Abstract
As a coarse-gained model, a super-thin elastic rod subjected to interfacial interactions is used to investigate the condensation of DNA in a multivalent salt solution. The interfacial traction between the rod and the solution environment is determined in terms of the Young–Laplace equation. Kirchhoff’s theory of elastic rod is used to analyze the equilibrium configuration of a DNA chain under the action of the interfacial traction. Two models are established to characterize the change of the interfacial traction and elastic modulus of DNA with the ionic concentration of the salt solution, respectively. The influences of the ionic concentration on the equilibrium configuration of DNA are discussed. The results show that the condensation of DNA is mainly determined by competition between the interfacial energy and elastic strain energy of the DNA itself, and the interfacial traction is one of forces that drive DNA condensation. With the change of concentration, the DNA segments will undergo a series of alteration from the original configuration to the condensed configuration, and the spiral-shape appearing in the condensed configuration of DNA is independent of the original configuration.
Keywords: DNA condensation, Interfacial traction, Ionic concentration
Introduction
In a salt solution, geometrical configuration of a DNA chain is determined by interaction between the DNA chain and the solution molecules. This interaction should be, in essence, characterized by von de Waals force or electrostatic force. However, if a DNA chain is regarded as a thin elastic rod, the interaction of the DNA chain with the solution molecules can be modeled by the interfacial traction between the rod and solution [1].
Studies of modeling of DNA as an elastic rod can be traced back to the 1970s. Benham [2, 3] and Le Bret [4, 5] firstly used the elastic model to analyze the equilibrium configuration of DNA. In the framework of this model, varied subjects related with the DNA configuration were investigated extensively [6–10]. Some geometrical configurations appearing in the DNA condensation have been explained perfectly. Recently, Cherstvy used the elastic model to further investigate the effect of counterions and attractive inter-segmental DNA–DNA interactions on molecular spatial conformations and the low-dielectric DNA core on the elastic DNA properties [9–11] in a micro field. Up to the present, it has been confirmed that the elastic rod model is adequate for characterizing the geometrical configuration and deformation of DNA. Some recent reviews can be found in references [12–16].
A series of experiments have shown that, as a polyelectrolyte, geometrical configuration of DNA in a multivalent salt solution is directly influenced by the ionic concentration of the salt solution [6, 17–28]. At low salt, the DNA chain manifests itself in a sparse interwound configuration; while at high salt, the DNA condenses into a toroidal structure. Recently, Huang used the elastic rod model to calculate the equilibrium configuration of DNA subjected to the rod-solution interfacial traction, and discussed the change of DNA configuration with the ionic concentration of solution [1]. However, in that work, the influences of the ionic concentration on the interfacial traction and elastic modulus of DNA are given in a vague way, being lack of a mathematical description. Therefore, the aim of this paper is to establish some models characterizing the influences of the ionic concentration on the interfacial traction and elastic modulus of DNA, and investigate the equilibrium configuration in the salt solution with different concentration.
The paper is divided into five parts. In the second section, basic equations given in [1] are summarized. In the third section, we propose two models that they characterize the rod-solution interfacial traction and the elastic modulus of DNA changing with the ionic concentration of the salt solution. In the fourth section, the influence of the ionic concentration on the equilibrium configuration of DNA is calculated and discussed in detail. Finally, some conclusions are given and the paper is closed.
Basic equations
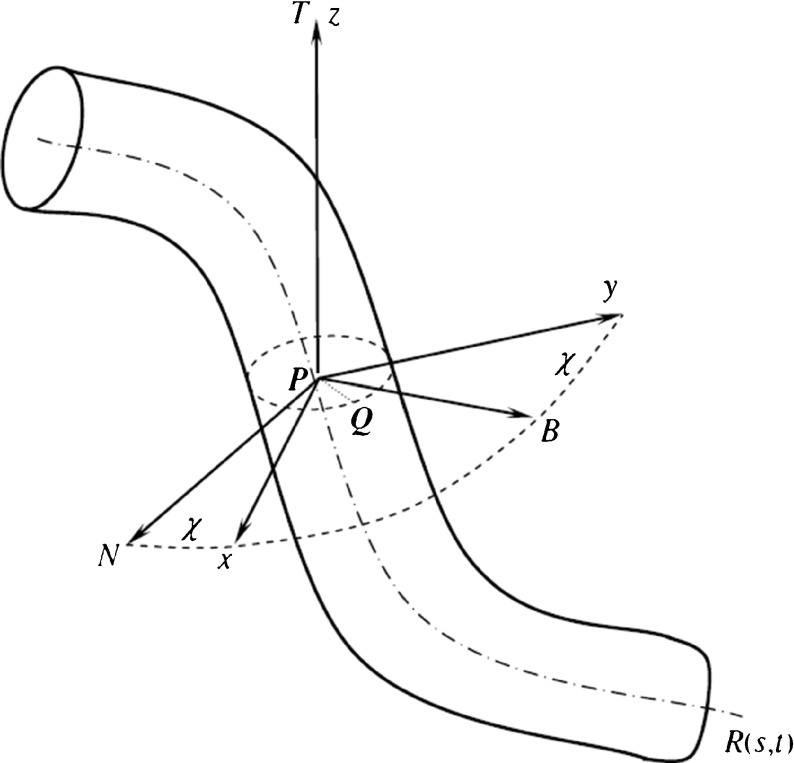
Consider a polymer filament immersed in a solution. The filament can be modeled by a thin flexible circular rod [2–8, 13, 19–26]. The central axis of the rod is a spatial curve R(s, t): R2→R3 parameterized by arc length s and time t. As shown in Fig. 1, the Frenet frame P-NBT and a local orthonormal basis P-xyz are simultaneously set on each cross section of the rod. P is a point on the curve R(s, t). In P-xyz, x-axis and y-axis are the two principal inertia axes of the cross section through the point P. The angle between the principal normal PN and x-axis (or the binormal PB and y-axis) is called the twisting angle, denoted by χ.
Fig. 1.
The thin rod model of DNA
In the initial state of rod, the curvature and torsion of the axis are κ0 and τ0, respectively. The initial twisting angle is χ0. Relative to the coordinates P-xyz, the Kirchhoff equations are written with the index symbols as follows [25]:
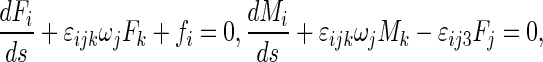 |
1 |
where Fi and Mi are the stress resultant and principal moment acting at the center of cross section, ωi the curvature-twisting vector, and εijk the permutation sign. fi refers to the interfacial traction distributed along the central axis R(s, t). In a solution, fi is a coarse-grained model to characterize molecular and electrostatic interactions between the DNA chain and solution. As a coarse-grained model, we suppose that fi is described by interfacial pressure between solution and the rod modeling the DNA chain. So it can be derived from the Young–Laplace equation
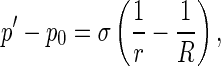 |
2 |
where R and r are two principal curvature radii in which r is the radius of the rod cross section, p0 is a constant pressure applied on the interfacial surface by solution, and p′ is the pressure on the interfacial surface by the rod. In (2), we have reckoned a principal curvature radius as positive if it is drawn into the interior of rod.
In terms of the action and reaction law, the pressure p applied on the rod by the interfacial surface is equal to p′ in magnitude, but they are opposite in direction. So, in coordinates P-xyz, p can be decomposed into
 |
3 |
where 𝜃 is the angle between PQ and PN. It has been proven that R can be written as [1]
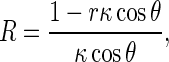 |
4 |
where κ is the curvature of the rod axis. Substituting (4) into (3) and integrating along the perimeter of the rod cross section leads to
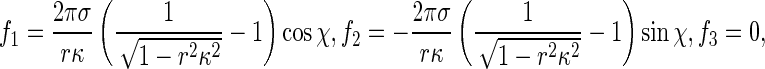 |
5 |
which characterize interfacial effects between the rod and solution. Clearly, −1 < rκ < 1.
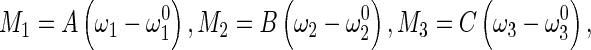 |
6 |
where 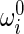 denotes the initial curvature-twisting vector. A and B are the bending stiffness with respect to x-axis and y-axis, respectively. C is the twisting stiffness. Assume the rod is isotropic. Then, A, B and C are written as
denotes the initial curvature-twisting vector. A and B are the bending stiffness with respect to x-axis and y-axis, respectively. C is the twisting stiffness. Assume the rod is isotropic. Then, A, B and C are written as
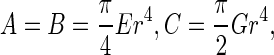 |
7 |
where r is the radius of the rod cross section. E is the elastic modulus, and G the shear modulus that is equal to E/2(1+ν). Here, ν is the Poisson ratio and it is a constant with ν=0.23 [25]. By the curvature κ, torsion τ and twisting angle χ, the curvature-twisting vector ωi can be represented as [25]
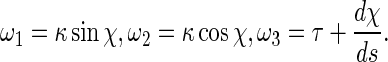 |
8 |
Inserting (5), (6), and (8) into (1) leads to:
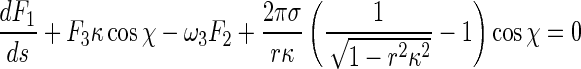 |
9 |
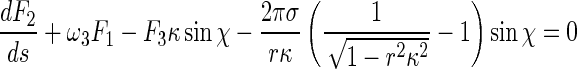 |
10 |
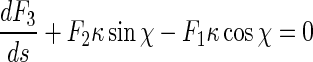 |
11 |
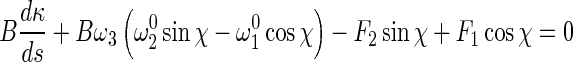 |
12 |
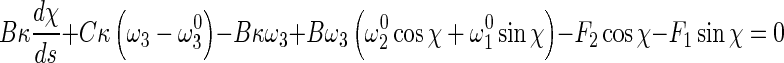 |
13 |
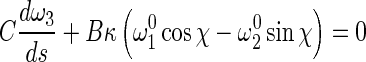 |
14 |
Let τ=ω3−dχ/ds, then (13) leads to
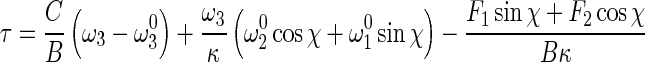 |
15 |
From (9–15), we can get the value of the curvature κ, torsion τ and twisting angle χ, and with the Frenet–Serret formula of curve as follows:
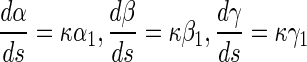 |
16 |
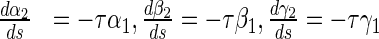 |
17 |
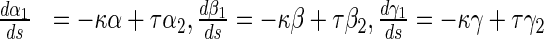 |
18 |
Relative to the coordinates P-xyz, we can solve the direction cosine ( α , β, γ) of the tangential vector with a point of the curve. So the configuration of the rod is determined by the equation below:
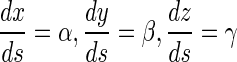 |
19 |
From (9–15), it is clear that the curvature κ, torsion τ, and twisting angle χ depend only on the interfacial energy and elastic properties of the rod. Therefore, the configuration of a rod is determined by competition between the interfacial energy and elastic strain energy of the rod.
Effects of solution concentration on interfacial traction and elastic modulus
As indicated in (5), the interfacial tractions are influenced by the interfacial tension σ between the rod and the solution, whereas σ depends directly on the concentration of the solution. Therefore, the interfacial tractions change with the concentration of solution. The relationship between them can be characterized by the Gibbs adsorption equation, which reads [37]
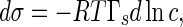 |
20 |
where c denotes the concentration, Γs is adsorbed amount, T and R are absolute temperature and the gas constant, respectively. On the other hand, Γs is also concentration-dependent. This dependence can be given by the Langmuir adsorption equation below [36, 37]:
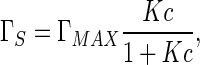 |
21 |
where ΓMAX is the maximum concentration when the solution arrives at saturation state, and K is the absorption constant. Substituting (21) into (20) and then calculating integration, we have
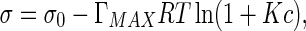 |
22 |
where σ0 is the interfacial tension of pure solvent. Equation (22) shows the interfacial tension decreases with the concentration increasing.
Besides effects on the interfacial tractions, the concentration has the influences on the elastic modulus of polymer filament. This is because the change of concentration will cause the Kuhn length of polymer chain altered, whereas the Kuhn length is a main factor to determine the elastic modulus of polymer material. In general, the correlation between the Kuhn length and concentration is described by the Poisson–Boltzmann theory [38, 39], which reads
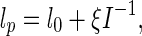 |
23 |
where lp denotes the persistence length, l0 and ξ are two constants, and I is the ionic strength that is defined as [38]
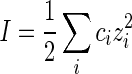 |
24 |
where ci is the concentration of ionic species i, and zi is its charge. For the convenience of analysis, in the following examples we choose the NaCl solution with monovalent Na +. So zi= 1. With help of the freely jointed chain model, the elastic modulus of polymer filament can be represented as [42]
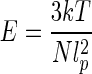 |
25 |
by the Kuhn length, where k is Boltzmann constant, T is the absolute temperature and N is the number of Kuhn length, which is the ratio of the contour length L of DNA and Kuhn segments length b[43]. Substituting (23) and (24) into (25) leads to
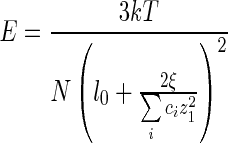 |
26 |
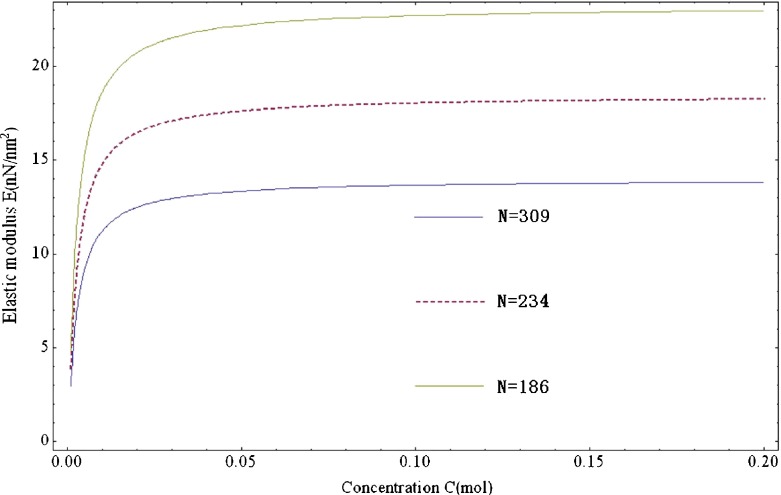
To summarize (22) and (26), we can find that the change of concentration will make elastic modulus and the interfacial tractions simultaneously varied. Figure 2 illustrates the change of elastic modulus with the ions concentration of solution. Therefore, we can implement the self-assembly of polymer filament by adjusting the concentration of solution. Some examples for DNA will be given in next section.
Fig. 2.
The change of elastic modulus with concentration from 1–2 mM for the freely jointed chain model with the parameter of k=1.38×10−23 J/K, T=293 K, ξ=0.324
Analysis and conclusions
In this section, we use (9–19) to solve numerically the equilibrium configuration of DNA under the action of interfacial traction.
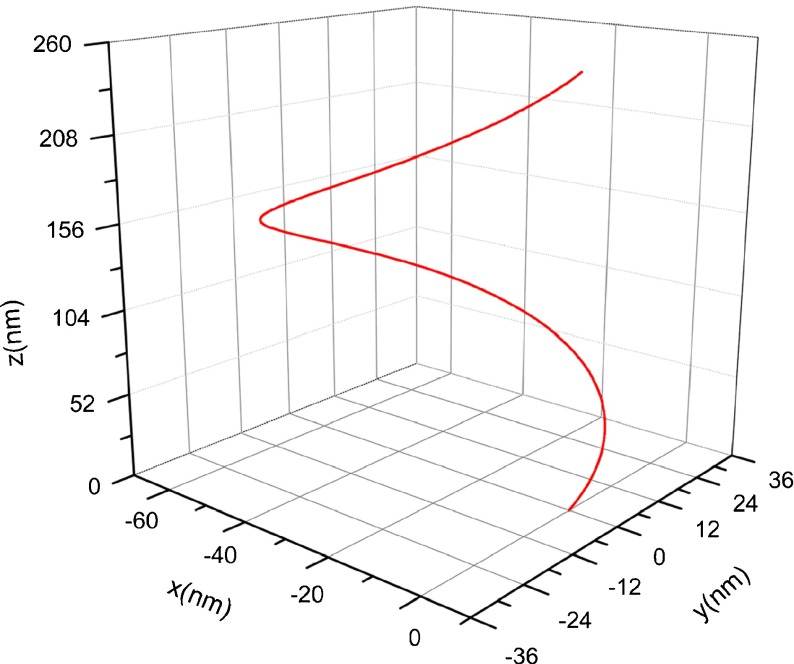
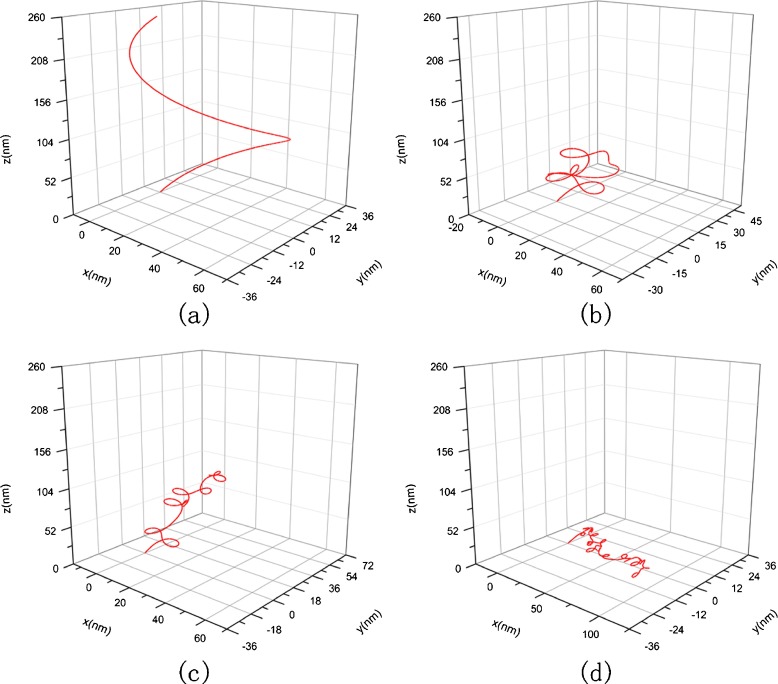
Consider a segment of DNA strand whose original configuration is a helix, as shown in Fig. 3. The length of the DNA segment is of nanometer order of magnitude. Relative to an orthogonal coordinates system, the helix curve is characterized by the equation below:
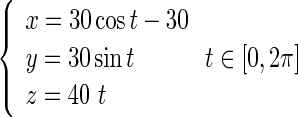 |
27 |
It is easy to give that the curvature and torsion of the helix curve are 0.012 nm−1 and 0.016 nm−1, respectively. When the DNA segment is immersed into a new solution environment, it will deform from the original configuration into the condensed configuration due to the change of interfacial traction.
Fig. 3.
Original configuration of DNA
As discussed in Section 3, the interfacial tension and elastic modulus of DNA are influenced by the concentration of solution. Therefore, the change of DNA configuration with the concentration of intracellular solution will be investigated in the following section. When calculation, we take r= 1.0 nm[33], ΓMAX=5.35×10−6mol/m2[41], K=100 [41], l0=53 nm, b=0.106 μm, ξ=0.324 [38], T=293 K, L=32.8 μm [43], and N=309(N=L/b) [43].
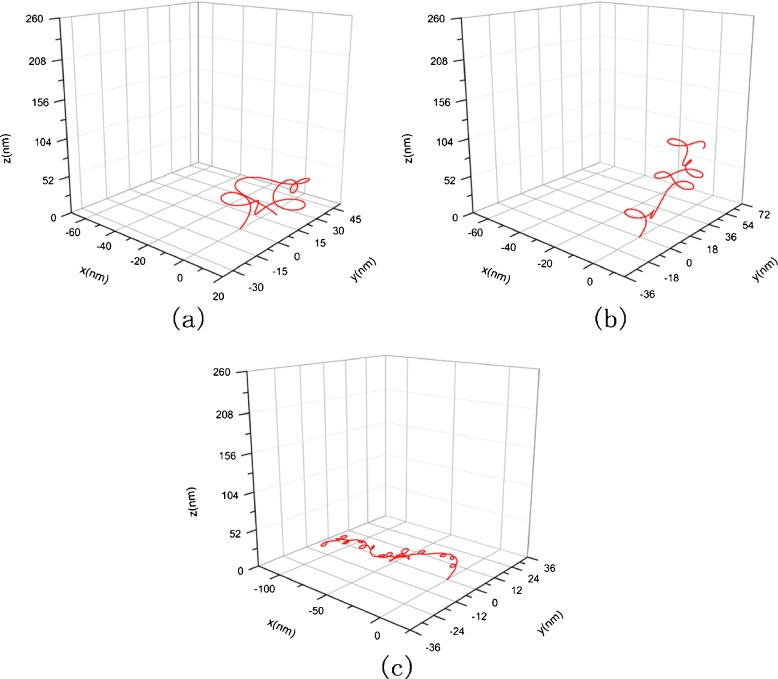
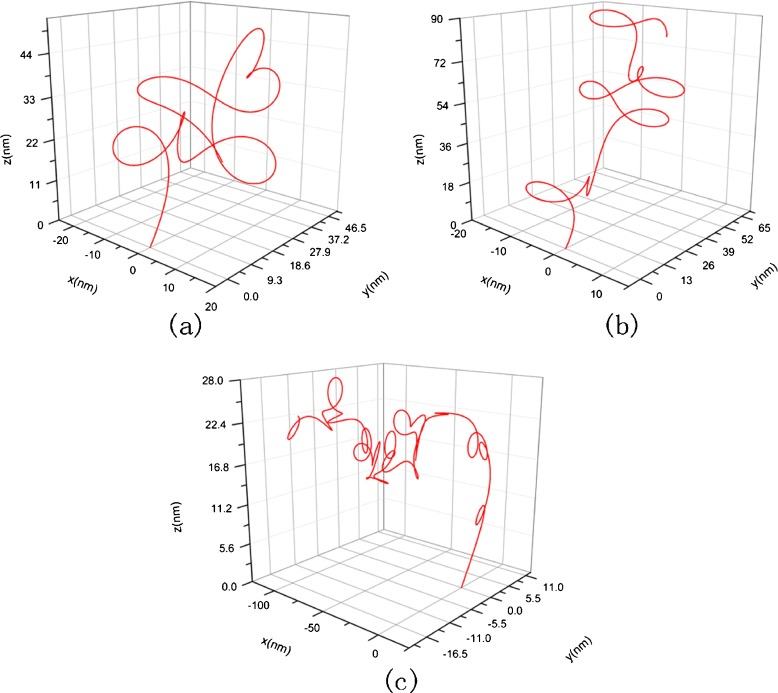
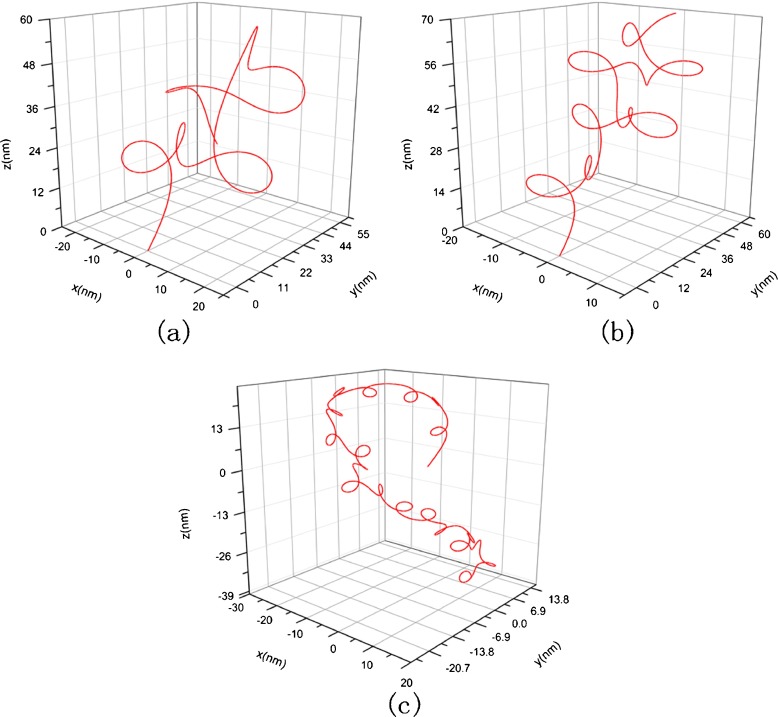
Using the classical Runge–Kutta algorithm, we calculate three condensed configurations corresponding to the original configuration in Fig. 3 in three solutions whose concentration are 1 mM, 10 mM, and 150 mM, respectively. The initial condition takes F1=F2=F3= 0, κ0= 0.012, τ0= 0.016, χ0=0 and x = y = z = 0. The results are shown in Figs. 4 and 5, where Fig. 5 is amplification to Fig. 4 in resolution. From them, one can see that the DNA segment coils into a compact “helix” spontaneously under the interfacial traction. The mechanism to produce this change can be attributed to the correlation between the interfacial traction and the curvature of the rod. When the interfacial traction is applied on the rod, the curvature of rod will alter; conversely, the change of the curvature will cause the interfacial traction to be altered. As thus, the coupling effects between the interfacial traction and the curvature of the rod make the rod (DNA) coil.
Fig. 4.
Condensed helical configuration corresponding to Fig. 3. a c = 1 nM, σ = 7.14×10−2 nN/nm, E =2.982 nN/nm2, G = 1.21 nN/nm2, b c = 10 nM, σ=6.38×10−2 nN/nm, E = 12.6 nN/nm2, G = 5.1 nN/nm2, c c = 150 nM, σ=3.61×10−2 nN/nm, E =15.42 nN/nm2, G = 6.27 nN/nm2
Fig. 5.
The amplification to Fig. 4 in resolution
The diameter of the helix in Fig. 4a is about 5 nm. This magnitude is nearly 120 times smaller than the diameter of the DNA segment in Fig. 3. Meanwhile, the total size of Fig. 4a reduces to about six times than that of Fig. 3. So, relative to the original configuration, the condensed configuration of the DNA segment seems like a cluster. The change of configuration with concentration is shown in Fig. 4a, b, and c. It can be seen that with the concentration rising, the shape of the DNA segment will change, and the spatial domain occupied by the DNA segment will increase in size. This result can be attributed to the fact that the increase of concentration makes elastic modulus and the interfacial traction simultaneously decrease. Just as implied by (5), the interfacial traction tends to reduce the curvature of a cured DNA segment. Therefore, a curved DNA segment will stretch due to the interfacial traction dropping. Meanwhile, the decrease of elastic modulus causes stretching more easily. Thus, when the concentration rises, the DNA segment will undergo a change from a compact state into a sparse state
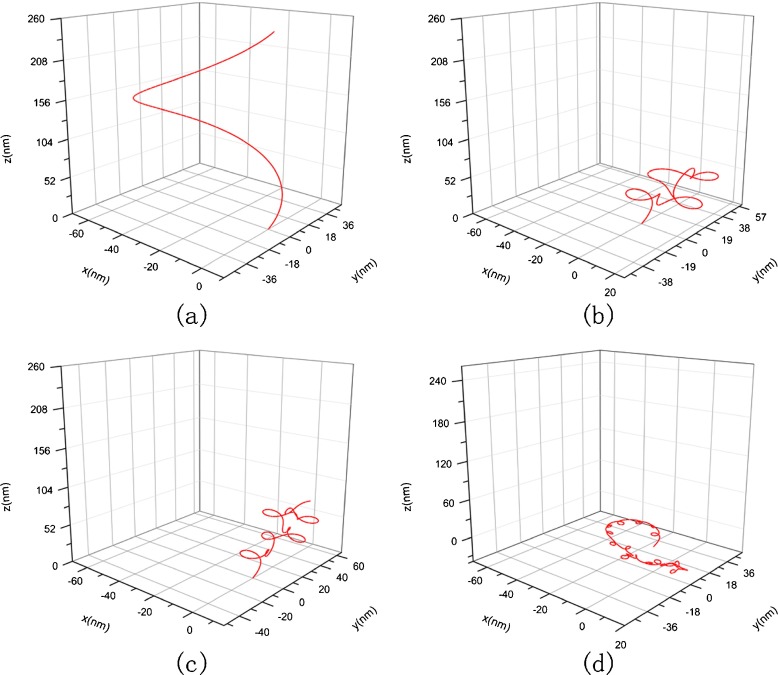
The DNA segments in Figs. 6 and 7 have the same physical properties and arc length as that in Fig. 3. Their differences consist only in their configurations. Figure 6a represents a helical line with spiral direction as opposed to Figs. 3 and 7a is an elliptical helix with κ0=0.0125, τ0=0.0125. The value of initial curvature and torsion are depicted in terms of the following equation [40]:
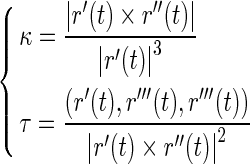 |
28 |
where r(t) is the parametric equation of a spiral line and r (t),r
(t),r (t),r
(t),r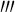 (t)are the first-, second-, third-order derivative of r(t), and t represents the arc length.
(t)are the first-, second-, third-order derivative of r(t), and t represents the arc length.
Fig. 6.
Original configuration as opposed to spiral direction of Fig. 3 and its condensed helical configuration. a Original conformation with κ 0=−0.012, τ 0=−0.016, b κ 0=−0.012, τ 0=−0.016, c = 1 nM, σ=7.14×10−2 nN/nm, E =2.982 nN/nm2, G =1.21 nN/nm2, c κ 0=−0.012, τ 0=−0.016, c = 10 nM, σ=6.38×10−2 nN/nm, E =12.6 nN/nm2, G =5.1 nN/nm2, d κ 0=−0.012, τ 0=−0.016, c = 150 nM, σ=3.61×10−2 nN/nm, E =15.42 nN/nm2, G =6.27 nN/nm2
Fig. 7.
Original and condensed helical configuration of DNA with κ 0=0.0125, τ 0=0.0125. a Original conformation, b c = 1 nM, σ=7.14×10−2 nN/nm, E = 2.982 nN/nm2, G = 1.21 N/nm2, c c = 10 nM, σ = 6.38 × 10−2 nN/nm, E = 12.6 nN/nm2, G =5.1 nN/nm2, d c = 150 nM, σ = 3.61 × 10−2 nN/nm, E =15.42 nN/nm2, G = 6.27 nN/nm2
The changes of two DNA segments with different original configuration with the concentration of solution are shown in Figs. 6 and 7, respectively. Comparing them with Fig. 3, we can see that three DNA segments with different original configuration all twist to a spiral curve in solution with the same concentration. This interesting result show that the condensed configuration of DNA is of spiral shape, being independent of the original configuration. However, the size and spinning direction of the spiral curve are correlated with the original configuration.
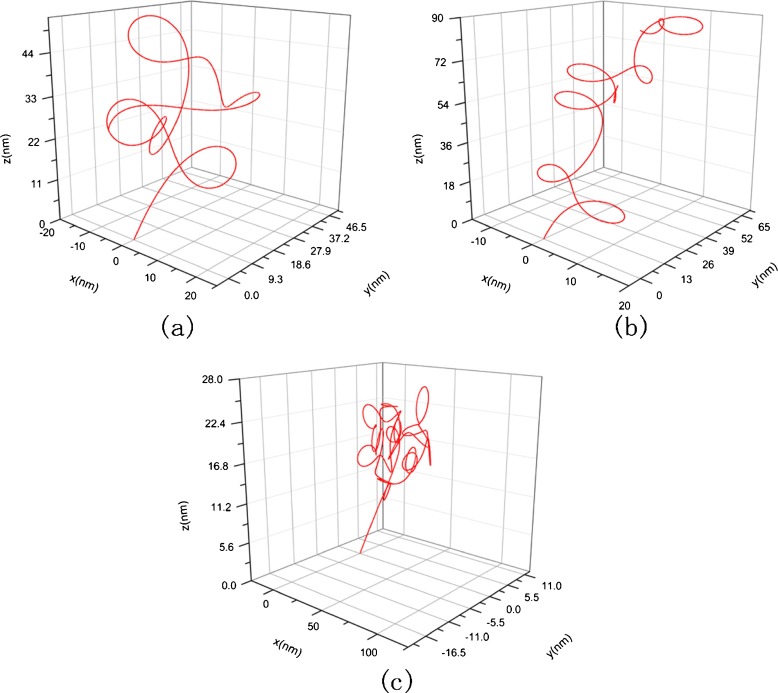
Figures 8 and 9 are amplifications to Figs. 6 and 7 in resolution, respectively.
Fig. 8.
The amplification to Fig. 6 in resolution, a κ 0 =-0.012, τ 0= 0.016, c = 1 nM, σ= 7.14 × 10 −2 nN/nm, E = 2.982 nN/nm 2, G = 1.21 nN/nm 2, b κ 0 =-0.012, τ 0= 0.016, c = 10 nM, σ= 6.38 × 10 −2 nN/nm, E = 12.6 nN/nm 2, G = 5.1 nN/nm 2, c κ 0 = -0.012,τ 0= 0.016, c = 150 nM, σ= 3.61 × 10 −2 nN/nm, E = 15.42 nN/nm 2, G = 6.27 nN/nm 2
Fig. 9.
The amplification to Fig. 7 in resolution, a c = 1 nM, σ = 7.14 × 10 −2 nN/nm, E = 2.982 nN/nm 2, G = 1.21 nN/nm 2, b c = 10 nM, σ = 6.38 × 10 −2 nN/nm, E = 12.6 nN/nm 2, G = 5.1 nN/nm 2, c c = 150 nM, σ = 3.61 × 10 −2 nN/nm, E = 15.42 nN/nm 2, G = 6.27 nN/nm 2
Figures 4, 6, and 7 reveal that the equilibrium configuration of DNA has non-uniform curvature. This is caused by competition between the interfacial traction and elastic strain energy. In general, the elastic strain energy tends to make the curvature of DNA axis become uniform, but the interfacial traction causes the DNA chain bending more easily due to that it is curvature-dependent. Since one end of DNA axis is fixed and another end is free, this constraint gives rise to that deformability is different along the axis of DNA. In the free end, the DNA is easy to be bent under the interfacial traction. So it is just the difference of deformability to cause curvature being non-uniform along the axis of DNA. The non-uniform curved DNA shapes can be observed in some electron micrographs from the DNA condensation experiments (e.g., [19, 32]), but no measured data on the DNA curvature have been given up to the present. In addition, Schlick gave some examples of DNA seguments with non-uniform curved deformation [44].
Figures 4, 6, and 7 also show that no preferred direction occurs in the configuration of the rod. This is because the free end of the rod is controlled by the interfacial traction dependent on the curvature. Under the action of different interfacial traction, the position and orientation of the free end are different, so the configuration of the rod manifests no preferred direction.
Conclusions
A three-dimensional elastic rod model is established to investigate the equilibrium configuration of DNA under the action of interfacial interactions induced by the changes of ion concentration. Using this model, we account for the effect of concentration on the equilibrium configuration of DNA. Some conclusions are summarized as follows:
Equations (22) and (26) characterize how the ion concentrations effect the interfacial tension and elastic modulus of DNA. With the concentration rising, the interfacial traction and elastic modulus of DNA tend to decrease.
The interfacial tractions act as a source to spin DNA. The equilibrium configuration of DNA is completely determined by competition between the interfacial energy and elastic strain energy of DNA.
Due to the change of concentration, the DNA segments will undergo a series of alteration from the original configuration to the condensed configuration.
When the concentration changes, the spiral shape appearing in the condensed configuration of DNA is independent of the original configuration. However, the size and spinning direction of the spiral curve are correlated with the original configuration.
Finally, it should be pointed out that although what we solve is an initial value problem of (9–15), this initial value problem is equivalent to a boundary value problem with one endpoint fixed and another endpoint free, because the stress resultant Fi acting on the beginning endpoint is zero.
Acknowledgements
Support of the National Nature Science Foundation of China through Grant No. 11172130 is gratefully acknowledged.
References
- 1.Huang Z. Modulating DNA configuration by interfacial traction: an elastic rod model to characterize DNA folding and unfolding. J. Biol. Phys. 2011;37:79–90. doi: 10.1007/s10867-010-9200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benham CJ. Elastic model of super-coiling. Proc. Natl. Acad. Sci. U.S.A. 1977;74:2397–2401. doi: 10.1073/pnas.74.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benham CJ. An elastic model of the large-scale structure of duplex DNA. Biopolymers. 1979;18:609–623. doi: 10.1002/bip.1979.360180310. [DOI] [PubMed] [Google Scholar]
- 4.Le Bret M. Relationship between the energy of superhelix formation, the shear modulus and the torsional Brownian motion of DNA. Biopolymers. 1978;17:1939–1955. doi: 10.1002/bip.1978.360170810. [DOI] [Google Scholar]
- 5.Le Bret M. Twist and writhing on short circular DNAs according to first-order elasticity. Biopolymers. 1984;23:1835–1867. doi: 10.1002/bip.360231004. [DOI] [PubMed] [Google Scholar]
- 6.Gelbart WM, Bruinsma RF, Pincus PA, Parsegian VA. DNA-inspired electrostatics. Phys. Today. 2000;53:38–44. doi: 10.1063/1.1325230. [DOI] [Google Scholar]
- 7.Shi Y, Borovik AE, Hearst JE. Elastic rod model incorporating shear and extension, generalized nonlinear Schrodinger equations, and novel closed-form solutions for supercoiled DNA. J. Chem. Phys. 1995;103:3166–3183. doi: 10.1063/1.470250. [DOI] [Google Scholar]
- 8.Manning RS, Maddocks JH, Kahn JD. A continuum rod model of sequence–dependent DNA structure. J. Chem. Phys. 1996;105:5626–5646. doi: 10.1063/1.472373. [DOI] [Google Scholar]
- 9.Cherstvy AG. Torque-induced deformations of charged elastic DNA rods: thin helices, loops, and precursors of DNA supercoiling. J. Biol. Phys. 2011;37:227–238. doi: 10.1007/s10867-010-9211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherstvy AG. Collapse of highly charged polyelectrolytes triggered by attractive dipole-dipole and correlation-induced electrostatic interactions. J. Phys. Chem. 2010;114:5241–5249. doi: 10.1021/jp910960r. [DOI] [PubMed] [Google Scholar]
- 11.Cherstvy AG. Effect of a low-dielectric interior on DNA electrostatic response to twisting and bending. J. Phys. Chem. 2007;111:12933–12937. doi: 10.1021/jp074166z. [DOI] [PubMed] [Google Scholar]
- 12.Tobias I, Swigon D, Coleman BD. Elastic stability of DNA configuration: I general theory. Phys. Rev. E. 2000;61:747–758. doi: 10.1103/PhysRevE.61.747. [DOI] [PubMed] [Google Scholar]
- 13.Coleman BD, Swigon D, Tobias I. Elastic stability of DNA configuration: II Supercoiled plasmids with self-contact. Phys. Rev. E. 2000;61:759–770. doi: 10.1103/PhysRevE.61.759. [DOI] [PubMed] [Google Scholar]
- 14.Munteanu MG, Vlahovicek K, Parthasarathy S, Simon I, Pongor S. Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci. 1998;23:341–347. doi: 10.1016/S0968-0004(98)01265-1. [DOI] [PubMed] [Google Scholar]
- 15.Eslami-Mossallam B, Ejtehadi MR. An asymmetric elastic rod model for DNA. Phys. Rev. E. 2009;80:011919. doi: 10.1103/PhysRevE.80.011919. [DOI] [PubMed] [Google Scholar]
- 16.Coleman BD, Olson WK, Swigon D. Theory of sequence-dependent DNA elasticity. J. Chem. Phys. 2003;118:7127–7140. doi: 10.1063/1.1559690. [DOI] [Google Scholar]
- 17.Slita AV, Kasyanenko NA, Nazarova OV, Gavrilova II, Eropkina EM, Sirotkin AK, Smirnova TD, Kiselev OI, Panarin EF. DNA–polycation complexes effect of polycation structure on physico-chemical and biological properties. J. Biotechnol. 2007;127:679–693. doi: 10.1016/j.jbiotec.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: a comparison of perspectives. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3987. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hud NV, Vilfan ID. Toroidal DNA condensates: Unraveling the fine structure and the role of nucleation in determining size. Ann. Rev. Biophys. Biomol. Struct. 2005;34:295. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- 20.Keyser UF, van Dorp S, Lemay SG. Tether forces in DNA electrophoresis. Chem. Soc. Rev. 2010;39:939. doi: 10.1039/b902072c. [DOI] [PubMed] [Google Scholar]
- 21.Leonard C, Gousle JAS. Compact form of DNA induced by supermidine. Nature. 1976;259:333. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Wang P-Y, Yan J, Li M. Impact of DNA twist accumulation on progressive helical wrapping of torsionally constrained DNA. Phys. Rev. Lett. 2012;109:218–102. doi: 10.1103/PhysRevLett.109.218102. [DOI] [PubMed] [Google Scholar]
- 23.Benham CJ, Mielke SP. DNA mechanics. Ann. Rev. Biomed. Eng. 2005;7:21–53. doi: 10.1146/annurev.bioeng.6.062403.132016. [DOI] [PubMed] [Google Scholar]
- 24.Travers AA, Thompson JM. An introduction to the mechanics of DNA. Philos. Trans. R. Soc. A. 2004;362:1265–1279. doi: 10.1098/rsta.2004.1392. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y-Z. Nonlinear Mechanics of Thin Elastic Rod: Theoretical Basis of Mechanical Model of DNA (in Chinese) Beijing: Tsinghua Press; 2006. [Google Scholar]
- 26.Bednar J, Furrer P, Stasiak A, Dubochet J. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. J. Mol. Biol. 1994;235:825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- 27.Brady G, Foos D, Benham CJ. Evidence for an interwound form of the superhelix in circular DNA. Biopolymers. 1984;23:2963–2966. doi: 10.1002/bip.360231219. [DOI] [PubMed] [Google Scholar]
- 28.Swigon, D.: The mathematics of DNA structure, mechanics, and dynamics. In: Benham C.J. et al. (eds.) Mathematics of DNA Structure, Function and Interactions, pp. 293–320. Springer, Berlin (2009)
- 29.Podgornik R. DNA off the Hooke. Nat. Nanotechnol. 2006;1:100–101. doi: 10.1038/nnano.2006.114. [DOI] [PubMed] [Google Scholar]
- 30.Gosule LC, Schellman JA. Compact form of DNA induced by spermidine. Nature. 1972;59:333–335. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 31.Hud NV, Downing KH, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc. Nat. Acad. Sci. USA. 1995;92:3581–3585. doi: 10.1073/pnas.92.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hud NV, Downing KH. Cryoelectron microscopy of λ phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Nat. Acad. Sci. USA. 2001;98:14925–14930. doi: 10.1073/pnas.261560398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westcott TP, Tobias I, Olson WK. Modeling self-contact forces in the elasticity of DNA supercoiling. J. Chem. Phys. 1997;107:3967–3980. doi: 10.1063/1.474752. [DOI] [Google Scholar]
- 34.Leforestier A, Livolant F. Structure of toroidal DNA collapsed inside the phage capsid. Proc. Nat. Acad. Sci. USA. 2009;106:9157–9162. doi: 10.1073/pnas.0901240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamley, W.I. : Introduction to soft matter : synthetic and biological self-assembling materials. Reading (2008)
- 37.Futian Z. Fundamentals of Molecular Interface Chemistry (in Chinese) Shanghai: Shanghai Scientific and Technology Literature Publishing House; 2006. [Google Scholar]
- 38.Daoud M, Williams CE. Soft Matter Physics. Berlin: Springer; 1999. [Google Scholar]
- 39.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Nat. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W. Differential geometry (in Chinese) Beijing: Beijing University Press; 2006. [Google Scholar]
- 41.Zhao Z. Adsorption Principle in Application (in Chinese) Beijing: Chemical Industry Press; 2005. [Google Scholar]
- 42.Liu F, Tang X. Polymer Physics (in Chinese) Beijing: Higher Education Press; 2004. [Google Scholar]
- 43.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:5085. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 44.Schlick T. Modeling Superhelical DNA: recent analytical and dynamic approaches. Current Opin. Struct. Biol. 1995;5:245–265. doi: 10.1016/0959-440X(95)80083-2. [DOI] [PubMed] [Google Scholar]