Abstract
All patients with metastatic lung, colorectal, pancreatic or head and neck cancers who initially benefit from epidermal growth factor receptor (EGFR)-targeted therapies eventually develop resistance. An increasing understanding of the number and complexity of resistance mechanisms highlights the Herculean challenge of killing tumors that are resistant to EGFR inhibitors. Our growing knowledge of resistance pathways provides an opportunity to develop new mechanism-based inhibitors and combination therapies to prevent or overcome therapeutic resistance in tumors. We present a comprehensive review of resistance pathways to EGFR-targeted therapies in lung, colorectal and head and neck cancers and discuss therapeutic strategies that are designed to circumvent resistance.
Oncologists are confronted with a formidable challenge in overcoming cancers with innate or acquired resistance to targeted therapies. This dilemma is especially acute in cancers that are dependent on EGFR activation: the initial enthusiasm over substantial clinical responses to EGFR tyrosine kinase inhibitors (TKIs) and monoclonal antibodies has now been tempered by the identification of an ever-increasing number of de novo and acquired resistance mechanisms.
EGFR addiction and signaling in cancer
EGFR (also known as ERBB1 or HER1) belongs to the ERBB family of cell-surface receptor tyrosine kinases that also includes HER2 (also known as NEU or ERBB2 (ref. 1)). EGF binding to EGFR triggers homodimerization or heterodimerization of this receptor with other ERBB members, namely HER2, receptor phosphorylation and activation of downstream effectors such as RAS–RAF–MEK–ERK–MAPK and PI3K–AKT–mTOR, leading to cell proliferation2 (Fig. 1). Other EGFR ligands include transforming growth factor-α (TGF-α), amphiregulin, epigen, betacellulin, heparin-binding EGF and epiregulin3. Wild-type EGFR signaling contributes to tumor cell proliferation, evasion of apoptosis, angiogenesis and metastasis2.
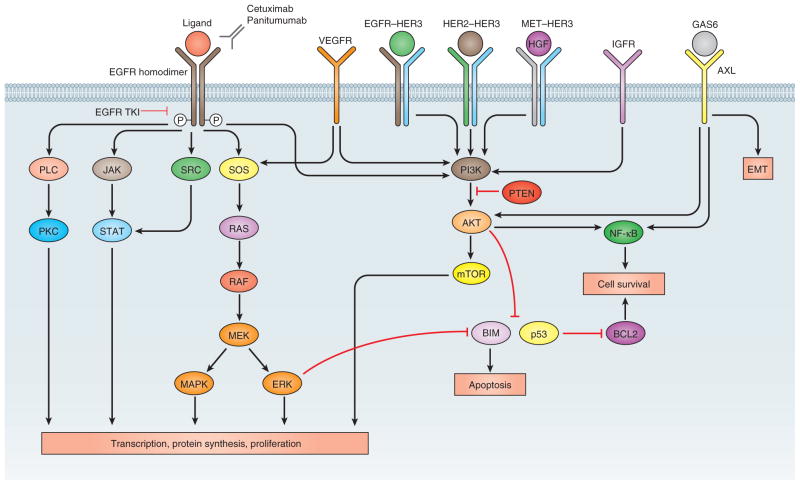
Figure 1.
EGFR signaling pathways. Activation of EGFR leads to downstream signaling pathways that ultimately drive tumor proliferation or impair apoptosis. These pathways mediate resistance through crosstalk or inappropriate activation but also provide targets for drugs to overcome resistance. IGFR, insulin-like growth factor receptor; PLC, phospholipase C; GAS6, growth arrest-specific 6.
The crucial importance of EGFR to tumor cell survival in lung adenocarcinoma highlights the concept of ‘oncogene addiction’ as defined by Weinstein in 2002 whereby a cancer cell becomes dependent on a specific oncogenic signaling pathway4. Drugs that inhibit mutant EGFR such as erlotinib turn off this key pathway and lead to tumor cell death through the BCL-2 family member BIM (also called BCL2L11). Since EGF was first identified by Stanley Cohen in 1962, considerable advances have been made in the understanding of EGF-mediated signaling and the therapeutic application of this knowledge (Fig. 2).
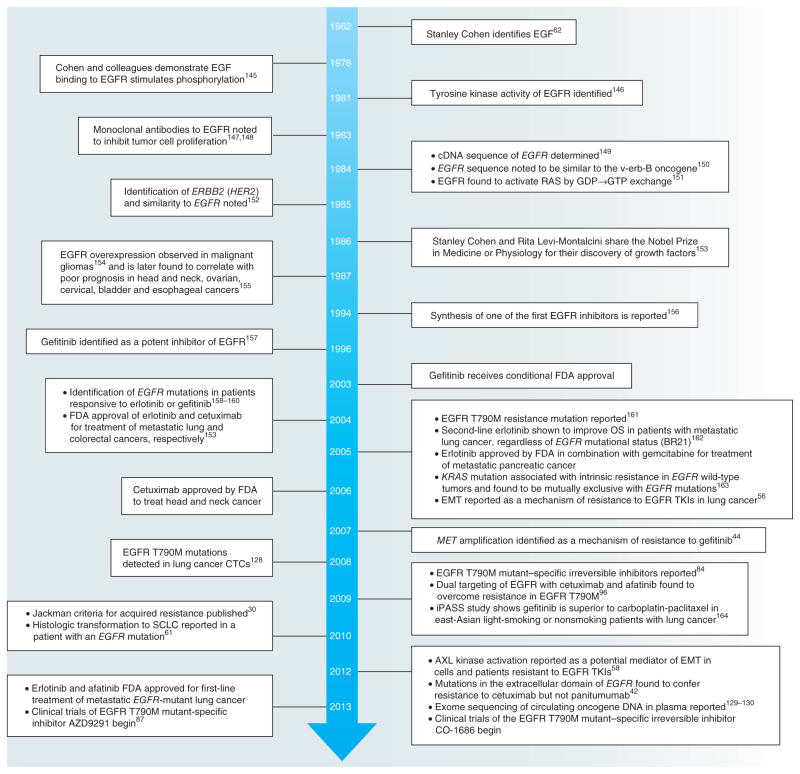
Figure 2.
Timeline of key discoveries in the EGFR field. The timeline charts important findings from basic and clinical research into EGFR and its role in cancer30,42,44,56,58,61,62,84,87,96,129,130,145–164 (adapted from ref. 153). FDA, US Food and Drug Administration; OS, overall survival; CTCs, circulating tumor cells; SCLC, small cell lung cancer; iPASS, Iressa Pan-Asia Study.
Whether there is addiction to EGFR signaling in cancers of the head and neck, colon and pancreas is less clear than in lung cancer: EGFR-targeted therapies are either combined with chemotherapy to be effective or are much less effective as single-agent therapies when compared to the initial response rates to EGFR TKIs in lung adenocarcinoma (Supplementary Table 1).
Therapeutic targeting of EGFR signaling
Therapies targeting EGFR signaling are part of the arsenal of agents that are used to treat lung, colorectal, pancreatic and head and neck cancers (Supplementary Table 1). Specific drugs used include erlotinib and gefitinib, which reversibly inhibit the EGFR tyrosine kinase domain by competitively binding with ATP, and the monoclonal antibodies (mAbs) cetuximab (a chimeric mouse-human IgG1 antibody) and panitumumab (a fully humanized IgG2 antibody). Cetuximab and panitumumab block ligand binding to the extracellular domain of EGFR, promote receptor internalization and mediate antibody- and complement-mediated cytotoxicity2. Antibody- or complement-mediated killing may be more effective with cetuximab as compared to panitumumab, as the IgG1 subclass is more effective than IgG2 at activating complement and the Fc receptor on immune effector cells5.
EGFR-activating mutations cluster in the catalytic kinase domain, are detected almost exclusively in adenocarcinomas of the lung, display up to a 50-fold increase in kinase activity by abrogating autoinhibition6 and are capable of oncogenic transformation of fibroblast and lung epithelial cells7. Activating mutations are heterozygous, and the mutant EGFR allele is frequently amplified. Although over 100 different mutations in the EGFR kinase domain have been identified in adenocarcinomas of the lung, the majority of patients harbor one of seven mutations8, the clinical properties of which are summarized in Table 1. The common EGFR-activating mutations, exon 19 deletions and L858R, which account for 85% of all EGFR mutations, predict sensitivity to the EGFR TKIs (gefitinib, erlotinib and afatinib) in preclinical models and in patients with lung cancer. With the exception of rare cases of familial lung adenocarcinoma9,10, most EGFR mutations in lung adenocarcinoma are somatic.
Table 1.
EGFR-activating and resistance mutations in adenocarcinoma of the lung
| Mutation | Frequency in EGFR-mutant lung adenocarcinoma (%) | Clinical properties
|
|||||
|---|---|---|---|---|---|---|---|
| Response rate to EGFR TKIs
|
Median PFS
|
Median OS
|
|||||
| % | Reference | Months | Reference | Months | Reference | ||
| Exon 19 deletions | 45 | 82.8 | 13 | 11.5 | 13 | 30.8 | 171 |
| 84.8 | 170 | 9.0 | 14 | 34 | 174 | ||
| 63 | 171 | 11 | 12 | 17.7 | 172 | ||
| 64 | 172 | 14.6 | 171 | 33.1 | 173 | ||
| 70 | 173 | 12 | 174 | ||||
| 9.3 | 172 | ||||||
| 9.8 | 173 | ||||||
| L858R (exon 21) | 40 | 67.3 | 13 | 10.8 | 13 | 14.8 | 171 |
| 60.9 | 170 | 9.6 | 14 | 8 | 174 | ||
| 50a | 171 | 8.4b | 12 | 20.5c | 172 | ||
| 62 | 172 | 9.7 | 171 | ||||
| 5 | 174 | ||||||
| 6.9 | 172 | ||||||
| Exon 20 insertions | 2–9 | The variable response to EGFR TKIs is thought to be related to the effect of varying insertion length on the drug-binding pocket175. Median OS of 16 months176 in one series and 4 years in another177. | |||||
| G719X | 3 | ~50 | 178 | 8.1 | 179 | 16.4 | 179 |
| L861X | 2 | 60 | 179 | 6 | 15.2 | ||
| Exon 19 insertions | 1 | Case series report responsiveness to erlotinib180 | |||||
| T790M | 0.5–3 (in some case series) | Associated with lack of response to EGFR TKIs in patients with EGFR-activating mutations41,179 | |||||
P = 0.39 compared to exon 19 deletions in this series.
P = 0.075 compared to exon 19 deletions in this series.
P = 0.65 compared to exon 19 deletions in this series.
PFS, progression-free survival; OS, overall survival.
The superiority of first-line gefitinib and erlotinib over conventional cytotoxic chemotherapy, both in terms of response rates and progression-free survival in patients with EGFR-mutant adenocarcinoma, has been well established in clinical trials in Western and Asian populations11–14. Similarities in overall survival between EGFR TKIs as compared to chemotherapy occur because patients who progress on either treatment cross over to the other; that is, a patient who progresses on an EGFR TKI will receive chemotherapy and vice versa. Trials of cetuximab in combination with chemotherapy in lung cancer have been more disappointing. Although the response rate with the addition of cetuximab is higher than that for chemotherapy alone, there was no statistically significant difference in progression-free survival and only an ~1 month improvement in overall survival in combination with cisplatin and vinorelbine15,16. Cetuximab in combination with chemotherapy or radiation in head and neck cancers has produced more encouraging results in terms of overall survival17,18. Cetuximab and panitumumab are also approved for use in colorectal cancer in combination with chemotherapy or, when other options are exhausted, as single agents19–24. Erlotinib produces an ~2 week increase in overall survival in pancreatic cancer when given in combination with gemcitabine as compared to gemcitabine monotherapy25, which is interesting given that EGFR signaling has been implicated in KRAS-mediated development of pancreatic cancer in preclinical mouse models26,27.
Mechanisms of resistance to EGFR-targeted therapies
Despite the demonstrated benefits of EGFR-targeting agents, not all patients with cancer respond to treatment, and any gain in the median progression-free survival with these therapies compared to chemotherapy is, rather disappointingly, still less than 1 year. Intrinsic, de novo or primary resistance is defined as the failure to respond to small-molecule or antibody inhibitors. Primary resistance is distinct from failure to respond due to insufficient drug exposure. This failure can occur when EGFR TKIs are coadministered with drugs such as fenofibrate, which induce CYP3A4 (thereby increasing erlotinib metabolism), or proton pump inhibitors and H2-receptor antagonists, which decrease pH-dependent drug solubility28,29. Acquired resistance occurs in patients who initially benefit from EGFR-targeted therapies. A clinical definition of acquired resistance to EGFR TKIs has been proposed and may also be expanded to also include EGFR-targeting mAbs: acquired resistance is systemic progression (by Response Evaluation Criteria in Solid Tumors (RECIST) or World Health Organization (WHO) criteria) after a complete or partial response or >6 months of stable disease after treatment with a targeted therapy30.
Resistance mechanisms to EGFR small-molecule inhibitors or antibodies that have been validated in patients may be grouped into four categories (Fig. 3, Table 2 and Supplementary Tables 1 and 2): mutation of EGFR to a drug-resistant state, for example, through the T790M or S492R mutations, which abrogate the activity of gefitinib or erlotinib and cetuximab, respectively, but do not diminish the kinase activity of the receptor; ‘oncogenic shift,’ or activation of a bypass signaling pathway (such as MET amplification, HER2 upregulation or KRAS activation)31; impairment of a pathway that is essential for EGFR TKI–mediated apoptosis, such as germline intronic deletions that remove the BH3 domain of BIM32; and histologic transformation to small cell lung cancer or an epithelial-mesenchymal transition33. All four resistance mechanisms have been observed to occur in patients with lung adenocarcinoma with resistance to EGFR TKIs, and some mechanisms, such as T790M, occur in both acquired and innate resistance34.
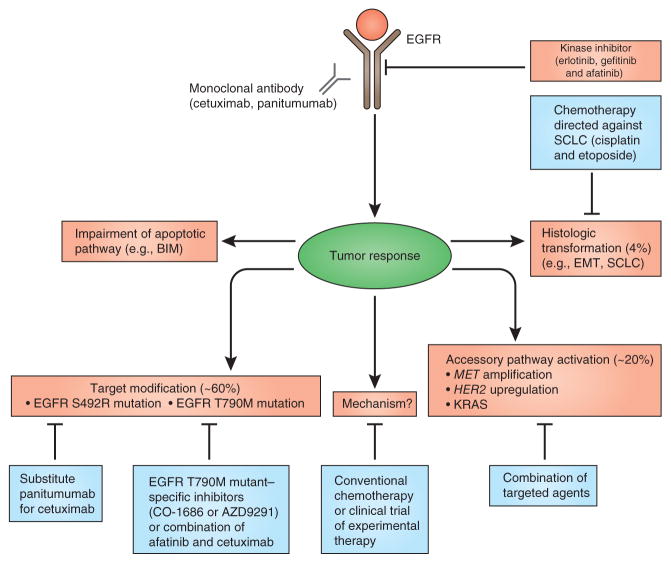
Figure 3.
Clinically validated resistance mechanisms to EGFR inhibitors. Treatment with EGFR-targeted therapy results in tumor responses, which are blunted by the selection or evolution of clones with resistant EGFR (T790M or S492R), oncogenic shift (activation, upregulation or amplification of a bypass pathway), inhibition of apoptosis or histologic transformation (figure adapted from ref. 79).
Table 2.
Overview of EGFR resistance mechanisms
| Lung (EGFR TKIs) | Colon (EGFR mAbs) | Head and neck (cetuximab) | |
|---|---|---|---|
| Primary resistance |
EGFR exon 20 insertion7,a BIM deletion32,a EGFR T790M41,179 |
KRAS47,48,a PIK3CA exon 20 (refs. 47,48)a BRAF mutation47,48,a PTEN deletion47,48,a |
|
| Acquired resistance | |||
| EGFR modification | T790M82,a | S492R42,a | EGFRvIII181 |
| Alternative pathway activation |
BRAF51,a CRKL65 DAPK182 FGF69–71 HER2 (ref. 45)a HER3 (ref. 68) IGF183,184 JAK2 (ref. 72) MED12 (ref. 185) MET44,a NF-κB186,a PTEN loss63,64 PUMA114 ROR1 (ref. 113) VEGF187 |
HER2 (ref. 46)a IGF189,a KRAS49,50,a MET190 |
Aurora191 HER2 HER3 MET192 |
| Histologic transformation | Acquisition of stem cell properties73 EMT (AXL, Notch-1 or TGF-β activation)57–60,185,a Small cell lung cancer transformation188,a |
EMT193,194 | |
Mechanisms have also been identified in patient tumors.
Secondary EGFR mutations
The most common mechanism of resistance to TKIs in EGFR-mutant lung cancer is the T790M ‘gatekeeper’ mutation, which is found in approximately 60% of patients with acquired resistance33. Secondary kinase mutations are a common mechanism of acquired resistance across other cancers that demonstrate oncogene addiction, and these mutations represent a form of oncogenic drift. Examples include ABL T315I in chronic myeloid leukemia (CML)35, KIT T670I in gastrointestinal stromal tumors36 and ALK L1196M in adenocarcinoma37. The EGFR T790M and ALK F1174L38 mutations are mechanistically similar in that they increase the kinase affinity for ATP by approximately fivefold, which decreases sensitivity to ATP-competitive reversible inhibitors such as erlotinib or crizotinib39. Germline T790M mutations have been reported in families with inherited lung cancer10. The T790M mutation lowers the growth kinetics of tumor cells40 and may be present before treatment with EGFR TKIs9,41. In colorectal cancer, an acquired mutation in the extracellular domain of EGFR (S492R) abrogates cetuximab binding, leading to clinical resistance42. Tumor cells with this mutation remain susceptible to panitumumab, and a patient with cetuximab resistance and an EGFR S492R mutation responded to panitumumab42. This difference in susceptibility between the two antibodies is likely due to differences in their interactions with EGFR, as the amino acid involved is located at the interface between antibody–receptor binding and affects the binding of cetuximab but not panitumumab.
In contrast to resistance to cytotoxic chemotherapy, which may be mediated by altered cellular import or efflux of drugs43, resistance to EGFR-targeted therapies may occur through multiple, interacting pathways (Fig. 1). Signaling pathways that stimulate cell growth may be thought of as a network in which loss of one node diverts prosurvival or proliferation stimuli through other nodes. Numerous pathways are clinically validated to trigger resistance to EGFR TKIs and monoclonal antibodies, including amplification of MET or HER2 (refs. 44–46), loss of PTEN47,48 and activation of KRAS, PIK3CA and BRAF49–52. These pathways lead to persistent activation of downstream signaling despite EGFR inhibition and hence block the apoptosis or decreased cellular proliferation that is normally mediated by EGFR inhibition. The activation of these pathways may be complementary and interchangeable across different cancers: for example, BRAF activation triggers resistance to EGFR TKIs in EGFR-mutant lung cancer51, and EGFR mediates resistance to vemurafenib in colon cancers with mutant BRAF V600E53,54.
How, when or why a tumor selects one pathway over another to overcome resistance remains unknown. For example, in lung cancer, EGFR-activating mutations are mutually exclusive to activating mutations in KRAS, whereas in colorectal cancer, KRAS activation is a mechanism of innate and acquired resistance to EGFR-targeting mAbs. Common culprits such as HER2 and MET have been implicated in resistance across lung, head and neck and colorectal cancers (Table 2). The bias of familiarity may also bend researchers’ search for resistance toward these and other well-studied pathways.
That the EGFR T790M mutation is by far the most common mechanism of resistance to EGFR TKIs in lung cancer highlights the ‘addiction’ of EGFR-mutant lung cancer: a simple amino acid substitution resurrects the ability of the tumor to proliferate when challenged with an EGFR TKI. In contrast, in colorectal cancer, resistance to EGFR-targeted mAbs is mediated primarily through other signaling networks such as KRAS, BRAF, PIK3CA and HER2 (Table 2 and Supplementary Tables 1 and 2). This distinction between EGFR functioning as a driver oncogene in a subset of lung cancers compared to its role as one of the many pathways that contribute to tumor growth in colorectal and other cancers is likely to be clinically relevant for three reasons. First, the durability and rate of response for EGFR-targeted monotherapy is much higher in lung cancers that are ‘addicted’ to EGFR signaling (response rate of 64–83% and progression-free survival of 9.7–13.1 months for erlotinib in EGFR- mutant lung cancer compared to a response rate of 12.8% and progression-free survival of 3.7 months for cetuximab monotherapy in colorectal cancer). Indeed, erlotinib behaves similarly to cetuximab in colorectal cancer in terms of response rate (9%) and progression-free survival (2.2 months) as a second-line treatment for patients with lung cancer unselected for EGFR mutation status (Supplementary Table 1). Second, the combination of erlotinib with chemotherapy in EGFR-mutant lung cancer does not improve the response rate, progression-free survival or overall survival compared to chemotherapy alone55, whereas the combination of cetuximab or panitumumab with chemotherapy in colorectal cancer results in a marked improvement in response rate and progression-free survival compared with chemotherapy alone (Supplementary Table 1). Third, it is much easier to pharmacologically overcome resistance due to a change in a driver oncogene (for example, EGFR T790M) than resistance due to activation of an accessory pathway. This is because overcoming accessory pathway activation requires combinations of targeted agents (with all the attendant toxicities, discussed below) and the activated pathway may be pharmacologically intractable (for example, KRAS).
Resistance through histologic transformation
The epithelial-to-mesenchymal transition (EMT), as manifest by loss of E-cadherin expression and increased expression of fibronectin and vimentin, was reported as an EGFR resistance mechanism in lung adenocarcinoma cell lines in 2005 (ref. 56) and in patients treated with EGFR TKIs independent of a T790M mutation in 2010 (ref. 57). This process may be mediated by the activation of AXL kinase as a potential resistance mechanism that is associated with an increase in the EMT, along with AKT activation, and the small-molecule AXL inhibitors MP-470 and XL-880 restore sensitivity to erlotinib58. Other pathways that have been reported to be involved in the histologic transformation of EGFR TKI–resistant tumors include Notch-1 and TGF-β59,60. Transformation of EGFR TKI–resistant adenocarcinoma to small cell lung cancer was reported in 2010 (ref. 61), and patients whose tumors have undergone such a change may benefit from treatment with etoposide and cisplatin, which is a standard chemotherapy regimen for small cell lung cancer. Although histologic transformation accounts for resistance in a minority of patients (~3% in some case series)33, this unique and rare phenomenon underscores the role that EGF signaling has in development, as was first observed by Stanley Cohen in 1962 (ref. 62).
Development of criteria to assess the clinical relevance of resistance mechanisms
An ever-increasing list of alternative signaling pathways and mechanisms has been reported to mediate resistance to EGFR TKIs in lung cancer cell–based models (Supplementary Table 3). These pathways and mechanisms include PTEN downregulation or loss63,64, CRKL amplification65, increased vascular endothelial growth factor (VEGF) production66, activation of HER2, HER3 or fibroblast growth factor receptor (FGFR)67–71, JAK signaling72, acquisition of stem cell–like properties73, involvement of tumor-associated fibro-blasts74 and EGFR ubiquitination75. Through modulation of microRNAs, MET expression has been proposed to upregulate genes that are involved in the EMT and downregulate the expression of genes that are involved in mediating apoptosis, such as BIM76. Liposomal transfection of lung adenocarcinoma cell lines and mouse xenograft models with microRNA targeting EGFR effectively suppresses growth of EGFR TKI–sensitive and –resistant (T790M) tumors77. EGFR-mutant cell lines propagated in the presence of an EGFR TKI demonstrate activation of other oncogenic pathways such as PI3K, AKT and HER2 and HER3—in effect, the ‘baton’ is passed from one oncogenic pathway to another. In 2010, another layer of complexity was added to the understanding of EGFR TKI resistance with the observation that chromatin modification mediated by insulin-like growth factor 1 (IGF-1) signaling confers resistance to erlotinib78. Cells that acquire resistance through chromatin modification are sensitive to histone deacetylase inhibitors, and treatment with IGF inhibitors prevents the development of resistance78.
Criteria to determine the clinical relevance of a possible resistance mechanism have been proposed and include that (i) the mechanism is necessary to generate resistance (that is, knockdown or inhibition of the resistance pathway restores sensitivity to EGFR inhibition); (ii) the mechanism is sufficient to confer resistance (that is, inappropriate activation of the mechanism confers the resistance phenotype); and (iii) the resistance mechanism is clinically observed in patients who progress on therapies that inhibit EGFR79. These criteria are helpful in distinguishing true, clinically relevant resistance mechanisms from ‘passenger’ mutations that are found in tumor evolution or mechanisms that are limited solely to in vitro observations79.
The criteria described above will be helpful in determining which combination treatments will progress to clinical trials. Additional practical criteria to move a combination treatment into the clinic include that (i) the pathway shown to mediate resistance must be detected by techniques currently used in molecular pathology (that is, DNA sequencing or immunohistochemistry); (ii) a sufficient number of patients will have the resistance mechanism to allow for statistical power in a clinical trial; and (iii) some clinical information will be available on the drug that targets the resistance pathway, for example, the drug will ideally have progressed through phase 1 as a single agent, and its toxicities should be clinically manageable. These criteria are drawn from experience in combining other targeted therapies with EGFR TKIs in lung cancer, which, as discussed below, has been difficult and disappointing.
Strategies to overcome resistance
The identification of various resistance mechanisms is essential to developing a strategy to overcome resistance and prolong the efficacy of EGFR-targeted therapies. Current clinical approaches to combat resistance in lung adenocarcinoma include irreversible and mutant-selective inhibitors of EGFR, combination of cetuximab and afatinib and combination of an EGFR inhibitor with a drug targeting a resistance pathway, such as the combination of erlotinib and a MET inhibitor (Figs. 1 and 3). As further research is performed on resistance to EGFR-targeted therapies in colorectal, head and neck and pancreatic cancers, we predict that even more therapeutic opportunities will arise.
Next-generation EGFR inhibitors
In the late 1990s, irreversible inhibitors of EGFR were developed to increase the potency of inhibition through covalent modification of Cys797 in the ATP binding cleft of EGFR, thereby reducing competition from millimolar concentrations of intracellular ATP80,81. In 2005, the activity of irreversible EGFR inhibitors against lung adenocarcinoma cells harboring an EGFR T790M mutation was noted in preclinical models82,83. This discovery was taken one step further in 2009 with the report of EGFR T790M mutant–specific inhibitors84. Irreversible EGFR inhibitors currently in clinical development include afatinib (BIBW 2992) and dacomitinib (PF00299804), which both also inhibit the kinase activity of HER2 and HER4 (ref. 85), and CO-1686, which specifically targets EGFR T790M86. AZD9291, another EGFR T790M–specific inhibitor, has shown promising activity in phase 1 trials of patients with acquired resistance through this mechanism87. The development of the irreversible EGFR inhibitors neratinib (HKI-272) and canertinib (CI-1033) in lung cancer was discontinued because of a lack of efficacy and dose-limiting diarrhea81,88. Midostaurin (PKC412), an inhibitor of protein kinase C (PKC), FLT and KIT that is currently in clinical development in acute myelogenous leukemia, was found to selectively target EGFR T790M with greater selectivity compared to wild-type EGFR than the irreversible inhibitors afatinib and neratinib89. Likewise, AP26113, an ALK inhibitor, was also found to selectively inhibit EGFR T790M90. These findings raise the intriguing possibility that midostaurin and AP26113 may have off-target effects that are similar to those of imatinib, a drug that was originally developed to inhibit BCR–ABL in chronic myelogenous leukemia but was later found to inhibit KIT in gastrointestinal stromal tumors91.
Clinical trials of patients with EGFR-mutant adenocarcinoma have demonstrated the efficacy of afatinib and dacomitinib in first-line treatment (Table 2 and Supplementary Table 4), and significant benefits in terms of response rate and progression-free survival have been seen for afatinib as compared to cytotoxic chemotherapy92. The results of second-line treatment with these inhibitors in patients with EGFR-mutant lung cancer who progress on an EGFR TKI are more disappointing, with afatinib showing an 8.2% response rate93. This result may be due to the fact that physiologic doses of current-generation irreversible EGFR TKIs do not fully inhibit EGFR T790M and in fact select for cells with amplification of this mutation94,95. Dose escalation of current irreversible EGFR inhibitors in the clinic is limited by on-target inhibition of wild-type EGFR, which leads to EGFR-mediated toxicity (skin rash). Whether mutant-selective EGFR inhibitors such as CO-1686 and AZD9291 are clinically effective in patients with EGFR T790M mutant lung cancers remains unknown, and we are still awaiting the results of ongoing clinical trials. An alternative strategy has been dual targeting of EGFR by combining afatinib and cetuximab. This approach has been found to be effective in a mouse model of EGFR T790M96, as well as in patients with EGFR-mutant lung cancer who developed acquired resistance to erlotinib97 (Supplementary Table 4). Interestingly, both patients with EGFR T790M and those without this mutation appeared to benefit from this therapeutic approach. The mechanistic basis for a benefit of afatinib and cetuximab in patients with EGFR-mutant or erlotinib-resistant lung adenocarcinoma that do not harbor a T790M mutation is not currently known. This benefit requires an irreversible EGFR inhibitor, as the combination of erlotinib and cetuximab in patients with acquired resistance to erlotinib failed to show any benefit in patients with lung cancer or in KRAS-mutant colorectal cancer98,99. Afatinib and dacomitinib have demonstrated efficacy in patients with metastatic or refractory head and neck cancer, with afatinib showing noninferiority compared to cetuximab in a phase 2 trial100,101.
Combination strategies
Preclinical studies identifying mechanisms of resistance to EGFR TKIs have been translated into several completed or ongoing clinical trials, including combinations of drugs that target MET or hepatocyte growth factor (HGF)102, dasatinib103, everolimus104, bortezomib105, bevacizumab106, sunitinib107 and cetuximab97,98. So far, however, none except the combination of afatinib and cetuximab in lung cancer98 and cetuximab and erlotinib in metastatic chemotherapy-refractory colorectal cancer99 has been very effective clinically. There are multiple potential reasons for these observations. Many clinical trials are undertaken without prospectively evaluating or targeting the specific subpopulation of drug-resistant patients when, on the basis of preclinical data, the addition of the new agent may be clinically effective. For example, MET amplification has been detected as an acquired resistance mechanism in ~10–20% of patients with lung cancer who progress on an EGFR TKI, yet clinical trials combining a MET inhibitor with erlotinib did not prospectively select for this population of patients108. A common clinical development strategy is to add a new agent to erlotinib in order to overcome erlotinib resistance. However, this strategy, although perhaps easier from a regulatory standpoint because erlotinib has been approved by the US Food and Drug Administration, does not consider that EGFR T790M is the most common mechanism of erlotinib resistance (detected in ~50–60% of patients), and it is unlikely that any erlotinib combination will overcome this specific drug resistance mechanism.
Another limitation of clinical trials involving combination therapies is the overlapping toxicities of agents that make up the combination. The majority of kinase inhibitors are administered daily, and the dose taken forward for clinical development is based on toxicity and not target inhibition. This approach predicts a high likelihood that any combination approach will be more toxic, and therefore the tolerable doses of each agent in the combination may be suboptimal. In a phase 1/2 trial of erlotinib and XL-184, which inhibits MET, vascular endothelial growth factor receptor 2 (VEGFR2) and RET, the recommended phase 2 doses of the combination were 50 mg of erlotinib and 125 mg of XL-184, both of which are below the registered single-agent doses of each109. Significant dose-limiting toxicities were encountered at higher doses of each drug when given in combination109. Given the lack of efficacy of the combinations of targeted therapies tested so far and the dose-limiting toxicities encountered, the ideal partner to an EGFR TKI may be a drug that acts by an entirely different mechanism. For example, promising candidates that have been proposed include antibodies to the T cell inhibitory receptor programmed death-1 (PD1) or its ligand (PD-L1), given the efficacy of this therapy in patients with cancer who have progressed on multiple lines of treatment110,111. However, whether the expression of PD1 or PD-L1 is upregulated in EGFR inhibitor–resistant cancers remains to be determined.
One potential solution to overcome multiple mechanisms of resistance is to target downstream pathways that mediate the balance between survival and apoptosis. ROR1 is a pseudokinase that is regulated by the homeodomain transcription factor NKX2-1, is essential for lung development112 and is thought to regulate the balance between survival and apoptosis. Knockdown of ROR1 is sufficient to inhibit the growth of lung cancer cell lines with multiple mechanisms of acquired resistance, including EGFR T790M, MET amplification and HGF overexpression113. PUMA is a BH3 BCL-2 effector of apoptosis that is induced along with BIM after EGFR inhibition and is essential for apoptosis induced by EGFR TKIs114. Inhibition of PI3K–AKT signaling leads to PUMA expression through nuclear translocation of the FOXO transcription factors114. A therapeutic strategy that directly activates the machinery that is necessary for apoptosis may circumvent multiple mechanisms of acquired resistance to EGFR-targeted therapies and may therefore be a broader approach than aiming to inhibit one resistance mechanism at a time. Key to the success of such an approach is ensuring specificity in targeting cancer cells while avoiding toxicity in the rapidly dividing cells found in the bone marrow and epithelium.
Ongoing research and challenges
Origins of resistance
Whether the resistance mechanisms to EGFR-targeted therapy highlighted in this article (Table 2 and Supplementary Tables 2 and 3) evolve under selective pressure from drug exposure or already exist before treatment is a matter of ongoing investigation. What is clear is that tumors that rely on EGFR signaling may draw on an arsenal of resistance mechanisms to overcome targeted therapy. This capability manifests in the clinic through response rates and times to progression that are disappointing compared to those seen with TKIs in the treatment of CML.
An analogy has been made between drug resistance in HIV and targeted therapies in cancer: both diseases are subject to high mutational rates, may benefit from combination treatment and may be difficult to eradicate entirely115,116. Re-biopsy studies in patients with EGFR-mutant, erlotinib-resistant lung adenocarcinoma at progression suggest that the number of resistance mutations may be limited to a handful of suspects, including the T790M mutation (63% of patients), small cell lung cancer transformation (3%), amplification of MET (5%) or HER2 (13%) or overlapping mechanisms (4%)33. However, such reports may underestimate the number of resistance mechanisms, as resistance pathways that have been reported more recently, such as nuclear factor-κB (NF-κB), were not examined. Upfront use of combination therapies with different mechanisms, as is done in HIV treatment, may circumvent the development of resistance. Unlike HIV, which has a defined set of foreign protein drug targets—reverse transcriptase, protease, integrase and gp41—cancer may appropriate numerous normal signaling pathways, and inhibiting these pathways may not be tolerated, as this inhibition affects the normal physiologic functions of these pathways. In this Review, in which we evaluated ~5,000 abstracts published on EGFR in cancer, we counted at least 20 different nodes in various signaling networks that have been reported to mediate resistance to EGFR-targeting strategies (Table 2). Given that each pathway is comprised of multiple protein members, each of which may undergo mutation, the number of potential resistance pathways is staggering.
Tumor heterogeneity and resistance
Because of genetic heterogeneity, tumors may exhibit different mechanisms of resistance at different sites within a patient. For example, the degree of MET amplification with or without a T790M mutation varied among metastatic sites sampled at autopsy from patients who died of TKI-resistant EGFR-mutant lung adenocarcinoma117, and MET amplification and the T790M mutation have been observed in the same tumor118. Discordance in EGFR mutation status within a tumor or at a metastatic site has been observed and was proposed to explain the mixed responses to EGFR TKI treatment119–121. As more sensitive sequencing methods are used, however, heterogeneity in EGFR mutations is noted to occur much less frequently than when less sensitive methods are used121,122. KRAS mutations are associated with decreased responsiveness to EGFR TKIs or monoclonal antibodies in patients unselected for EGFR mutational status123. Discrepancy between KRAS mutational status between the primary tumor and a metastatic site has been noted in a patient with EGFR-mutant lung cancer124. Tumor heterogeneity may also be of therapeutic benefit after resistance occurs, as clinical responses to the reintroduction of EGFR TKIs have been noted125. The T790M mutation confers a growth disadvantage, and after removal of selective pressure from an EGFR TKI, further tumor growth may be driven by clones lacking T790M, leaving the tumor vulnerable to retreatment by an EGFR TKI40.
The lack of complete understanding of genetic heterogeneity in patients with cancer can limit the ability to develop and/or interpret the efficacy of therapies and select patients for clinical trials that are biomarker driven. Because only one site of drug resistance is typically biopsied and the mechanism of resistance is determined from that biopsy, we may often assume that all of the non-biopsied sites of disease harbor the same mechanism of resistance. It is crucial to bear in mind that sequence data from a tumor biopsy represents the genetic makeup of that isolated piece of tissue. More data from clinical trials and sequential biopsies from multiple sites are needed to distinguish whether a drug fails because of lack of efficacy against one particular resistance mechanism or because of resistance heterogeneity.
Monitoring the evolution of drug resistance
How can a strategy that pairs a resistance mechanism with a therapy directed at overcoming resistance be applied clinically? One can imagine application of an adaptive clinical trial design to combat resistance to EGFR TKIs126: patients with EGFR-mutant lung cancer who progress on primary TKI therapy undergo either re-biopsy or the detection of circulating tumor DNA or tumor cells to analyze mechanisms of resistance. These patients may then be adaptively randomized to receive additional therapy that specifically targets their resistance mechanism (for example, treatment with a MET or IκB kinase inhibitor). The success of this approach depends on the ability to rapidly detect resistance mechanisms using small amounts of DNA. The EGFR T790M mutation has been successfully detected in serum127 and circulating tumor cells128, and sequencing of EGFR, TP53, BRAF and RAS in circulating tumor DNA has been reported129,130. Proteomic attempts to determine serum biomarkers that predict susceptibility and resistance to EGFR TKIs have shown some promise in lung and head and neck cancers131,132. In the next decade, patients who progress on an EGFR-targeted therapy may undergo circulating tumor or tumor cell DNA analysis and be paired with a therapy that is tailored to overcome resistance. Coupled with advances in sequencing technologies, including targeted next-generation sequencing and whole-exome sequencing, these technologies may be able to detect drug resistance mechanisms noninvasively before clinical resistance or clinical consequences develop.
Preventing compared to treating drug resistance
Another method to delay the development of resistance involves alteration of EGFR TKI dose and schedule on the basis of the observation that drug-sensitive cells grow more rapidly than those with an acquired EGFR T790M mutation40. This phenomenon has also been reported in a study of patient-derived mouse xenografts of BRAF V600E melanoma, which demonstrated that discontinuous dosing strategies may prolong the duration of vemurafenib response as a result of drug dependency in resistant cells133. Tumor flares have been reported in patients who discontinue an EGFR TKI because of disease progression or inability to tolerate treatment104,134,135, as well as in patients with progression on an EGFR TKI who have responded after a treatment ‘holiday’136,137. In both cases, disease progression on EGFR TKI treatment is likely due to slow growth of a resistant clone (such as T790M) followed by fast growth of sensitive clones once the selective pressure of the EGFR TKI is removed. Mathematical and evolutionary modeling support the idea that high-dose pulses of EGFR TKI therapy along with low-dose maintenance may prevent the emergence of resistance40,138. In a case report, a patient who experienced progression on erlotinib and afatinib demonstrated a remarkable response to weekly high-dose (1,500 mg) erlotinib139. A relationship between drug plasma concentration and imatinib efficacy is observed in the treatment of CML: patients who achieve higher imatinib plasma concentrations have an increased response rate that is more rapid and durable compared to patients with lower plasma concentrations140,141. Meta-analyses show that the presence of a rash after EGFR TKI treatment, which indicates inhibition of cutaneous EGFR signaling, correlates with improved progression-free and overall survival142,143.
Preventing the emergence of drug resistance is an alternative strategy to developing a treatment approach for each of the potential resistance mechanisms. If a strategy could be developed that prevented not just one but a broad array of drug resistance mechanisms, it could have substantial clinical impact. However, it is not currently clear which combinations of agents need to be combined with EGFR TKIs or monoclonal antibodies to achieve this effect. In addition, any such combination needs to be tolerable, as the anticipation and hope would be for a longer duration of treatment than what is currently achievable.
Combining targeted therapies by understanding differences in effectiveness across various cancers
Oncologists hope to cure cancer; if that is not possible, they hope to turn it into a chronic disease, and if that is not possible, they aim to provide a longer progression-free survival. This spectrum is seen in targeted agents that cure certain cancers (retinoic acid in combination with chemotherapy in acute promyelocytic leukemia), turn other cancers into chronic diseases (imatinib in CML) and increase progression-free survival in others (imatinib in gastrointestinal stromal tumors, crizotinib, erlotinib and gefitinib in lung cancer and vemurafenib in melanoma) (Fig. 4). It is not clear whether differences in the biology of the tumors (that is, liquid as compared to solid) or the activating pathways underlie the variations in effectiveness of different targeted agents in different cancers. One potential explanation lies in the high mutation rates described as ‘genomic chaos’ that are observed in solid tumors144. Other explanations may include a higher degree of clonality and greater drug exposure in hematologic as compared to solid tumors. The situation we are currently in with creating combinations of targeted therapies may parallel the early days of cytotoxic chemotherapy before the advent of combination treatments, such as 5-fluorouracil, oxaliplatin and leucovorin (FOLFOX), cyclophosphamide, doxorubicin, vincristine and prednisone (CHOP), bleomycin, etoposide and cisplatin (BEP) and doxorubicin, cyclophosphamide and paclitaxel (AC→T), that are now well known to patients and oncologists. Determination of optimal therapeutic sequences and combinations will likely take dozens of clinical trials involving thousands of patients. One can hope that knowledge of resistance mechanisms will accelerate clinical translation by guiding trial design.
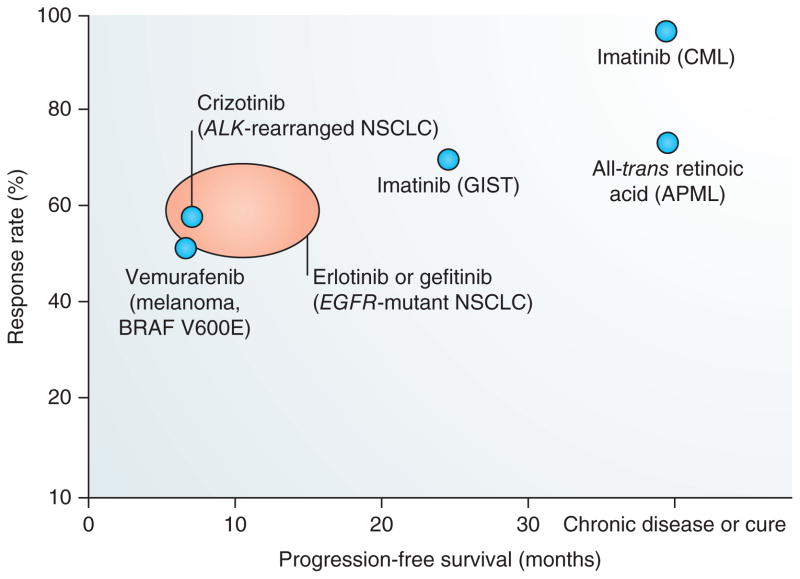
Figure 4.
Efficacy of targeted therapies in cancer treatment. Response rates and impact on progression-free survival vary widely among targeted therapies. The orange oval represents the reported response rates and progression-free survival for EGFR TKIs in EGFR-mutant lung adenocarcinoma (adapted from Supplementary Table 1). Imatinib targets BCR–ABL, has a ~95% response rate and has turned CML into a chronic disease for many patients165. All-trans retinoic acid targets the PML–RAR fusion protein and may cure acute promyelocytic leukemia (APML) when given in combination with chemotherapy166. For solid tumors such as melanoma and lung cancer, targeted therapies have a lower response rate and less of an impact on progression-free survival167–169. GIST, gastrointestinal stromal tumors.
Conclusions
Oncogene addiction was described by Weinstein as being the “Achilles heel of cancer”4. A decade later, therapies targeted to some oncogenes grant a reprieve to patients lucky enough to have select driver mutations, and remissions last years in some cases (Fig. 4). For patients with EGFR-mutant lung cancer, the outlook, although better than in patients without such a mutation, is guarded, with tumor responses lasting months if they occur at all. Cancer may be more like the mythical, multiheaded Hydra battled by Hercules because of its ability to resist targeted therapies through evolution and tumor heterogeneity.
One lesson for cancer researchers and clinicians is the importance of perseverance, creativity and collaboration. Whether we defeat tumor resistance using combinations of drugs, immunotherapy, new dosing strategies or an as-yet-undiscovered approach, our success cannot come soon enough for our patients.
Supplementary Material
Acknowledgments
This study is supported by US National Institutes of Health grants RO1CA114465 (P.A.J.), R01CA135257 (P.A.J.) and P01CA154303 (P.A.J.) and by an American Society of Clinical Oncology Young Investigator Award (C.R.C.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Cataldo VD, Gibbons DL, Perez-Soler R, Quintas-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364:947–955. doi: 10.1056/NEJMct0807960. [DOI] [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 3.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 5.Campoli M, Ferris R, Ferrone S, Wang X. Immunotherapy of malignant disease with tumor antigen-specific monoclonal antibodies. Clin Cancer Res. 2010;16:11–20. doi: 10.1158/1078-0432.CCR-09-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yun CH, et al. Structures of lung cancer–derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greulich H, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh P, et al. DNA-mutation Inventory to Refine and Enhance Cancer Treatment (DIRECT): a catalog of clinically relevant cancer mutations to enable genome-directed anticancer therapy. Clin Cancer Res. 2013;19:1894–1901. doi: 10.1158/1078-0432.CCR-12-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oxnard GR, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7:1049–1052. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell DW, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 11.Zhou C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 12.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 13.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 14.Mitsudomi T, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 15.Pirker R, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373:1525–1531. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 16.Lynch TJ, et al. Cetuximab and first-line taxane/carboplatin chemotherapy in advanced non-small-cell lung cancer: results of the randomized multicenter phase III trial BMS099. J Clin Oncol. 2010;28:911–917. doi: 10.1200/JCO.2009.21.9618. [DOI] [PubMed] [Google Scholar]
- 17.Vermorken JB, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 18.Bonner JA, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 19.Amado RG, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham D, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 21.Douillard JY, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 22.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 23.Peeters M, et al. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:4706–4713. doi: 10.1200/JCO.2009.27.6055. [DOI] [PubMed] [Google Scholar]
- 24.Van Cutsem E, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 25.Moore MJ, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 26.Ardito CM, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navas C, et al. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budha NR, et al. Drug absorption interactions between oral targeted anticancer agents and PPIs: is pH-dependent solubility the Achilles heel of targeted therapy? Clin Pharmacol Ther. 2012;92:203–213. doi: 10.1038/clpt.2012.73. [DOI] [PubMed] [Google Scholar]
- 29.Mir O, Blanchet B, Goldwasser F. Drug-induced effects on erlotinib metabolism. N Engl J Med. 2011;365:379–380. doi: 10.1056/NEJMc1105083. [DOI] [PubMed] [Google Scholar]
- 30.Jackman D, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng KP, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 33.Yu HA, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JK, et al. Primary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory study. Ann Oncol. 2013;24:2080–2087. doi: 10.1093/annonc/mdt127. [DOI] [PubMed] [Google Scholar]
- 35.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 36.Heinrich MC, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 37.Choi YL, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 38.Bresler SC, et al. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med. 2011;3:108ra114. doi: 10.1126/scitranslmed.3002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chmielecki J, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inukai M, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 42.Montagut C, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 43.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 44.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 45.Takezawa K, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yonesaka K, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laurent-Puig P, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 48.Sartore-Bianchi A, et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS ONE. 2009;4:e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diaz LA, Jr, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohashi K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci USA. 2012;109:E2127–E2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sartore-Bianchi A, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 53.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prahallad A, et al. Unresponsiveness of colon cancer to BRAFV600E inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 55.Janne PA, et al. Randomized phase II trial of erlotinib alone or with carboplatin and paclitaxel in patients who were never or light former smokers with advanced lung adenocarcinoma: CALGB 30406 trial. J Clin Oncol. 2012;30:2063–2069. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomson S, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 57.Uramoto H, et al. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res. 2010;30:2513–2517. [PubMed] [Google Scholar]
- 58.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie M, et al. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem. 2012;113:1501–1513. doi: 10.1002/jcb.24019. [DOI] [PubMed] [Google Scholar]
- 60.Yao Z, et al. TGF-β IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alam N, et al. Small-cell carcinoma with an epidermal growth factor receptor mutation in a never-smoker with gefitinib-responsive adenocarcinoma of the lung. Clin Lung Cancer. 2010;11:E1–E4. doi: 10.3816/CLC.2010.n.046. [DOI] [PubMed] [Google Scholar]
- 62.Cohen S. Isolation of a mouse submaxillary gland protein accelerating incisor eruption and eyelid opening in the new-born animal. J Biol Chem. 1962;237:1555–1562. [PubMed] [Google Scholar]
- 63.Sos ML, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suda K, et al. Conversion from the “oncogene addiction” to “drug addiction” by intensive inhibition of the EGFR and MET in lung cancer with activating EGFR mutation. Lung Cancer. 2012;76:292–299. doi: 10.1016/j.lungcan.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 65.Cheung HW, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1:608–625. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Viloria-Petit A, et al. Acquired resistance to the antitumor effect of epidermal growth factor receptor-blocking antibodies in vivo: a role for altered tumor angiogenesis. Cancer Res. 2001;61:5090–5101. [PubMed] [Google Scholar]
- 67.Wang SE, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 68.Zhou BB, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terai H, et al. Activation of the FGF2-FGFR1 autocrine pathway: a novel mechanism of acquired resistance to gefitinib in NSCLC cells. Mol Cancer Res. 2013;11:759–767. doi: 10.1158/1541-7786.MCR-12-0652. [DOI] [PubMed] [Google Scholar]
- 70.Ware KE, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ware KE, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS ONE. 2010;5:e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harada D, et al. JAK2-related pathway induces acquired erlotinib resistance in lung cancer cells harboring an epidermal growth factor receptor-activating mutation. Cancer Sci. 2012;103:1795–1802. doi: 10.1111/j.1349-7006.2012.02363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shien K, et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell–like properties in cancer cells. Cancer Res. 2013;73:3051–3061. doi: 10.1158/0008-5472.CAN-12-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mink SR, et al. Cancer-associated fibroblasts derived from EGFR-TKI–resistant tumors reverse EGFR pathway inhibition by EGFR-TKIs. Mol Cancer Res. 2010;8:809–820. doi: 10.1158/1541-7786.MCR-09-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu Y, et al. Epidermal growth factor receptor (EGFR) ubiquitination as a mechanism of acquired resistance escaping treatment by the anti-EGFR monoclonal antibody cetuximab. Cancer Res. 2007;67:8240–8247. doi: 10.1158/0008-5472.CAN-07-0589. [DOI] [PubMed] [Google Scholar]
- 76.Garofalo M, et al. EGFR and MET receptor tyrosine kinase–altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Rai K, et al. Liposomal delivery of microRNA-7–expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther. 2011;10:1720–1727. doi: 10.1158/1535-7163.MCT-11-0220. [DOI] [PubMed] [Google Scholar]
- 78.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garraway LA, Janne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 80.Bridges AJ. The rationale and strategy used to develop a series of highly potent, irreversible, inhibitors of the epidermal growth factor receptor family of tyrosine kinases. Curr Med Chem. 1999;6:825–843. [PubMed] [Google Scholar]
- 81.Kwak E. The role of irreversible HER family inhibition in the treatment of patients with non-small cell lung cancer. Oncologist. 2011;16:1498–1507. doi: 10.1634/theoncologist.2011-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 83.Kwak EL, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dienstmann R, De Dosso S, Felip E, Tabernero J. Drug development to overcome resistance to EGFR inhibitors in lung and colorectal cancer. Mol Oncol. 2012;6:15–26. doi: 10.1016/j.molonc.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu HA, Riely GJ. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in lung cancers. J Natl Compr Canc Netw. 2013;11:161–169. doi: 10.6004/jnccn.2013.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ranson M, et al. Preliminary results from a Phase I study with AZD9291: an irreversible inhibitor of epidermal growth factor receptor (EGFR) activating and resistance mutations in non-small cell lung cancer (NSCLC). European Cancer Conference; Amsterdam. 2013. [Google Scholar]
- 88.Ou SH. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): a better mousetrap? A review of the clinical evidence. Crit Rev Oncol Hematol. 2012;83:407–421. doi: 10.1016/j.critrevonc.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 89.Lee HJ, et al. Noncovalent wild-type-sparing inhibitors of EGFR T790M. Cancer Discov. 2013;3:168–181. doi: 10.1158/2159-8290.CD-12-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rivera VM, et al. AP26113 is a dual ALK/EGFR inhibitor: characterization against EGFR T790M in cell and mouse models of NSCLC. Cancer Res. 2012;72(suppl 1):abstract 1794. [Google Scholar]
- 91.Red Brewer M, Pao W. Maximizing the benefits of off-target kinase inhibitor activity. Cancer Discov. 2013;3:138–140. doi: 10.1158/2159-8290.CD-12-0581. [DOI] [PubMed] [Google Scholar]
- 92.Yang JC, Schuler MH, Yamamoto N, O’Byrne KJ, Hirsh V. LUX-Lung 3: a randomized, open-label, phase III study of afatinib versus pemetrexed and cisplatin as first-line treatment for patients with advanced adenocarcinoma of the lung harboring EGFR-activating mutations. J Clin Oncol. 2012;30:LBA7500. [Google Scholar]
- 93.Atagi S, Katakami N, Hida T, Goto K, Horai T. LUX-Lung 4: a phase II trial of afatinib (BIBW 2992) in advanced NSCLC patients previously treated with erlotinib or gefitinib. 14th World Conference on Lung Cancer; Amsterdam. 2011. [Google Scholar]
- 94.Ercan D, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Godin-Heymann N, et al. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 96.Regales L, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Janjigian Y, et al. Activity and tolerability of afatinib (BIBW 2992) and cetuximab in NSCLC patients with acquired resistance to erlotinib or gefitinib. J Clin Oncol. 2011;29:7525. [Google Scholar]
- 98.Janjigian YY, et al. Phase I/II trial of cetuximab and erlotinib in patients with lung adenocarcinoma and acquired resistance to erlotinib. Clin Cancer Res. 2011;17:2521–2527. doi: 10.1158/1078-0432.CCR-10-2662. [DOI] [PubMed] [Google Scholar]
- 99.Weickhardt AJ, et al. Dual targeting of the epidermal growth factor receptor using the combination of cetuximab and erlotinib: preclinical evaluation and results of the phase II DUX study in chemotherapy-refractory, advanced colorectal cancer. J Clin Oncol. 2012;30:1505–1512. doi: 10.1200/JCO.2011.38.6599. [DOI] [PubMed] [Google Scholar]
- 100.Abdul Razak AR, et al. A phase II trial of dacomitinib, an oral pan-human EGF receptor (HER) inhibitor, as first-line treatment in recurrent and/or metastatic squamous-cell carcinoma of the head and neck. Ann Oncol. 2013;24:761–769. doi: 10.1093/annonc/mds503. [DOI] [PubMed] [Google Scholar]
- 101.Seiwert T, et al. A randomized, open-label, phase II study Of afatinib (bibw 2992) versus cetuximab in recurrent or metastatic squamous cell carcinoma of the head and neck—final data. Multidisciplinary Head and Neck Symposium presentation LBPV10; 2012. [Google Scholar]
- 102.Robinson KW, Sandler AB. The role of MET receptor tyrosine kinase in non-small cell lung cancer and clinical development of targeted anti-MET agents. Oncologist. 2013;18:115–122. doi: 10.1634/theoncologist.2012-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johnson ML, et al. Phase II trial of dasatinib for patients with acquired resistance to treatment with the epidermal growth factor receptor tyrosine kinase inhibitors erlotinib or gefitinib. J Thorac Oncol. 2011;6:1128–1131. doi: 10.1097/JTO.0b013e3182161508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Riely GJ, et al. Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clin Cancer Res. 2007;13:5150–5155. doi: 10.1158/1078-0432.CCR-07-0560. [DOI] [PubMed] [Google Scholar]
- 105.Lynch TJ, et al. A randomized phase 2 study of erlotinib alone and in combination with bortezomib in previously treated advanced non-small cell lung cancer. J Thorac Oncol. 2009;4:1002–1009. doi: 10.1097/JTO.0b013e3181aba89f. [DOI] [PubMed] [Google Scholar]
- 106.Herbst RS, et al. Efficacy of bevacizumab plus erlotinib versus erlotinib alone in advanced non-small-cell lung cancer after failure of standard first-line chemotherapy (BeTa): a double-blind, placebo-controlled, phase 3 trial. Lancet. 2011;377:1846–1854. doi: 10.1016/S0140-6736(11)60545-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scagliotti GV, et al. Sunitinib plus erlotinib versus placebo plus erlotinib in patients with previously treated advanced non-small-cell lung cancer: a phase III trial. J Clin Oncol. 2012;30:2070–2078. doi: 10.1200/JCO.2011.39.2993. [DOI] [PubMed] [Google Scholar]
- 108.Sequist LV, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 109.Wakelee HA, et al. A phase Ib/II study of XL184 (BMS 907351) with and without erlotinib in patients (pts) with non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28(suppl 15):abstract 3017. [Google Scholar]
- 110.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Topalian SL, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. Ror1 is a pseudokinase that is crucial for Met-driven tumorigenesis. Cancer Res. 2011;71:3132–3141. doi: 10.1158/0008-5472.CAN-10-2662. [DOI] [PubMed] [Google Scholar]
- 113.Yamaguchi T, et al. NKX2–1/TITF1/TTF-1–induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell. 2012;21:348–361. doi: 10.1016/j.ccr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 114.Bean GR, et al. PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci Signal. 2013;6:ra20. doi: 10.1126/scisignal.2003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bock C, Lengauer T. Managing drug resistance in cancer: lessons from HIV therapy. Nat Rev Cancer. 2012;12:494–501. doi: 10.1038/nrc3297. [DOI] [PubMed] [Google Scholar]
- 116.Chmielecki J, Pao W. Highly active antitumor therapy (HAATT) for epidermal growth factor receptor–mutant lung cancer. Clin Cancer Res. 2010;16:5371–5373. doi: 10.1158/1078-0432.CCR-10-2405. [DOI] [PubMed] [Google Scholar]
- 117.Suda K, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res. 2010;16:5489–5498. doi: 10.1158/1078-0432.CCR-10-1371. [DOI] [PubMed] [Google Scholar]
- 118.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Park S, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4:809–815. doi: 10.1097/JTO.0b013e3181a94af4. [DOI] [PubMed] [Google Scholar]
- 121.Chen ZY, et al. EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist. 2012;17:978–985. doi: 10.1634/theoncologist.2011-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 123.Linardou H, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 124.Sun L, et al. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Cancer Res. 2011;30:30. doi: 10.1186/1756-9966-30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim ES, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuang Y, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Forshew T, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 130.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 131.Box C, et al. A novel serum protein signature associated with resistance to epidermal growth factor receptor tyrosine kinase inhibitors in head and neck squamous cell carcinoma. Eur J Cancer. 2013;49:2512–2521. doi: 10.1016/j.ejca.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 132.Taguchi F, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 133.Das Thakur M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Milton DT, et al. Molecular on/off switch. J Clin Oncol. 2006;24:4940–4942. doi: 10.1200/JCO.2006.06.4410. [DOI] [PubMed] [Google Scholar]
- 135.Chaft JE, et al. Disease flare after tyrosine kinase inhibitor discontinuation in patients with EGFR-mutant lung cancer and acquired resistance to erlotinib or gefitinib: implications for clinical trial design. Clin Cancer Res. 2011;17:6298–6303. doi: 10.1158/1078-0432.CCR-11-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Becker A, et al. Retreatment with erlotinib: regain of TKI sensitivity following a drug holiday for patients with NSCLC who initially responded to EGFR-TKI treatment. Eur J Cancer. 2011;47:2603–2606. doi: 10.1016/j.ejca.2011.06.046. [DOI] [PubMed] [Google Scholar]
- 137.Oxnard GR, et al. Maintained sensitivity to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer recurring after adjuvant erlotinib or gefitinib. Clin Cancer Res. 2011;17:6322–6328. doi: 10.1158/1078-0432.CCR-11-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Foo J, Chmielecki J, Pao W, Michor F. Effects of pharmacokinetic processes and varied dosing schedules on the dynamics of acquired resistance to erlotinib in EGFR-mutant lung cancer. J Thorac Oncol. 2012;7:1583–1593. doi: 10.1097/JTO.0b013e31826146ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kuiper JL, Smit EF. High-dose, pulsatile erlotinib in two NSCLC patients with leptomeningeal metastases—one with a remarkable thoracic response as well. Lung Cancer. 2013;80:102–105. doi: 10.1016/j.lungcan.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 140.Guilhot F, et al. Plasma exposure of imatinib and its correlation with clinical response in the Tyrosine Kinase Inhibitor Optimization and Selectivity Trial. Haematologica. 2012;97:731–738. doi: 10.3324/haematol.2011.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Larson RA, et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood. 2008;111:4022–4028. doi: 10.1182/blood-2007-10-116475. [DOI] [PubMed] [Google Scholar]
- 142.Fausto P, Karen B, Mary C, Lonati V, Sandro B. Relationship between skin rash and outcome in non-small-cell lung cancer patients treated with anti-EGFR tyrosine kinase inhibitors: a literature-based meta-analysis of 24 trials. Lung Cancer. 2012;78:8–15. doi: 10.1016/j.lungcan.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 143.Liu HB, et al. Skin rash could predict the response to EGFR tyrosine kinase inhibitor and the prognosis for patients with non-small cell lung cancer: a systematic review and meta-analysis. PLoS ONE. 2013;8:e55128. doi: 10.1371/journal.pone.0055128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sledge GW., Jr The challenge and promise of the genomic era. J Clin Oncol. 2012;30:203–209. doi: 10.1200/JCO.2011.39.0088. [DOI] [PubMed] [Google Scholar]
- 145.Carpenter G, King L, Jr, Cohen S. Epidermal growth factor stimulates phosphorylation in membrane preparations in vitro. Nature. 1978;276:409–410. doi: 10.1038/276409a0. [DOI] [PubMed] [Google Scholar]
- 146.Hunter T, Cooper JA. Epidermal growth factor induces rapid tyrosine phosphorylation of proteins in A431 human tumor cells. Cell. 1981;24:741–752. doi: 10.1016/0092-8674(81)90100-8. [DOI] [PubMed] [Google Scholar]
- 147.Kawamoto T, et al. Growth stimulation of A431 cells by epidermal growth factor: identification of high-affinity receptors for epidermal growth factor by an anti-receptor monoclonal antibody. Proc Natl Acad Sci USA. 1983;80:1337–1341. doi: 10.1073/pnas.80.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sato JD, et al. Biological effects in vitro of monoclonal antibodies to human epidermal growth factor receptors. Mol Biol Med. 1983;1:511–529. [PubMed] [Google Scholar]
- 149.Ullrich A, et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature. 1984;309:418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 150.Downward J, et al. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 151.Kamata T, Feramisco JR. Epidermal growth factor stimulates guanine nucleotide binding activity and phosphorylation of ras oncogene proteins. Nature. 1984;310:147–150. doi: 10.1038/310147a0. [DOI] [PubMed] [Google Scholar]
- 152.King CR, Kraus MH, Aaronson SA. Amplification of a novel v-erbB–related gene in a human mammary carcinoma. Science. 1985;229:974–976. doi: 10.1126/science.2992089. [DOI] [PubMed] [Google Scholar]
- 153.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 154.Wong AJ, et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37 (suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 156.Fry DW, et al. A specific inhibitor of the epidermal growth factor receptor tyrosine kinase. Science. 1994;265:1093–1095. doi: 10.1126/science.8066447. [DOI] [PubMed] [Google Scholar]
- 157.Wakeling AE, et al. Specific inhibition of epidermal growth factor receptor tyrosine kinase by 4-anilinoquinazolines. Breast Cancer Res Treat. 1996;38:67–73. doi: 10.1007/BF01803785. [DOI] [PubMed] [Google Scholar]
- 158.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 159.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 160.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shepherd FA, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 163.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 165.Kantarjian H, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 166.Tallman MS, et al. All-trans-retinoic acid in acute promyelocytic leukemia. N Engl J Med. 1997;337:1021–1028. doi: 10.1056/NEJM199710093371501. [DOI] [PubMed] [Google Scholar]
- 167.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Blanke CD, et al. Long-term results from a randomized phase II trial of standard-versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J Clin Oncol. 2008;26:620–625. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 169.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fukuoka M, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 171.Jackman DM, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15:5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Won YW, et al. Comparison of clinical outcome of patients with non-small-cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations. J Clin Pathol. 2011;64:947–952. doi: 10.1136/jclinpath-2011-200169. [DOI] [PubMed] [Google Scholar]
- 173.Chung KP, et al. Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res. 2012;18:3470–3477. doi: 10.1158/1078-0432.CCR-11-2353. [DOI] [PubMed] [Google Scholar]
- 174.Riely GJ, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 175.Arcila ME, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Oxnard GR, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Arcila ME, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wu JY, et al. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 180.He M, et al. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res. 2012;18:1790–1797. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sok JC, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 182.Ogawa T, et al. Methylation of death-associated protein kinase is associated with cetuximab and erlotinib resistance. Cell Cycle. 2012;11:1656–1663. doi: 10.4161/cc.20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cortot AB, et al. Resistance to irreversible EGF receptor tyrosine kinase inhibitors through a multistep mechanism involving the IGF1R pathway. Cancer Res. 2013;73:834–843. doi: 10.1158/0008-5472.CAN-12-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang S, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bivona TG, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Naumov GN, et al. Combined vascular endothelial growth factor receptor and epidermal growth factor receptor (EGFR) blockade inhibits tumor growth in xenograft models of EGFR inhibitor resistance. Clin Cancer Res. 2009;15:3484–3494. doi: 10.1158/1078-0432.CCR-08-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 189.Scartozzi M, et al. Correlation of insulin-like growth factor 1 (IGF-1) expression and clinical outcome in K-RAS wild-type colorectal cancer patients treated with cetuximab-irinotecan. J Clin Oncol. 2009;27(suppl 15):abstract 4017. [Google Scholar]
- 190.Liska D, Chen CT, Bachleitner-Hofmann T, Christensen JG, Weiser MR. HGF rescues colorectal cancer cells from EGFR inhibition via MET activation. Clin Cancer Res. 2011;17:472–482. doi: 10.1158/1078-0432.CCR-10-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hoellein A, et al. Aurora kinase inhibition overcomes cetuximab resistance in squamous cell cancer of the head and neck. Oncotarget. 2011;2:599–609. doi: 10.18632/oncotarget.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wheeler DL, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Holz C, et al. Epithelial-mesenchymal-transition induced by EGFR activation interferes with cell migration and response to irradiation and cetuximab in head and neck cancer cells. Radiother Oncol. 2011;101:158–164. doi: 10.1016/j.radonc.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 194.La Fleur L, Johansson AC, Roberg KA. CD44high/EGFRlow subpopulation within head and neck cancer cell lines shows an epithelial-mesenchymal transition phenotype and resistance to treatment. PLoS ONE. 2012;7:e44071. doi: 10.1371/journal.pone.0044071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.