacerbated antigen-induced oral anaphylaxis in comparison to SPF mice, implying that dysregulated immune reactions such as food allergies may be highly dependent on adequate acquisition of bacterial consortia at early age. The fact that hygiene-mediated IgE does not require an underlying genetic immunodeficiency suggests that exposure to a diverse microbial population during a critical time window early in life is absolutely required in order to set the baseline immune regulatory state for life. It is interesting that isotype switch to IgE has been recently shown to be favored in immature B cells (Wesemann et al., 2011). Although the maturation state of the B cells undergoing IgE class switch was not investigated in the current study, B cells were found to switch to IgE in mucosal tissues, especially PP, and not in B cell developmental sites, such as bone marrow or spleen. Additional studies are required to carefully investigate the impact of microbial exposure on lymphocyte development and function.
Although IgE alone does not constitute allergy and the simplicity of a bicolonization does not necessarily represent the restricted microbiota resulting from improved human hygiene, our results do show that diverse microbial colonization during a critical time window in neonatal life is required in order to limit default immune pathways, resulting in excessive IgE levels, mirroring one possible immune component for rationalizing the hygiene hypothesis.
Experimental Procedures
Mouse Strains, Hygiene Status, and Colonization
Germ-free Mice
C57BL/6, TCRβδ−/−, JH−/−, Rag1−/−, TSLPR−/− (a kind gift from N. Harris [École Polytechnique Fédérale de Lausanne]) (all on a C57BL/6 background), NIH-Swiss, BALB/c, Swiss Webster, and NMRI mice were rederived to germ-free status via two-cell embryo transfer and bred and maintained in flexible-film isolators as described previously (Smith et al., 2007).
Antigen-free Mice
Germ-free mice were fed an irradiated (5M Rad Co-60 for 20 hr) elemental antigen-free diet consisting of extensively hydrolyzed proteins supplemented with fats, vitamins, and minerals (Pregestimil, Enfamil). Bedding consisted of endotoxin-free sand (baked at 250°C for 30 min).
Gnotobiotic Mice
Gnotobiotic mice associated with altered Schaedler flora (ASF) (Dewhirst et al., 1999) were originally obtained from germ-free mice cohoused with an ASF colonizer. For the generation of a bicolonized colony, germ-free C57BL/6 breeding pairs were orally gavaged with pure cultures of P. distasonis and L. murinus. Serum samples from low-complexity microbiota mice (Stecher et al., 2010; Endt et al., 2010) were kindly provided by W.-D. Hardt (Eidgenössische Technische Hochschule Zürich). E.coli HA107 was cultured and prepared for gavage of germ-free mice as described previously (Hapfelmeier et al., 2010), and 2 × 109−1010 CFU HA107 was administered by oral gavage. Maintenance of gnotobiotic mice is described in the Supplemental Information.
SPF Mice
C57BL/6, BALB/c, and TCRβδ−/− mice were purchased from Taconic or maintained in the central animal facilities at McMaster University or the University of Bern. MHC II−/− and IL-4Rα−/− mice were a kind gift from N. Harris. For SPF colonization experiments, germ-free mice were cohoused with an SPF mouse in individually ventilated cages.
All animal experiments were carried out in accordance with the McMaster University animal utilization protocols and the Canadian Council on Animal Care guidelines or in accordance with Swiss federal regulations.
Isotype-Specific ELISA for the Detection of Serum Antibodies
Blood was collected in serum-separating tubes and total serum IgE, IgA, IgM, IgG1, IgG2a, IgG2c, and IgG3 concentrations determined by sandwich ELISA as described in the Supplemental Information.
Cell Isolation and Quantitative Real Time PCR
In order to obtain single-cell suspensions, lymph nodes (pLNs, PPs, and MLNs) were digested with 0.14 Wünsch U/ml Liberase C1 or Liberase TL (Roche Applied Science). Then, lymphoid tissues (spleen, pLNs, MLNs, and PPs) were homogenized and filtered through a 40 μm cell strainer. Lymphocytes from cLP were isolated as described previously (Geuking et al., 2011). Cells were resuspended in TRIzol reagent (Invitrogen) and RNA isolated according to the manufacturer’s instructions. qPCR was performed with SYBR Green Supermix or SsoFast EvaGreen Supermix (Bio-Rad) with specific primer pairs (see Table S1 and the Supplemental Information).
Genomic DNA was isolated from cecal contents or fecal pellets with the QIAamp DNA Stool Mini Kit (Qiagen) and analyzed with SsoFast EvaGreen Supermix (Bio-Rad) with the use of primers specific for the 16S rRNA genes of the individual ASF species (see Table S2).
PCR and analysis were performed on an iQ5 or CFX384 (Bio-Rad) platform and software.
Flow Cytometry and Flow Cytometry Sorting
Antibodies and corresponding clones used for flow cytometry are listed in the Supplemental Information. Dead cells were excluded with live/dead fixable dye eFluor 506 (eBioscience) and Fc receptors blocked with CD16/CD32 Fc blocking antibody. Data were acquired on a FACSCalibur (BD Biosciences) or LSRII (BD Biosciences) and analyzed with FlowJo (Tree Star). For FACS sorting, splenocytes were enriched for CD4+ T cells or CD19+ B cells (>80% of lymphocytes) with CD19 or CD4 magnetic beads (Miltenyi Biotec) and sorted for IgM+CD19+B220+CD3− (naive B cells) or CD3+CD25−CD45RBhigh (naive CD4+ T cells) populations on a FACSAria (BD Biosciences).
Axenic In Vivo Administration of mAb
Twice a week, germ-free mice (28 days old) were administered (200 μg intraperitoneal injected) one of the following sterile mAbs: anti-CD4 (clone YTS191.1.2), anti-iL-4 (clone 11B11), anti-TSLP (clone 28F12, a kind gift from A. Farr [University of Washington]), or isotype control (35.61). In order to abrogate PP ontogeny in vivo, anti-iL7Rα (3 mg, clone A7R34, Bio X Cell) was administered (intravenous [i.v.] injection) to germ-free pregnant mice at E14.5–E15.5. The absence of PP and germ-free status was confirmed at the end of each experiment.
Axenic Adoptive Cell Transfer
For cotransfer of purified naive B and T cells, 1 × 106 FACS-sorted splenic B and T cells were i.v. injected into germ-free Rag1−/− recipients. B cells were purified from WT C57BL/6, IL-4Rα−/−, or MHC II-deficient mice.
Mast Cell Quantification
Samples were fixed in 10% formalin and processed by standard histological techniques prior to paraffin embedding. Then, 6 μm sections were deparaffinized and stained with an α-Naphthyl Chloroacetate Esterase Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The sections were counterstained with Gill’s No.3 Hematoxylin Solution for 30 s.
Antigen-Induced Oral and Systemic Anaphylaxis
Germ-free, bicolonized, or SPF mice were subcutaneously injected with 50 μg of ovalbumin (Sigma-Aldrich) in the presence of 2 mg of Alum adjuvant (Sigma- Aldrich) in sterile PBS or with Alum alone. Two weeks later, mice received either an oral (50 mg/250 μl PBS) or systemic (100 μg ovalbumin in 500 μl PBS) ovalbumin challenge. For the oral challenge, mice were deprived of food for 4 hr prior to gavage. Rectal temperatures were monitored every 5–10 min for 90 min with a TCAT-2LV animal temperature controller (Physitemp Instruments).
Acknowledgments
The research leading to these results received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP/2007-2013) and ERC grant agreement 281785 (K.D.M.), the Swiss National Science Foundation (K.D.M.), and the Canadian Institute of Health Research (K.D.M.). M.B.G. is a recipient of an Ambizione fellowship from the Swiss National Science Foundation. The Clean Mouse Facility is supported by the Genaxen Foundation, Inselspital, and the University of Bern. We thank A. Macpherson for scientific input and M. Lawson and L. Hai for technical assistance.
Supplemental Information
References
- Ahrens R., Osterfeld H., Wu D., Chen C.Y., Arumugam M., Groschwitz K., Strait R., Wang Y.H., Finkelman F.D., Hogan S.P. Intestinal mast cell levels control severity of oral antigen-induced anaphylaxis in mice. Am. J. Pathol. 2012;180:1535–1546. doi: 10.1016/j.ajpath.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.E., Maizels R.M. Diversity and dialogue in immunity to helminths. Nat. Rev. Immunol. 2011;11:375–388. doi: 10.1038/nri2992. [DOI] [PubMed] [Google Scholar]
- Antón I.M., de la Fuente M.A., Sims T.N., Freeman S., Ramesh N., Hartwig J.H., Dustin M.L., Geha R.S. WIP deficiency reveals a differential role for WIP and the actin cytoskeleton in T and B cell activation. Immunity. 2002;16:193–204. doi: 10.1016/s1074-7613(02)00268-6. [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach J.F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- Bacharier L.B., Geha R.S. Molecular mechanisms of IgE regulation. J. Allergy Clin. Immunol. 2000;105:S547–S558. doi: 10.1016/s0091-6749(00)90059-9. [DOI] [PubMed] [Google Scholar]
- Boyce J.A. The biology of the mast cell. Allergy Asthma Proc. 2004;25:27–30. [PubMed] [Google Scholar]
- Braun-Fahrländer C., Gassner M., Grize L., Neu U., Sennhauser F.H., Varonier H.S., Vuille J.C., Wüthrich B., Swiss Study on Childhood Allergy and Respiratory Symptoms with Respect to Air Pollution Prevalence of hay fever and allergic sensitization in farmer’s children and their peers living in the same rural community. SCARPOL team. Clin. Exp. Allergy. 1999;29:28–34. doi: 10.1046/j.1365-2222.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- Cahenzli J., Balmer M.L., McCoy K.D. Microbial-immune cross-talk and regulation of the immune system. Immunology. 2013;138:12–22. doi: 10.1111/j.1365-2567.2012.03624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash H.L., Whitham C.V., Behrendt C.L., Hooper L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin C., Kocur M., Pott J., Duerr C.U., Gütle D., Lotz M., Hornef M.W. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8:358–368. doi: 10.1016/j.chom.2010.09.005. [DOI] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S., Collini S., Pieraccini G., Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F.E., Chien C.C., Paster B.J., Ericson R.L., Orcutt R.P., Schauer D.B., Fox J.G. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., Flavell R.A. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endt K., Stecher B., Chaffron S., Slack E., Tchitchek N., Benecke A., Van Maele L., Sirard J.C., Mueller A.J., Heikenwalder M. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010;6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J.J., McNulty N.P., Rey F.E., Gordon J.I. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman F.D., Katona I.M., Urban J.F., Jr., Holmes J., Ohara J., Tung A.S., Sample J.V., Paul W.E. IL-4 is required to generate and sustain in vivo IgE responses. J. Immunol. 1988;141:2335–2341. [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Ganal S.C., Sanos S.L., Kallfass C., Oberle K., Johner C., Kirschning C., Lienenklaus S., Weiss S., Staeheli P., Aichele P., Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S., Glickman J.N., Glimcher L.H. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geha R.S., Jabara H.H., Brodeur S.R. The regulation of immunoglobulin E class-switch recombination. Nat. Rev. Immunol. 2003;3:721–732. doi: 10.1038/nri1181. [DOI] [PubMed] [Google Scholar]
- Geuking M.B., Cahenzli J., Lawson M.A., Ng D.C., Slack E., Hapfelmeier S., McCoy K.D., Macpherson A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Giblin W., Chatterji M., Westfield G., Masud T., Theisen B., Cheng H.L., DeVido J., Alt F.W., Ferguson D.O., Schatz D.G., Sekiguchi J. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. 2009;113:2965–2975. doi: 10.1182/blood-2008-07-165167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould H.J., Sutton B.J. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008;8:205–217. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S., Lawson M.A., Slack E., Kirundi J.K., Stoel M., Heikenwalder M., Cahenzli J., Velykoredko Y., Balmer M.L., Endt K. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst T., Sichelstiel A., Schär C., Yadava K., Bürki K., Cahenzli J., McCoy K., Marsland B.J., Harris N.L. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- Hill D.A., Siracusa M.C., Abt M.C., Kim B.S., Kobuley D., Kubo M., Kambayashi T., Larosa D.F., Renner E.D., Orange J.S. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt P.G. Programming for responsiveness to environmental antigens that trigger allergic respiratory disease in adulthood is initiated during the perinatal period. Environ. Health Perspect. 1998;106(Suppl 3):795–800. doi: 10.1289/ehp.98106795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalesnikoff J., Huber M., Lam V., Damen J.E., Zhang J., Siraganian R.P., Krystal G. Monomeric IgE stimulates signaling pathways in mast cells that lead to cytokine production and cell survival. Immunity. 2001;14:801–811. doi: 10.1016/s1074-7613(01)00159-5. [DOI] [PubMed] [Google Scholar]
- Kieper W.C., Troy A., Burghardt J.T., Ramsey C., Lee J.Y., Jiang H.Q., Dummer W., Shen H., Cebra J.J., Surh C.D. Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J. Immunol. 2005;174:3158–3163. doi: 10.4049/jimmunol.174.6.3158. [DOI] [PubMed] [Google Scholar]
- Kitaura J., Song J., Tsai M., Asai K., Maeda-Yamamoto M., Mocsai A., Kawakami Y., Liu F.T., Lowell C.A., Barisas B.G. Evidence that IgE molecules mediate a spectrum of effects on mast cell survival and activation via aggregation of the FcepsilonRI. Proc. Natl. Acad. Sci. USA. 2003;100:12911–12916. doi: 10.1073/pnas.1735525100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R., Angenent L.T., Ley R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz D., Ziętara N., Uzel G., Weidemann T., Braun C.J., Diestelhorst J., Krawitz P.M., Robinson P.N., Hecht J., Puchałka J. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J. Exp. Med. 2013;210:433–443. doi: 10.1084/jem.20111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz C.S., Yamaguchi M., Oettgen H.C., Katona I.M., Miyajima I., Kinet J.P., Galli S.J. IgE regulates mouse basophil Fc epsilon RI expression in vivo. J. Immunol. 1997;158:2517–2521. [PubMed] [Google Scholar]
- Lin A., Bik E.M., Costello E.K., Dethlefsen L., Haque R., Relman D.A., Singh U. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS ONE. 2013;8:e53838. doi: 10.1371/journal.pone.0053838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Enders A., Siggs O.M. Unravelling the association of partial T-cell immunodeficiency and immune dysregulation. Nat. Rev. Immunol. 2008;8:545–558. doi: 10.1038/nri2336. [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., Harris N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., Smith K. Mesenteric lymph nodes at the center of immune anatomy. J. Exp. Med. 2006;203:497–500. doi: 10.1084/jem.20060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J.G., Frank D.N., Mortin-Toth S., Robertson C.E., Feazel L.M., Rolle-Kampczyk U., von Bergen M., McCoy K.D., Macpherson A.J., Danska J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Martinez F.D., Holt P.G. Role of microbial burden in aetiology of allergy and asthma. Lancet. 1999;354(Suppl 2):SII12–SII15. doi: 10.1016/s0140-6736(99)90437-3. [DOI] [PubMed] [Google Scholar]
- McCoy K.D., Harris N.L., Diener P., Hatak S., Odermatt B., Hangartner L., Senn B.M., Marsland B.J., Geuking M.B., Hengartner H. Natural IgE production in the absence of MHC Class II cognate help. Immunity. 2006;24:329–339. doi: 10.1016/j.immuni.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Muegge B.D., Kuczynski J., Knights D., Clemente J.C., González A., Fontana L., Henrissat B., Knight R., Gordon J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M.C., Huffnagle G.B. The ‘microflora hypothesis’ of allergic diseases. Clin. Exp. Allergy. 2005;35:1511–1520. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- Okada H., Kuhn C., Feillet H., Bach J.F. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin. Exp. Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T., An D., Zeissig S., Vera M.P., Richter J., Franke A., Glickman J.N., Siebert R., Baron R.M., Kasper D.L., Blumberg R.S. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterfeld H., Ahrens R., Strait R., Finkelman F.D., Renauld J.C., Hogan S.P. Differential roles for the IL-9/IL-9 receptor alpha-chain pathway in systemic and oral antigen-induced anaphylaxis. J. Allergy Clin. Immunol. 2010;125:469–476. doi: 10.1016/j.jaci.2009.09.054. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan E., Notarangelo L.D., Geha R.S. Primary immune deficiencies with aberrant IgE production. J. Allergy Clin. Immunol. 2008;122:1054–1062. doi: 10.1016/j.jaci.2008.10.023. quiz 1063–1064. [DOI] [PubMed] [Google Scholar]
- Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul W.E., Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., McCoy K.D., Macpherson A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Smith M.I., Yatsunenko T., Manary M.J., Trehan I., Mkakosya R., Cheng J., Kau A.L., Rich S.S., Concannon P., Mychaleckyj J.C. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Chaffron S., Käppeli R., Hapfelmeier S., Freedrich S., Weber T.C., Kirundi J., Suar M., McCoy K.D., von Mering C. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan D.P. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton B.J., Gould H.J. The human IgE network. Nature. 1993;366:421–428. doi: 10.1038/366421a0. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Quince C., Faith J.J., McHardy A.C., Yatsunenko T., Niazi F., Affourtit J., Egholm M., Henrissat B., Knight R., Gordon J.I. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D., Shinkai K., Locksley R.M. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- Walter J., Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wesemann D.R., Magee J.M., Boboila C., Calado D.P., Gallagher M.P., Portuguese A.J., Manis J.P., Zhou X., Recher M., Rajewsky K. Immature B cells preferentially switch to IgE with increased direct Sμ to Sε recombination. J. Exp. Med. 2011;208:2733–2746. doi: 10.1084/jem.20111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Honda K., Shinkura R., Adachi S., Nishikawa S., Maki K., Ikuta K., Nishikawa S.I. IL-7 receptor alpha+ CD3(-) cells in the embryonic intestine induces the organizing center of Peyer’s patches. Int. Immunol. 1999;11:643–655. doi: 10.1093/intimm/11.5.643. [DOI] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.