Abstract
Whereas many genes associated with intellectual disability (ID) encode synaptic proteins, transcriptional defects leading to ID are less well understood. We studied a large, consanguineous pedigree of Arab origin with seven members affected with ID and mild dysmorphic features. Homozygosity mapping and linkage analysis identified a candidate region on chromosome 17 with a maximum multipoint logarithm of odds score of 6.01. Targeted high-throughput sequencing of the exons in the candidate region identified a homozygous 4-bp deletion (c.169_172delCACT) in the METTL23 (methyltransferase like 23) gene, which is predicted to result in a frameshift and premature truncation (p.His57Valfs*11). Overexpressed METTL23 protein localized to both nucleus and cytoplasm, and physically interacted with GABPA (GA-binding protein transcription factor, alpha subunit). GABP, of which GABPA is a component, is known to regulate the expression of genes such as THPO (thrombopoietin) and ATP5B (ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide) and is implicated in a wide variety of important cellular functions. Overexpression of METTL23 resulted in increased transcriptional activity at the THPO promoter, whereas knockdown of METTL23 with siRNA resulted in decreased expression of ATP5B, thus revealing the importance of METTL23 as a regulator of GABPA function. The METTL23 mutation highlights a new transcriptional pathway underlying human intellectual function.
INTRODUCTION
Intellectual disability (ID) is a significant public health issue with an estimated prevalence of 1–3% worldwide (1,2). Although ID can be caused by environmental factors such as traumatic injury, toxin exposure, and radiation, genetic causes are thought to account for a large proportion of ID cases (3). The molecular mechanisms underlying genetic causes of ID are diverse (4). Whereas many ID genes function at the synapse (5), the role of transcriptional regulation in intellectual function has been increasingly realized, with the identification of mutations in many genes encoding transcription factors [TCF4 (6), zinc finger genes (7–9), SOX5 (10)] and DNA-binding or modifying proteins [MECP2, encoding a methyl-CpG-binding protein (11), and DNMT3B, encoding a DNA methyltransferase (12)] in individuals with ID.
Recently an increasing number of genes associated with ID have been found to encode transcriptional regulators with functions that relate to chromatin structure. These include histone acetyltransferases [CREBBP (13,14)], histone deacetylases [HDAC4 (15)], histone methyltransferases [EHMT1 (16)], and histone demethylases [PHF8 (17), JARID1C (18)]. These proteins directly modify histones through the addition or the removal of chemical moieties and affect transcription by altering chromatin structure and the availability of transcription factor binding sites, as well as by interacting in multiprotein complexes with pertinent transcription factors. Other genes encoding chromatin-remodeling proteins and implicated in ID include ARID1B (19) and ATRX (20), which encode members of the SWI/SNF complex involved in the manipulation of nucleosome positions and consequent transcriptional activity. Thus, transcriptional regulation by proteins encoded by ID genes can take many different forms, and genetic forms of ID have provided important insights into mechanisms of chromatin regulation in the brain.
Here, we identify a new transcriptional regulator essential for normal human cognition, METTL23 (methyltransferase like 23), and show that it interacts with the transcription factor subunit GABPA (GA-binding protein transcription factor, alpha subunit; encoded by GABPA), with functional effects on the expression of the GABP target genes THPO (thrombopoietin) (21) and ATP5B (ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide) (22). This study identifies METTL23 as the causative gene in a family with ID and implicates the GABP transcriptional pathway in human cognitive function, adding to the growing body of evidence supporting a significant role for transcriptional regulation in cognition.
RESULTS
Clinical presentation
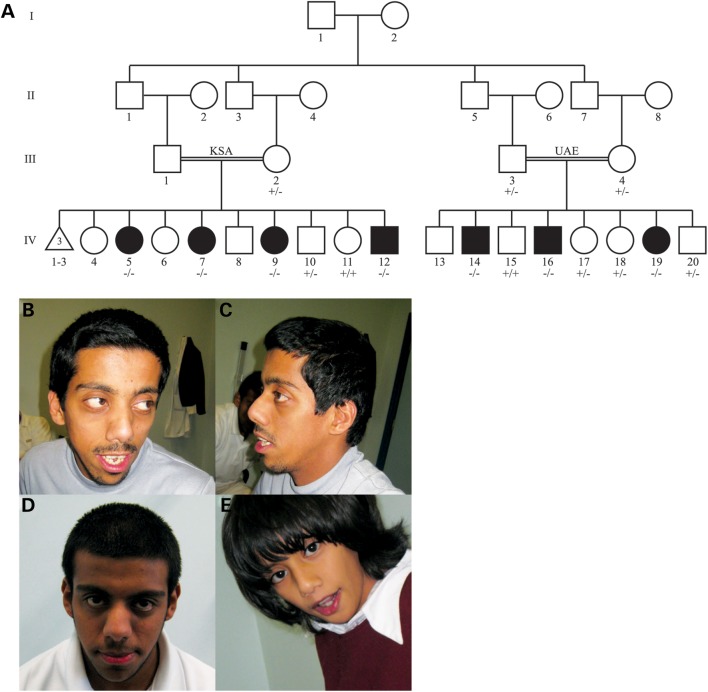
We identified a large, consanguineous pedigree of Yemeni origin with seven individuals affected by ID and dysmorphic features (Fig. 1A). One branch of the family resides in the United Arab Emirates (UAE) and has three affected individuals (UAE branch), and another branch resides in the Kingdom of Saudi Arabia (KSA) and has four affected individuals (KSA branch). The clinical findings for the UAE branch are summarized in Table 1 and characteristic dysmorphic features can be seen in Figure 1B–E. The affected members of the KSA branch exhibited ID without seizures, though a detailed medical history of these individuals was not available.
Figure 1.
Family pedigree and overview of dysmorphic features. (A) Pedigree of the KSA (left) and UAE (right) branches of the studied family. Filled symbols denote affected individuals. Genotypes for all individuals sequenced for the identified METTL23 variant are indicated (+, wild-type allele; –, mutant allele). (B–E) Facial features of the affected individuals. Individuals IV-14 (B and C), IV-16 (D) and IV-19 (E) show a flat occiput, large eyes, a depressed nasal bridge, a short upturned nose, a long philtrum and thin lips.
Table 1.
Summary of clinical findings of affected individuals in the UAE branch of the studied family
| Individual | IV-14 | IV-16 | IV-19 |
|---|---|---|---|
| Gender | Male | Male | Female |
| Age at evaluation | 26 years | 18 years | 8 years |
| Birth head circumference | Not available | 36 cm (55th percentile) | 37 cm (90th percentile) |
| Head circumference at evaluation | 55 cm (47th percentile) | 54 cm (23rd percentile) | 51.4 cm (43rd percentile) |
| Cognitive impairment | Severe, with autistic symptoms | Moderate | Moderate |
| Early motor development | Walked at 1.5 years | Walked at 1.5 years | Walked at 1.5 years |
| Muscle tone | Normal | Normal | Mild hypotonia |
| Deep tendon reflexes | Normal | Normal | Normal |
| Seizures | Yes, right-sided focal (developed at age 5 years) | Yes, generalized tonic-clonic (resolved) | No |
| Brain imaging | Not available | Not available | Normal head CT |
| Flat occiput | Yes | Yes | Yes |
| Large eyes | Yes | Yes | Yes |
| Depressed nasal bridge | Yes | Yes | Yes |
| Short, upturned nose | Yes | Yes | Yes |
| Long philtrum | Yes | Yes | Yes |
| Thin lips | Yes | Yes | Yes |
| Incomplete syndactyly | Yes | Yes | Yes |
| Other findings | Hydrocephalus (complication of meningitis at age 8 months) | Shawl scrotum | Cleft uvula, submucosal cleft palate, flat epiphyses (distal femur, proximal and distal tibia) |
Identification of the causative mutation
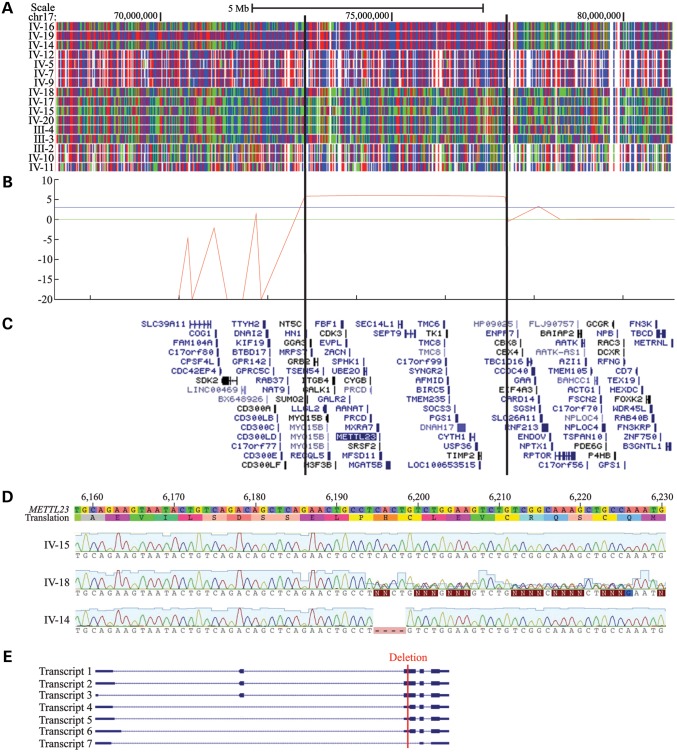
Based on the pedigree and clinical analysis, an autosomal recessive condition due to an ancestral mutation common to both branches of the studied family was suspected. Following genome-wide single-nucleotide polymorphism (SNP) genotyping and linkage analysis of 16 members of the family, only one genomic region was identified as homozygous in all affected individuals. The candidate interval was defined by markers rs820388 and rs4313838 on chromosome 17 (17q25.1–25.3) and was 14.2 cM (4.0 Mb) in size (Fig. 2A). A maximum multipoint logarithm of odds (LOD) score of 6.01 was obtained within this region (Fig. 2B). There were 72 annotated RefSeq genes in this interval, and only one gene in this interval, ACOX1, was associated with ID (MIM 264470; peroxisomal acyl-CoA oxidase deficiency). However, the reported clinical phenotype associated with ACOX1 mutations was distinct from that of the family studied herein.
Figure 2.
Mapping and sequencing of the causative mutation. (A) SNP genotyping results on chromosome 17. A block of homozygosity shared only by the affected individuals is evident and delineated by the vertical lines. Red and blue represent homozygous SNP markers and green represents heterozygous SNP markers. Individual ID numbers correspond to those in Figure 1. (B) logarithm of odds (LOD) score graph of the candidate region. A maximum multipoint LOD score of 6.01 was obtained. The blue horizontal line indicates a statistically significant LOD score of 3. (C) Genes within the candidate region, shown as the RefSeq track of the UCSC Genome Browser (GRCh build 37/hg19). Not all genes in the candidate region are shown. (D) Representative METTL23 chromatograms from wild-type, heterozygous, and homozygous affected individuals. IV-15 is a wild-type unaffected sibling, IV-18 is a heterozygous unaffected sibling and IV-14 is affected and homozygous for the deletion. A deletion of 4 bp corresponding to bases 74 729 144 through 74 729 147 on chromosome 17 (hg19) was identified in the affected individuals. (E) All seven known transcriptional variants of human METTL23 are shown, with the causative deletion indicated by a red line (Transcript 1 = NM_001080510.3, Transcript 2 = NM_001206983.1, Transcript 3 = NM_001206984.1, Transcript 4 = NM_001206985.1, Transcript 5 = NM_001206986.1, Transcript 6 = NM_001206987.1, Transcript 7 = NR_038193.1). Transcripts 1, 2 and 3 encode isoform 1, and transcripts 4, 5 and 6 encode isoform 2. According to the UCSC genome browser conventions, horizontal lines represent introns, thin blocks represent non-coding exons and thick blocks represent coding exons.
Array capture and high-throughput sequencing were performed on all exons within the candidate interval using a genomic DNA sample from a single affected individual (Fig. 2C), yielding 404 candidate variants. Among these, 274 variants were found to be homozygous, and 51 were located in exons or splice sites (first and last two bases of introns) with respect to known RefSeq genes. Only 24 of the homozygous variants were predicted to be protein-altering, and of these, only two were present at a minor allele frequency of 1% or less in the Exome Variant Server (EVS) and 1000 Genomes (1000G) database (Table 2). The remaining two candidate variants in UBALD2 and METTL23 were subjected to validation by Sanger sequencing. The variant in UBALD2 was absent by Sanger sequencing, whereas the 4-bp deletion in the METTL23 gene was confirmed (Table 3; Fig. 2D). This mutation is expected to lead to a frameshift and premature truncation (c.169_172delCACT; p.His57Valfs*11; RefSeq NM_001080510.3). Genotyping of 16 individuals confirmed that the mutation segregated with the condition in this pedigree (Fig. 1A). This deletion was absent from >800 exomes sequenced in our lab, many of which are from Middle Eastern individuals with autism and brain malformations. Additionally, it was absent from a set of 357 Saudi exomes with diverse Mendelian disorders, 112 of which have no neurological involvement. These Saudi exomes serve as a good ethnic control for the Yemini genetic background, since there has historically been tremendous mobilization of individuals of Bedouin descent across the Arabian Peninsula. This deletion is present in EVS at a low frequency (1/11615), and another nearby 2-bp deletion (c.171_172delCT; p.His57Glnfs*13) is also present at a low frequency (2/11622). No other nonsense, frameshift, or canonical splice site mutations in the METTL23 coding sequence are listed in the EVS.
Table 2.
Variant summary
| Criterion | Number of variants |
|---|---|
| Total | 404 |
| Homozygous | 274 |
| In RefSeq exon or splice sitea | 51 |
| Predicted to be protein-alteringb | 24 |
| ≤1% minor allele frequency in EVS and 1000G | 2 |
aSplice site is defined as first and last two bases of an intron.
bVariants predicted to be protein-altering were stop-gain, missense, insertion/deletion, splice site (first or last two bases of an intron), stop-loss or start-loss.
Table 3.
Validation of candidate variants on chromosome 17
| Gene symbol | Variant (cDNA; protein) | RefSeq | Sanger sequencing |
|---|---|---|---|
| METTL23 | c.169_172delCACT; p.His57Valfs*11 | NM_001080510.3 | Validated |
| UBALD2 | c.203_204insGG; p.Pro69Alafs*80 | NM_182565.3 | Not validated |
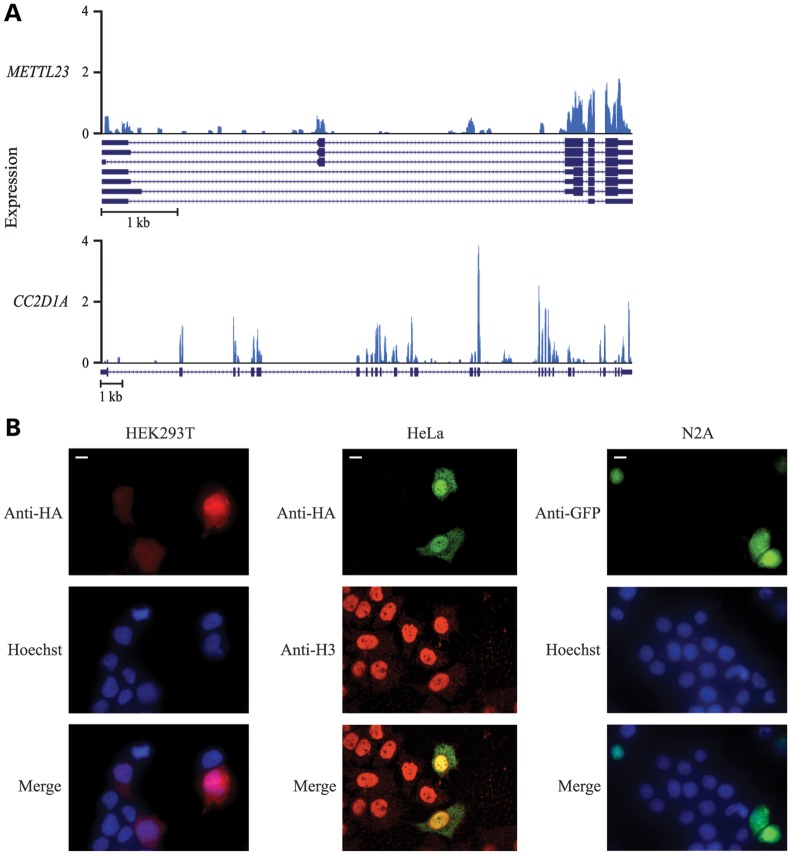
There are seven known transcription variants of METTL23 in humans, encoding two isoforms of the protein (Fig. 2E). The identified mutation falls within a coding region of three transcripts (isoform 1), a 5′ untranslated region of three other transcripts (isoform 2) and an intronic region of another presumably non-coding transcript (Fig. 2E). Transcripts in which the mutation falls within a coding region are characterized by the presence of an alternative first coding exon. This alternative exon is expressed in the developing human brain at moderate but slightly lower levels than the downstream exons (Fig. 3A), indicating that the transcripts containing this exon most likely make up a sizeable but not complete portion of the transcripts expressed in the developing human brain. Thus, the mutation could be hypomorphic in nature. Notably, only isoform 1 is conserved in most other species, and therefore, the mutation we identified affects the coding sequence of this evolutionally conserved isoform.
Figure 3.
Expression of METTL23 in the developing human brain and subcellular localization. (A) Expression of METTL23 (top) and a comparable ID gene CC2D1A (bottom) in a Carnegie stage 18 developing human brain. Reference sequences of known variants are shown below each expression plot (METTL23 transcripts = NM_001080510.3, NM_001206983.1, NM_001206984.1, NM_001206985.1, NM_001206986.1, NM_001206987.1, NR_038193.1; CC2D1A = NM_017721.4). Low signals of initial exons are an artifact of reverse transcription from the 3′ end of mRNA. (B) Immunofluorescence of HEK293T cells (left), HeLa cells (middle) and N2A cells (right) expressing HA-tagged (HEK293T and HeLa) or GFP-tagged (N2A) METTL23 protein. HEK293T cells were stained for HA (red) and the nuclear marker Hoechst (blue), HeLa cells were stained for HA (green) and the nuclear marker histone H3 (red) and N2A cells were stained for GFP (green) and Hoechst (blue). METTL23 localized throughout the nucleus and cytoplasm and showed clear nuclear enrichment. Scale bar = 10 μm.
METTL23 expression and protein localization
Our RNA-seq data set revealed that METTL23 is expressed at low-to-moderate levels in the developing human brain (Fig. 3A). This level of expression is comparable with other genes that have been implicated in ID, such as CC2D1A (Fig. 3A) (23), and equal to or greater than the expression of the most closely related family members of METTL23 (data not shown). Using confocal microscopy, we found that transfected METTL23 localized to both the cytoplasm and the nucleus in HEK293T, HeLa, and N2A cells and was enriched in the nucleus (Fig. 3B).
Physical and functional interaction between METTL23 and GABPA
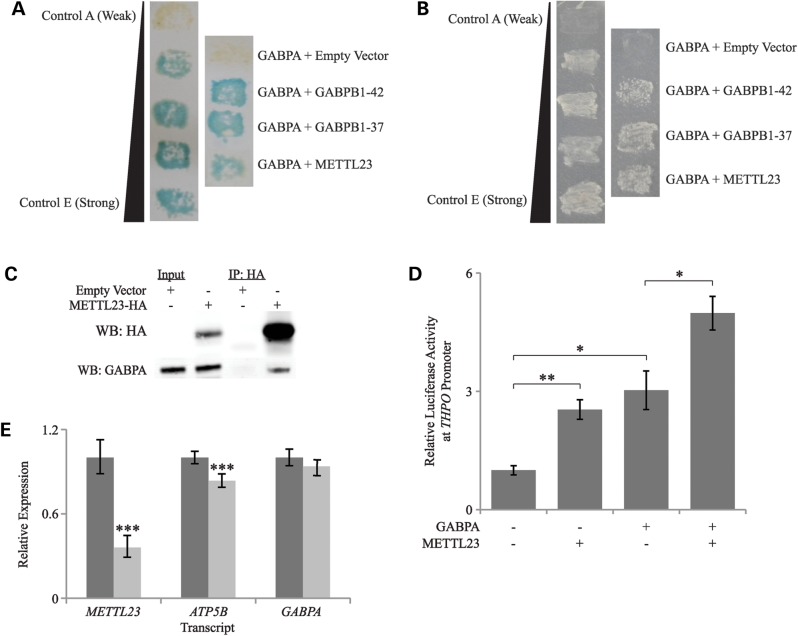
METTL23 was identified as an interacting partner of GABPA via a yeast two-hybrid screen using GABPA as bait (Fig. 4A and B). This experiment was performed independent of the human genetic studies, in order to identify novel interactors of GABPA. This screen yielded multiple positive colonies for three separate cDNAs encoding METTL23, as well as positive results for several known interactors of GABPA, including isoforms of GABPB. We confirmed the interaction between GABPA and METTL23 through co-immunoprecipitation and found that endogenous GABPA was recovered from immunoprecipitates of overexpressed tagged METTL23 in N2A cells (Fig. 4C). The co-immunoprecipitation was performed successfully in multiple cell types (HEK293T and N2A) and with or without a chemical cross-linker. To determine whether the METTL23/GABPA interaction is required for GABP function, we measured how manipulation of METTL23 expression alters the expression of GABP target genes. Through a luciferase assay, we found that the overexpression of METTL23 significantly increased transcription at the promoter of THPO, a gene that is known to be regulated by GABP. Furthermore, the positive effects of overexpressed GABPA and METTL23 were additive, indicating that METTL23 has a positive modulatory effect on GABP function via its interaction with GABPA (Fig. 4D). Next, we assessed the effects of METTL23 knockdown on the expression of the GABP target gene ATP5B using real-time quantitative reverse transcription polymerase chain reaction with gene-specific probes. In HEK293T cells, METTL23 siRNA treatment resulted in a 64% reduction in METTL23 expression, no change in GABPA expression and a modest (16.4%) but highly significant (p = 0.00052) reduction in ATP5B expression (Fig. 4E). This finding suggests that the interaction between METTL23 and GABPA is functionally significant and that the METTL23/GABP complex, rather than GABPA levels alone, regulates the expression of crucial genes such as ATP5B.
Figure 4.
Interaction of METTL23 with GABPA and functional effects on THPO and ATP5B expression. (A) Yeast two-hybrid LacZ/β-galactosidase assay, showing positive interactions of GABPA with two GABPB isoforms (GABPB1–42 and GABPB1–37) and with METTL23. Colony growth with blue color represents a positive interaction. Control interactions in the left panel, as provided by Invitrogen in the ProQuest Two-Hybrid Gateway Kit, are A (empty vectors, no interaction), B (human RB/E2F1, weak interaction), C (Drosophila DP/E2F, moderately strong interaction), D (rat c-Fos/mouse c-Jun, strong interaction) and E (GAL4/none, very strong interaction). (B) Yeast two-hybrid HIS3 assay, showing a positive interaction between GABPA and METTL23. Colony growth represents a positive interaction. Controls are the same as in part A. (C) Co-immunoprecipitation of GABPA with HA-tagged METTL23 in N2A cells, suggesting a physical interaction between the two proteins. (D) Transcriptional activity at the THPO promoter was significantly increased in the presence of overexpressed METTL23, and the positive effects of overexpressed GABPA and METTL23 were additive. Values are represented as mean ± SEM. N = 3; *P < 0.05, **P < 0.01 (Student's two-tailed t-test). (E) Real-time qRT-PCR showed a significant decrease in ATP5B mRNA expression with METTL23 knockdown in HEK293T cells. GABPA mRNA expression levels were not affected. Darkly shaded bars represent negative control siRNA and lightly shaded bars represent METTL23 siRNA. Values are represented as the mean ± pooled standard deviation. N = 4–5 for each data point; ***P < 0.001 (Student's two-tailed t-test).
Having established METTL23 as a functional interactor of GABPA, we queried our cohort of >800 exomes, mainly from consanguineous families with autism and brain malformations, for other recessive mutations in the following known GABP pathway genes: GABPA, GABPB1, GABPB2, ATF1, CREB1, SP1, SPI1, MITF, YAF2, YY1 and E2F1. Although this query did not identify any plausible candidate mutations in these genes, the sample size is too small to make definitive determination of the importance of these genes to ID.
DISCUSSION
We have described a novel cause of ID and identified METTL23 as a key gene involved in human cognition. METTL23 encodes a 190- (isoform 1) or 123- (isoform 2) amino acid protein that is predicted to contain an adenosine-methionine or lysine methyltransferase domain, but about whose function almost nothing was previously known. METTL23 belongs to a small family of similar predicted methyltransferases, which have been ascribed a variety of potential functions, including the regulation of chaperone proteins (24), protein folding, DNA repair, histone modification, splicing factor regulation and signal transduction (25). There is a precedent for other METTL genes being involved in human cognition, as METTL2B has been implicated in neurodevelopmental disorder with cognitive impairment (26).
Importantly, we have identified METTL23 as a functional interacting partner of GABPA with significant effects on transcription. The potential role of METTL23 as a transcription factor regulator is consistent with its presence in the nucleus. GABPA is a subunit of the obligate heteromeric E twenty six (ETS) transcription factor GA-binding protein [also known as nuclear respiratory factor 2 (NRF-2) and E4TF1]. The GABPA subunit contains the ETS DNA-binding motif (27) as well as a protein–protein interaction domain for binding to the GABP beta subunit (28). Briefly, GABPA binds with its near-C-terminal ETS domain to specific DNA sequences that are rich in guanine and adenine and subsequently uses its C terminus to recruit one of several GABPB isoforms to the target gene to form a tetrameric α2β2 complex. There are multiple forms of GABPB, encoded by two separate genes, and spliced into five distinct isoforms (GABPB1-42, GABPB1-41, GABPB1-38, GABPB1-37 and GABPB2) with different expression patterns. All GABPB proteins bind GABPA via a common N-terminal ankyrin domain and induce transcription via transcriptional activation domains (29). Although no functions of the N-terminal and middle regions of GABPA have been confirmed, the pointed (PNT) domain in the middle region of GABPA is predicted to mediate interactions with other proteins (29). GABPA has been found to interact with several other proteins, including Sp1 (30) and p300 (31), and mutations in the EP300 gene encoding the transcriptional co-activator p300 are known to cause Rubinstein–Taybi syndrome (MIM 613684) (32), a multiple congenital anomaly syndrome with ID. Furthermore, there is reason to believe that GABP might be important in ID, as the regulation of its dosage has been implicated in the pathophysiology of Down syndrome (33). GABP was originally identified as a regulator of viral genes, but it is now known to also activate transcription of genes that control a wide variety of cellular functions including energy metabolism, cell cycle progression, apoptosis and differentiation (29).
Among the specific genes that GABP has been shown to regulate are THPO and ATP5B, and through overexpression and knockdown experiments, we have shown that METTL23 has a functional effect on the expression of these genes via its interaction with GABPA. Both of these genes have possible roles in cognition and neurodevelopment, as THPO expressed in the brain is thought to be involved in neuroprotection, apoptosis, development and neural cell differentiation (34), whereas ATP5B has been found to be down-regulated in the thalamic region of the brains of autistic individuals (35).
Although the importance of METTL23 as a regulator of GABPA function is apparent, it is not yet clear whether METTL23 directly impacts the ability of GABPA to regulate transcription (e.g. by altering its conformation and DNA-binding affinity), or whether GABPA recruits METTL23 to methylate other proteins or DNA, thus impacting the expression of GABP target genes indirectly. The latter scenario would be similar to the case of the known GABP interactor Ying Yang 1 (YY1), which has been demonstrated to physically recruit the histone methyltransferase PRMT1 to specific promoters (36), or the case of the ETS transcription factor ETS-related gene (ERG), which has been found to regulate transcription via interaction with the histone H3-specific methyltransferase ESET (37). Although in a recent report, METTL23 did not show methyltransferase activity toward histones or amino acid homopolymers (38), it is possible that the target of METTL23 methylation simply has not yet been found. As more details about the function of METTL23 are discovered, the exact mechanism by which METTL23 and GABPA work together to affect transcription will be revealed.
In summary, we have identified a novel autosomal recessive cause of ID in a large consanguineous family and shown that the causative gene, METTL23, is expressed in the brain during development. The METTL23 gene encodes a protein that localizes throughout the cell with particular enrichment in the nucleus and interacts with a subunit of the essential transcription factor GABP. Disruption of METTL23 expression has functional effects on GABP, leading to altered expression of the GABP target genes THPO and ATP5B. Thus, the ID in this family might be caused by alterations in the expression of genes that are normally regulated by GABP.
MATERIALS AND METHODS
Subjects
Subjects were identified and evaluated in a clinical setting for medical history, cognitive impairment and physical abnormalities. We collected peripheral blood samples from the affected individuals and their family members after obtaining written informed consent according to protocols approved by the participating institutions. All research procedures were in accordance with the ethical standards of the responsible national and institutional committees on human subject research.
Genome-wide SNP genotyping and linkage
A total of 16 individuals were genotyped by either the Illumina Human610-Quad BeadChip array (the UAE branch) or Affymetrix GeneChip Human Mapping 250 K Sty Array (the KSA branch) according to the manufacturers' instructions. Genome-wide LOD scores were calculated using high-quality SNPs shared between the Illumina and Affymetrix arrays. PLINK (39) was used to reduce linkage disequilibrium between markers and MERLIN (40) was used to remove genotyping errors and calculate LOD scores, assuming an autosomal recessive mode of inheritance with 100% penetrance and a disease allele prevalence of 0.0001. Due to computational limitations, only affected individuals and their parents were included in the LOD score calculations.
Array capture and exon sequencing
We performed array capture followed by high-throughput Illumina sequencing of all exons within the candidate interval identified by homozygosity mapping. Array capture and sequencing were performed as previously described (41) in one affected individual, IV-16. Briefly, a custom array was designed with oligonucleotide probes targeting all exons within the candidate interval. DNA bound to the array was amplified and used to generate sequencing libraries. The average depth of target coverage was 1961X, with 98.1% of bases covered by ≥10 reads. A total of 404 potential sequence variants were called and filtered for quality and mapping confidence.
Variant analysis and sequencing confirmation
Variants were annotated with ANNOVAR (http://www.openbioinformatics.org/annovar/, last accessed on 11 February 2014) (42). Candidate variants were then selected using the following criteria: (1) homozygous, (2) in an exon or splice site (first or last two bases of an intron) of a RefSeq gene, (3) protein-impacting [stop-gain, missense, insertion/deletion, splice site (first or last two bases of an intron), stop-loss, start-loss], (4) minor allele frequency of 1% or less in EVS (http://evs.gs.washington.edu/EVS/, last accessed on 11 February 2014) and 1000G (http://www.1000genomes.org, last accessed on 11 February 2014). Variants that could not be verified by Sanger sequencing were also ruled out. Sequences of the primers for coding exons of METTL23 were designed with Primer3 (http://frodo.wi.mit.edu/primer3/, last accessed on 11 February 2014) (43) and are presented in Supplementary Material, Table S1. Reference sequence NM_001080510.3 was used for METTL23 mutation analysis.
RNA-Seq
RNA was isolated from the telencephalon of a Carnegie stage 18 human embryo using the mirVana Kit (Ambion). Poly-A tailed mRNA was purified using the Oligotex mRNA Mini Kit (Qiagen), and a barcoded sequencing library preserving strand information was prepared with the SOLiD Whole Transcriptome Analysis Kit (Applied Biosystems). The library was sequenced on the SOLiD version 3 Plus sequencing system. Reads were mapped with standard settings using Bioscope software v1.2 (Applied Biosystems) to the human genome reference and splice junctions were obtained from the UCSC Genes annotation track (http://genome.ucsc.edu/, last accessed on 11 February 2014). Coverage graphs were generated by normalizing the coverage of uniquely mapping reads to the number of million mapped reads.
Subcellular localization
HEK293T cells were grown on cover slips in Hyclone DMEM/High Glucose medium (Thermo Scientific) with 100 U/ml of penicillin/streptomycin (Fisher Scientific) and 10% fetal bovine serum (Sigma-Aldrich) for 24 h before being transfected with an N-terminally HA-tagged human METTL23 plasmid using Lipofectamine 2000 (Invitrogen). Approximately 30 h after transfection cells were washed once in phosphate-buffered saline (PBS, Cell Signaling) and fixed in 4% paraformaldehyde (Affymetrix) for 10 min at room temperature. Cells were then washed in PBS, permeabilized in PBS plus 0.04% Tween 20 (Sigma-Aldrich) for 30 min and blocked with 5% donkey serum (Jackson ImmunoResearch) in PBS/Tween for 1 h. Cells were incubated with a primary anti-HA antibody (1:500, Covance mouse monoclonal, clone 16B12) in PBS/Tween/donkey serum overnight at 4°C, washed three times in PBS/Tween and incubated with the secondary donkey anti-mouse antibody (1:500, Life Technologies Alexa Fluor 568) plus the nuclear stain Hoechst 33342 (10 μg/ml, Invitrogen) for 1 h at room temperature. Slides were washed three more times in PBS, mounted with Fluoromount-G (Southern Biotech) and examined under a confocal microscope. Similar subcellular localization experiments were performed in HeLa cells transfected with a C-terminally HA-tagged human METTL23 plasmid and in N2A cells transfected with a C-terminally GFP-tagged METTL23 plasmid. Antibodies used in HeLa cell staining were anti-HA (1:200, Cell Signaling mouse monoclonal), anti-H3 (1:100, Cell Signaling rabbit polyclonal), Alexa Fluor 488 goat anti-mouse (1:200, Invitrogen) and Alexa Fluor 555 donkey anti-rabbit (1:200, Invitrogen). Antibodies and stains used in N2A cell staining were anti-GFP (1:500, Abcam chicken polyclonal ab13970), donkey anti-chicken (1:500, Life Technologies Alexa Fluor 488) and the nuclear stain Hoechst 33342 (10 μg/ml, Invitrogen).
Yeast two-hybrid screen
Bait (GABPA) and prey (GABPB; GA-binding protein transcription factor, beta subunit, encoding the GABPB1-42 isoform) clones were generated using the Clonase II Kit (Invitrogen) and primers listed in Supplementary Material, Table S2, according to the manufacturer's instructions. Products were inserted into the donor vector pDONR221 using the Gateway BP Recombination Reaction (Invitrogen) and subsequently into pDEST32 (bait) and pDEST22 (prey) using the Gateway LR Recombination Reaction (Invitrogen). Constructs were amplified in the competent E. coli strain DH5α using appropriate antibiotic selection, and plasmid DNA was isolated from selected colonies using the AccuPrep Plasmid MiniPrep DNA Extraction Kit (Bioneer). Competent MaV203 yeast cells were transformed with GABPA bait according to the ProQuest Two-Hybrid System with Gateway Technology (Invitrogen) instructions (user manual version C), with OD600 = 1.5 instead of the suggested 0.3. Non-specific or self-activation of GABPA with empty prey vector was eliminated by using 10 mm 3-amino-1,2,4-triazole (3AT, an inhibitor of the HIS3 gene product).
A HeLa ProQuest pre-made cDNA library (Invitrogen) in pPC86 vector (1.5 kb average insert size) was used for the two-hybrid assay. The known interaction of GABPA and GABPB was used as a positive control, whereas the empty vector pEXP-AD502 was used as a negative control. Strong and weak positive control interactions provided by the manufacturer were also used. MaV203 cells containing GABPA bait were transformed with 5 μg of HeLa library cDNA and grown on plates lacking leucine, tryptophan and histidine with 10 mm 3AT. Obtained clones were tested using the LacZ (β-galactosidase) assay with colonies grown on an YPAD (yeast extract-peptone-adenine-dextrose) plate covered by a nylon membrane and using the HIS3 (synthetic complete medium minus leucine, tryptophan, and uracil plate) reporter gene system. Plasmid DNA was isolated for selected yeast interaction clones using Zymolyase (Zymo Research) according to the protocol provided by the manufacturer. The isolated cDNAs were sequenced and identified using the online Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed on 11 February 2014). Genbank (http://www.ncbi.nlm.nih.gov/genbank/, last accessed on 11 February 2014) data were used to confirm that full-length cDNAs had been isolated.
Co-immunoprecipitation
N2A cells were grown as described above for HEK293T cells and transfected with pcDNA3.1 (empty vector) or a C-terminally HA-tagged human METTL23 construct. After 24–48 h, cells were washed one time in PBS on ice then incubated with the chemical crosslinker dithiobis (succinimidyl propionate) (Thermo Scientific) at a 1.5 mm concentration in PBS for 30 min at room temperature. The cross-linking reaction was quenched with 15 mm Tris base (Sigma-Aldrich), pH 7.5, at room temperature for 15 min. Cells were then collected in a lysis buffer consisting of 150 mm NaCl (Sigma-Aldrich), 20 mm [4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, Sigma-Aldrich (HEPES)] at a pH of 7.4, 5 mm ethylenediaminetetraacetic acid, Sigma-Aldrich (EDTA) at a pH of 8.0, 1% Triton X-100 (Sigma-Aldrich), benzamidine hydrochloride hydrate (Sigma-Aldrich) and one protease inhibitor tablet (Roche) per 50 ml of total buffer. Cells were scraped into Eppendorf tubes and lysed end-over-end for 1 h at 4°C. Lysates were then centrifuged for 20 min at 16 000 × g at 4°C, and small amounts of the supernatants were reserved (input fractions). The remaining portions of the supernatants were incubated with pre-washed monoclonal anti-HA agarose beads (Sigma-Aldrich) end-over-end for 90 min at 4°C. Beads were then washed four times in remaining lysis buffer and resuspended in 1× Laemmli sample buffer (Sigma-Aldrich). All samples were then boiled at 100°C for 5 min before being run on a 4–12% Bis-Tris gel (Invitrogen) and transferred to a polyvinylidene fluoride membrane (Millipore). Western blotting was performed using primary antibodies anti-HA (1:5000, Abcam rabbit polyclonal ab9110) and anti-GABPA (1:500, Santa Cruz rabbit polyclonal, 22810), followed by the secondary antibody donkey anti-rabbit IRDye 800CW (1:4000, Licor).
Luciferase assay
The THPO promoter was inserted into the pGL3-Enhancer vector (Promega) using the primers listed in Supplementary Material, Table S3, and Mlul and Xhol restriction enzymes. HeLa cells were transfected with 5 ng pRL-SV40 and 200 ng pGL3-THPO promoter vectors plus 400 ng GABPA, METTL23 and/or empty pCL-Neo vectors at 2 μl Lipofectamine 2000 (Invitrogen) per 1 μg plasmid DNA per 2 × 105 cells in a 24-well plate. A Dual-Luciferase Reporter Assay (Promega) was carried out 48 h after transfection, according to the manufacturer's instructions. Firefly luciferase values were normalized to Renilla luciferase internal controls and averaged across experiments, and relative values were obtained by dividing by the mean value of the firefly reporter in cells transfected with only the reporter constructs and the empty vector.
Real-time qRT-PCR
HEK293T cells were grown as described above and transfected with 50 nm siGENOME Non-Targeting siRNA Control Pool #2 (Thermo Scientific) or 50 nm siGENOME Human METTL23 siRNA SMARTpool (Thermo Scientific). After 48 h, RNA was isolated from cells using the PureLink RNA Mini Kit (Life Technologies). Genomic DNA was removed using the PureLink DNAse Set (Life Technologies). RNA was reverse transcribed into cDNA with the SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Real-time qRT-PCR was performed with StepOne software v2.1 (Applied Biosystems) using KAPA Probe Fast ABI Prism Master Mix (Kapa Biosystems) and FAM-dyed Taqman Gene Expression Assay probes for human METTL23 (HS01047752_g1), GABPA (HS01022016_m1) and ATP5B (HS00969569_m1, Life Technologies). A VIC-dyed ACTB probe (HS01060665_g1, Life Technologies) was used as a loading control. Raw threshold cycle (CT) values were exported and ΔCT values were calculated by normalizing each sample to the loading control. ΔΔCT values were obtained by normalizing knockdown conditions to negative control siRNA conditions. Data were pooled across multiple experiments and relative expression levels were calculated.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institute of Neurological Disorders and Stroke (2R01NS035129 to C.A.W.); the National Institute of Mental Health (R01MH083565 to C.A.W.); the Fogarty International Center (R21TW008223 to C.A.W.) and also by the Simons Foundation, the Dubai Harvard Foundation for Medical Research and the Manton Center for Orphan Disease Research. R.E.R. and G.D.E. were supported by award T32GM007753 from the National Institute of General Medical Sciences. T.W.Y. was supported by the National Institutes of Health (T32NS007484-08), the Clinical Investigator Training Program at Harvard-MIT Health Sciences and Technology and Beth Israel Deaconess Medical Center in collaboration with Pfizer, Inc. and Merck & Co., Inc. and the Nancy Lurie Marks Junior Faculty MeRIT Fellowship. M.E.C. was supported by award 1F30MH702909-01 from the National Institute of Mental Health.
Supplementary Material
ACKNOWLEDGMENTS
We thank the individuals and their families reported herein for their participation in this research. SNP genotyping of the UAE branch was performed at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University through the National Institutes of Health Neuroscience Microarray Consortium. SNP genotyping of the KSA branch was performed at the Genotyping Core Facility laboratory at the King Faisal Specialist Hospital and Research Centre. Human embryonic material was provided by the joint MRC/Wellcome Trust (grant 099175/Z/12/Z) Human Developmental Biology Resource (www.HDBR.org). C.A.W. is an Investigator of the Howard Hughes Medical Institute. The content of this project is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Conflict of interest statement. T.W.Y.: personal fee and stock options, Claritas Genomics (founder and consultant); patent pending, PCT/US2011/63489. C.A.W.: grant/grant pending, National Institute of Mental Health; consulting fees, Allen Institute for Brain Science, Autism Consortium, F. Hoffmann-La Roche Ltd; patent pending, PCT/US2011/63489. G.H.M.: grants/grants pending, National Institute of Neurological Disorders and Stroke, Fogarty International Center, Simons Foundation, Dubai Harvard Foundation for Medical Research, F. Hoffmann-La Roche Ltd; personal fee/other, UCB Japan (honorarium for non-promotional lecture); personal fee, Otsuka Pharmaceutical Company Ltd (honorarium for non-promotional lecture); patent pending, PCT/US2011/63489.
REFERENCES
- 1.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Roeleveld N., Zielhuis G.A., Gabreels F. The prevalence of mental retardation: a critical review of recent literature. Dev. Med. Child Neurol. 1997;39:125–132. doi: 10.1111/j.1469-8749.1997.tb07395.x. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman L., Ayub M., Vincent J.B. The genetic basis of non-syndromic intellectual disability: a review. J. Neurodev. Disord. 2010;2:182–209. doi: 10.1007/s11689-010-9055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 5.Ropers H.H. X-linked mental retardation: many genes for a complex disorder. Curr. Opin. Genet. Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Hamdan F.F., Daoud H., Patry L., Dionne-Laporte A., Spiegelman D., Dobrzeneicka S., Rouleau G.A., Michaud J.L. Parent-child exome sequencing identifies a de novo truncating mutation in TCF4 in non-syndromic intellectual disability. Clin. Genet. 2013;83:198–200. doi: 10.1111/j.1399-0004.2012.01890.x. [DOI] [PubMed] [Google Scholar]
- 7.Schoichet S.A., Hoffmann C., Menzel U., Trautmann U., Moser B., Hoeltzenbein M., Echenne B., Partington M., Van Bokhoven H., Moraine C., et al. Mutations in the ZNF41 gene are associated with cognitive deficits: identification of a new candidate for X-linked mental retardation. Am. J. Hum. Genet. 2003;6:1341–1354. doi: 10.1086/380309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleefstra T., Yntema H.G., Oudakker A.R., Banning M.J., Kalscheuer V.M., Chelly J., Moraine C., Ropers H.H., Fryns J.P., Janssen I.M., et al. Zinc finger 81 (ZNF81) mutations assocated with X-linked mental retardation. J. Med. Genet. 2004;5:394–399. doi: 10.1136/jmg.2003.016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lugtenberg D., Yntema H.G., Banning M.J., Oudakker A.R., Firth H.V., Willatt L., Raynaud M., Kleefstra T., Fryns J.P., Ropers H.H., et al. ZNF674: a new kruppel-associated box-containing zinc-finger gene involved in nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 2006;2:265–278. doi: 10.1086/500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schanze I., Schanze D., Bacino C.A., Douzgou S., Kerr B., Zenker M. Haploinsufficiency of SOX5, a member of the SOX (SRY-related HMG-box) family of transcription factors is a cause of intellectual disability. Eur. J. Med. Genet. 2013;56:108–113. doi: 10.1016/j.ejmg.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Dragich J., Houwink-Manville I., Schanen C. Rett syndrome: a surprising result of mutation in MECP2. Hum. Mol. Genet. 2000;9:2365–2375. doi: 10.1093/hmg/9.16.2365. [DOI] [PubMed] [Google Scholar]
- 12.Jin B., Tao Q., Peng J., Soo H.M., Wu W., Ying J., Fields C.R., Delmas A.L., Liu X., Qiu J., et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 13.Petrij F., Giles R.H., Dauwerse H.G., Saris J.J., Hennekam R.C., Masuno M., Tommerup N., van Ommen G.J., Goodman R.H., Peters D.J., et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 14.Ogryzko V.V., Schiltz R.L., Russanova V., Howard B.H., Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 15.Williams S.R., Aldred M.A., Der Kaloustian V.M., Halal F., Gowans G., McLeod D.R., Zondag S., Toriello H.V., Magenis R.E., Elsea S.H. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 2010;87:219–229. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleefstra T., Smidt M., Banning M.J., Oudakker A.R., Van Esch H., de Brouwer A.P., Nillesen W., Sistermans E.A., Hamel B.C., de Bruijn D., et al. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J. Med. Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loenarz C., Ge W., Coleman M.L., Rose N.R., Cooper C.D., Klose R.J., Ratcliffe P.J., Schofield C.J. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum. Mol. Genet. 2010;19:217–222. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen L.R., Amende M., Gurok U., Moser B., Gimmel V., Tzschach A., Janecke A.R., Tariverdian G., Chelly J., Fryns J.P., et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyer J., Ekici A.B., Endele S., Popp B., Zweier C., Wiesener A., Wohlleber E., Dufke A., Rossier E., Petsch C., et al. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am. J. Hum. Genet. 2012;90:565–572. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbons R.J., Picketts D.J., Villard L., Higgs D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 21.Kamura T., Handa H., Hamasaki N., Kitajima S. Characterization of the human thrombopoietin gene promoter. A possible role of an Ets transcription factor, E4TF1/GABP. J. Biol. Chem. 1997;272:11361–11368. doi: 10.1074/jbc.272.17.11361. [DOI] [PubMed] [Google Scholar]
- 22.Villena J.A., Vinas O., Mampel T., Iglesias R., Giralt M., Villarroya F. Regulation of mitochondrial biogenesis in brown adipose tissue: nuclear respiratory factor-2/GA-binding protein is responsible for the transcriptional regulation of the gene for the mitochondrial ATP synthase beta subunit. Biochem. J. 1998;331:121–127. doi: 10.1042/bj3310121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basel-Vanagaite L., Attia R., Yahav M., Ferland R.J., Anteki L., Walsh C.A., Olender T., Straussberg R., Magal N., Taub E., et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J. Med. Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cloutier P., Lavallee-Adam M., Faubert D., Blanchette M., Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9:e1003210. doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf S.S. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol. Life Sci. 2009;66:2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talkowski M.E., Rosenfeld J.A., Blumenthal I., Pillalamarii V., Chiang C., Heilbut A., Ernst C., Hanscom C., Rossin E., Lindgren A.M., et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batchelor A.H., Piper D.E., de la Brousse F.C., McKnight S.L., Wolberger C. The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 28.Brown T.A., McKnight S.L. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 29.Rosmarin A.G., Resendes K.K., Yang Z., McMillan J.N., Fleming S.L. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol. Dis. 2004;32:143–154. doi: 10.1016/j.bcmd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Galvagni F., Capo S., Oliviero S. Sp1 and Sp3 physically interact and co-operate with GABP for the activation of the utrophin promoter. J. Mol. Biol. 2001;306:985–996. doi: 10.1006/jmbi.2000.4335. [DOI] [PubMed] [Google Scholar]
- 31.Bush T.S., St Coeur M., Resendes K.K., Rosmarin A.G. GA-binding protein (GABP) and Sp1 are required, along with retinoid receptors, to mediate retinoic acid responsiveness of CD18 (beta 2 leukocyte integrin): a novel mechanism of transcriptional regulation in myeloid cells. Blood. 2003;101:311–317. doi: 10.1182/blood.V101.1.311. [DOI] [PubMed] [Google Scholar]
- 32.Roelfsema J.H., White S.J., Ariyurek Y., Bartholdi D., Niedrist D., Papadia F., Bacino C.A., den Dunnen J.T., van Ommen G.J., Breuning M.H., et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Leary D.A., Pritchard M.A., Xu D., Kola I., Hertzog P.J., Ristevski S. Tissue-specific overexpression of the HSA21 gene GABPalpha: implications for DS. Biochim. Biophys. Acta. 2004;1739:81–87. doi: 10.1016/j.bbadis.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Diederich K., Schabitz W.R., Minnerup J. Seeing old friends from a different angle: novel properties of hematopoietic growth factors in the healthy and diseased brain. Hippocampus. 2012;22:1051–1057. doi: 10.1002/hipo.20904. [DOI] [PubMed] [Google Scholar]
- 35.Anitha A., Nakamura K., Thanseem I., Matsuzaki H., Miyachi T., Tsujii M., Iwata Y., Suuki K., Sugiyama T., Mori N. Downregulation of the expression of mitochondrial electron transport complex genes in autism brains. Brain Pathol. 2013;23:294–302. doi: 10.1111/bpa.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y.D., Yao Y.L., Gyory I., Wright K., Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L., Xia L., Wu D.Y., Wang H., Chansky H.A., Schubach W.H., Hickstein D.D., Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- 38.Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Maelandsmo G.M., Egge-Jacobsen W., Falnes P.O. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 41.Yu T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topcu M., McDonald M.T., Barry B.J., Felie J.M., et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecure. Nat. Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.