Abstract
Rattlesnake venom can differ in composition and in metalloproteinase-associated activities. The molecular basis for this intra-species variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) remains an enigma. To understand the molecular basis for intra-species variation of metalloproteinase-associated activities, we modeled the three-dimensional structures of four metalloproteinases based on the amino acid sequence of four variations of the proteinase domain of the C. s. scutulatus metalloproteinase gene (GP1, GP2, GP3, and GP4). For comparative purposes, we modeled the atrolysin metalloproteinases of C. atrox as well. All molecular models shared the same topology. While the atrolysin metalloproteinase molecular models contained highly conserved substrate binding sites, the Mojave rattlesnake metalloproteinases showed higher structural divergence when superimposed onto each other. The highest structural divergence among the four C. s. scutulatus molecular models was located at the northern cleft wall and the S’1-pocket of the substrate binding site, molecular regions that modulate substrate selectivity. Molecular dynamics and field potential maps for each C. s. scutulatus metalloproteinase model demonstrated that the non-hemorrhagic metalloproteinases (GP2 and GP3) contain highly basic molecular and field potential surfaces while the hemorrhagic metalloproteinases GP1 and atrolysin C showed extensive acidic field potential maps and shallow but less dynamic active site pockets. Hence, differences in the spatial arrangement of the northern cleft wall, the S’1-pocket, and the physico-chemical environment surrounding the catalytic site contribute to differences in metalloproteinase activities in the Mojave rattlesnake. Our results provide a structural basis for variation of metalloproteinase-associated activities in the rattlesnake venom of the Mojave rattlesnake.
Electronic Supplementary Material
The online version of this article (doi:10.1007/s10867-013-9339-3) contains supplementary material, which is available to authorized users.
Keywords: Molecular models, Rattlesnake venom metalloproteinases, Structural basis of hemorrhagic activity
Introduction
Zn2+-dependent metalloproteinases are very important toxins in the venom of most rattlesnakes. Upon envenomation, these enzymes produce deleterious reactions in the bite victim including profuse hemorrhage, coagulation, and immunological reactions due to the hydrolysis of blood components and the blood vasculature, and the inactivation/activation of complement proteins [1–5]. Despite recent progress made in determining the biochemical activities and in elucidating the primary amino acid sequences and crystal structures of cloned or purified venom metalloproteinases [6, 7], there is scarce knowledge regarding the relationship between metalloproteinases’ tertiary structure and their physiological function. Snake venom metalloproteinases (SVMPs) have evolved from an ancient mammalian ADAM gene [8–10]. Hence, SVMPs and ADAMs share a similar secondary structural organization [11–16]. These metalloproteinases are complex multi-domain proteins that are classified as P-I, P-II, and P-III based on the presence or absence of the non-catalytic ancillary domains that extend beyond the mature proteinase domain (P-I) [12, 13, 17]. There is a growing consensus that the diverse activities of these metalloproteinases are attributed to the non-catalytic ancillary domains that may target the proteinase domain (P-I) to specific substrates such as extracellular matrix proteins [12, 13, 18–23]. For instance, the disintegrin and cysteine-rich domains inserted into the C-terminal portion of the proteinase domain physically block integrin receptors and subsequently inhibit platelet aggregation, leading to disrupted hemostasis and profuse hemorrhage in the affected victim [6, 7, 18, 20, 21]. The ancillary domains of SVMPs significantly increase hemorrhagic potency of the catalytic P-I domain [24].
The P-I domains of SVMPs have also been found to show a high variation in substrate specificity and catalytic activity and they have been further divided into two subclasses, namely the P-IA and P-IB [25, 26]. Although both types of P-I domains are sensitive to EDTA treatment, the P-IA subclass has strong hemorrhagic activity, whereas the P-IB subclass has weaker or no hemorrhagic activity, which includes adamalysin II from C. adamanteus, the Eastern diamondback rattlesnake [25]. The observation that P-IA and P-IB show variation in proteolytic and hemorrhagic activities implies that the P-I proteinase domain has additional structural features which are involved in modulating hemorrhagic and proteolytic activities [25, 26].
Rattlesnake venom belonging to the Crotalus genus exhibits high variability in snake venom composition and shows complex pharmacological profiles and in biochemical-associated activities [27–30]. Recently, a series of elegant transcriptome sequencing-based approaches have shown that rattlesnake venom glands from C. adamanteus are composed mainly of serine proteases, while a small fraction of venom consists of Zn+2-dependent metalloproteinases [31]. Our lab and other groups have previously shown that different venoms from C. s. scutulatus exhibit variability in metalloproteinase-associated proteolytic activities, neurotoxic activity, and in the production of hemorrhage [32, 33]. Based on these metalloproteinase activities, we previously categorized venom from 14 C. s. scutulatus into four groups. Venom from the first group (GP1) was highly hemorrhagic and proteolytic whereas the other three groups (GP2, GP3, GP4) either lacked or contained modest hemorrhagic activity and varied in their proteolytic activities. In that same study, we categorized the genomic DNA sequences into four groups according to nucleotide sequence differences. This genomic DNA categorization completely correlated with the metalloproteinase-associated hemorrhagic and proteolytic activities of rattlesnake venom. Although the coding regions of all four groups showed a high degree of sequence homology to known snake venom metalloproteinases, there were single-nucleotide polymorphisms found within zymogen and proteinase regions for sequences of the four groups of C. s. scutulatus metalloproteinases. We have previously suggested that single-nucleotide polymorphisms within the coding regions of the metalloproteinase gene may give rise to intra-species variation in metalloproteinase-associated activities in C. s. scutulatus [32].
In this study, we hypothesized that the P-I venom metalloproteinases from C. s. scutulatus possess specific tertiary structural differences, which may explain the variability in metalloproteinase-associated hemorrhagic and proteolytic activities. To address this hypothesis, we generated molecular models based on the translated amino acid sequences derived from the coding nucleotide regions of the mature proteinase domain for each of the four C. s. scutulatus metalloproteinase groups. The amino acid sequence of each isoform was folded in silico and compared to the known X-ray crystal structures of the hemorrhagic metalloproteinase atrolysin C (Ht-d) [35] and the non-hemorrhagic metalloproteinase adamalysin II [34].
Overall, we found that the molecular models of three groups, GP1, GP3 and GP4, superimposed well with each other with the exception of GP2, which showed the highest structural divergence. A comparative analysis of the spatial arrangement of different structural domains revealed that the S’1 pocket of the GP1 model, a metalloproteinase group associated with high hemorrhagic activity, structurally superimposed better to atrolysins, hemorrhagic metalloproteinases derived from C. atrox. However, we did not find any specific amino acid sequences or secondary structural elements that were conserved between the atrolysins and the C. s. scutulatus molecular models that was associated with high hemorrhagic (GP1) or weak hemorrhagic (GP4) activities. This observation suggests that the presence or absence of specific secondary structural elements may not confer hemorrhagic activity and other factors such as the chemical environment surrounding the active site may be involved. Hence, by studying the molecular potential surface of the metalloproteinase molecular models of each of the four groups, we observed that GP2 metalloproteinase model had a severely decreased electrostatic potential near the zinc binding site, whereas the surface potential map of the GP1 metalloproteinase model showed a narrower catalytic binding groove and a wide acidic patch, which may attract positively charged extracellular matrix substrates such as collagen type IV through long-range electrostatic interactions.
Materials and methods
Secondary structural alignment and molecular modeling
Each of the gene groups of the C. s. scutulatus metalloproteinases were translated into mature proteinase domains using the Contig-Express module of the Vector NTI Suite program (InforMax, Inc., North Bethesda, MD, USA). This gene translation software package is currently marketed and licensed by Life Technologies (Grand Island, NY, USA). The deduced C. s. scutulatus amino acid sequences and the atrolysin (Ht-a, Ht-b, and Ht-c) amino acid sequences were initially aligned with the amino acid sequences of adamalysin II and atrolysin C (Ht-d) by employing the PAM250 algorithms.
Atrolysin C (Ht-d; PDB: 1HTD) and adamalysin II (PDB: 4AIG) were used as template proteins for the first round of modeling using the “Homology” module of the Insight2000 program (MSI, San Diego, CA, USA). These template crystal structures were chosen based on the following factors: (1) similar phylogenetic classification of the rattlesnakes for which the metalloproteinases (models or crystal structures) are derived (C. s. scutulatus, C. atrox, and C. adamanteus); (2) a good amino acid sequence homology (34–38 % homology of atrolysin C and adamalysin II to Mojave rattlesnake sequences); (3) variability in hemorrhagic activity of template structures—adamalysin II is non-hemorrhagic whereas atrolysin C (Ht-d) is hemorrhagic—which are ideal template structures to model the structures of groups of metalloproteinases possessing variable in vivo hemorrhagic activities from the same rattlesnake species; and (4) good structural alignment of target protein sequences to template sequences as judged by a lack of major structural/domain gaps in the alignment and the presence of similar clusters of conserved amino acids (zinc binding sites, methionine turns, α-helix C, and C-terminal region). The initial structural alignment of the templates and the target C. s. scutulatus amino acid sequences was done by using the AS1 algorithm of the Align2D program of the Insight2000 program. Small C-terminal and N-terminal overhangs (2–3 amino acids) within the amino acid alignment of the template and target amino acid sequences were cut off from the final version of the amino acid alignment of the metalloproteinases and thus were not used for generating the molecular models. Following the generation of the draft versions of each of the eight molecular models, it was necessary to “fill-in” small, unresolved structural gaps (<12 % of the amino acid sequence) that were generated after the first round of molecular modeling. To address the unresolved gaps, we re-submitted the draft versions of the molecular models for a second round of molecular modeling to build missing loops using a loop database of Insight2000.
Energy minimization of the molecular models
The final versions of the molecular models were subject to energy minimization protocols as explained below using the Discovery module of Insight2000. All molecular models were globally and locally optimized by applying at least 300 iterations using the Step Gradient and Steepest Decent minimization methods of the Discovery 3 module of the Insight2000 molecular simulation software package and further revalidated using the SPDB Viewer software to correct for molecular constrains such as unusual bond angles of the side chains, fixing unrealistic spatial strains within the Cα trace, unusual geometry, bond lengths, stereochemistry, and unusual β-factors. To validate the quality of all the molecular models, we analyzed the Ramachandran plots and structural and energy profiles for all of the molecular models as shown in supplementary Fig. 1 for C. s. scutulatus models. In brief, all the molecular models contained 1–3 glycine amino acid residues outside the allowable psi/phi ranges of the Ramachandran plots, which are similar glycine residues as seen for the crystal structure templates and showed comparable structural and energy profiles to that of the template crystal structures adamalysin II and atrolysin C (Supplementary Table 1). Moreover, none of these residues were located in secondary structural elements but were found in expected interconnecting loops (Supplementary Table 1). All models were further refined for atomic defects in their structure including incorporating missing hydrogen bonds within certain amino acid side chains, ligating breaks in the carbon traces of the structure (more than 3 Å), and adding a solvation layer (water). Final Gibbs free-energy calculations for all models were calculated using Insight2000 and SPDV Viewer, which yielded results that were comparable to the template crystal structures. The final molecular conformations were rendered in Kabsch–Sander mode as depicted in Figs. 3 and 4 of this manuscript. The molecular models were superimposed and analyzed for the number of disulfide bonds, root mean square (RMS) deviation values for the complete Cαtrace, and for important molecular regions using the Swiss PDB Viewer freeware (http://spdbv.vital-it.ch/download.html) [36–39], PyMol and Insight 2000 Viewer packages (MSI, San Diego, CA, USA).
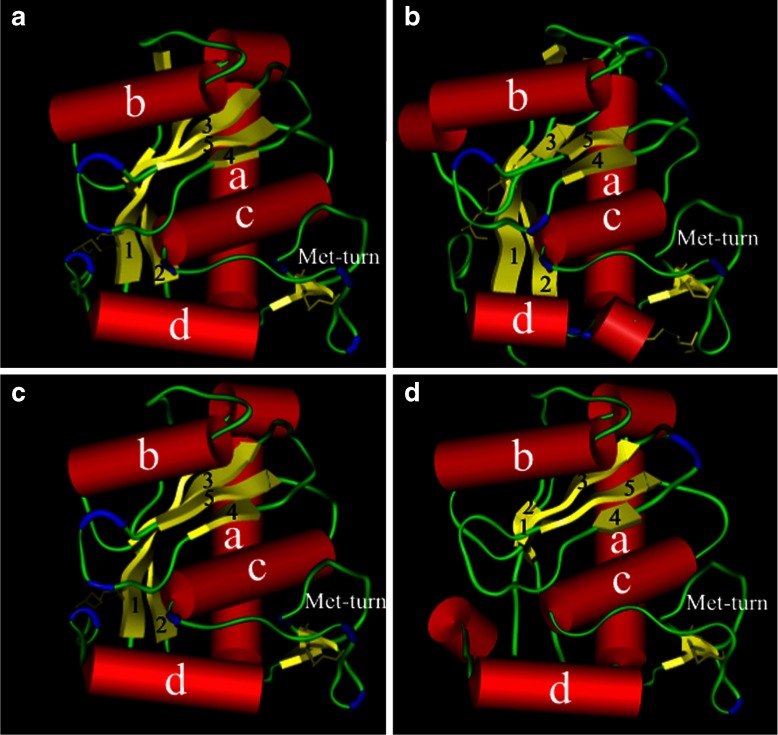
Fig. 3.
Molecular models of the four groups of C. s. scutulatus metalloproteinases. The three-dimensional molecular models of the four putative Crotalus s. scutulatus rattlesnakes metalloproteinases (quadrants I, II, III, and IV show models for GP1, GP2, GP3, and GP4 Mojave metalloproteinase models, respectively) as rendered in Kabsch–Sander mode. The a-helices have been rendered as red cylinders while the β-sheets illustrated as yellow flattened elongated arrows. The β-turns are rendered in blue and the disulfide bonds are illustrated as thick yellow lines bridging several regions of the enzyme
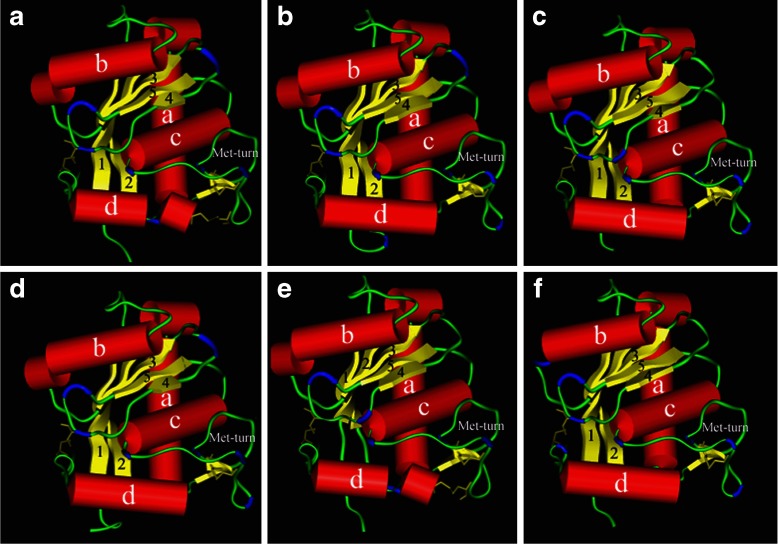
Fig. 4.
Molecular models of atrolysin and adamalysin II metalloproteinases. The three-dimensional molecular models of Crotalus atrox Atrolysins Ht-a, Ht-b, Ht-d, Ht-c, and Ht-e, and adamalysin II (crystal structure) are rendered in Kabsch–Sander mode and illustrated as quadrants I, II, III, IV, V, and VI, respectively. The a-helices illustrated are rendered red cylinders and the β-sheets illustrated as flat yellow arrows. The β-turns are in colored in blue and the disulfide bonds are illustrated as thick yellow lines
Surface potential and field potential maps and catalytic active-site calculations
In order to further elucidate a mechanism of variability in metalloproteinase-associated activities in the venom of C. s. scutulatus, the molecular surface potential maps of the catalytic region (β-strand IV, Met-turn, zinc binding domain, and α-helix C) were generated using the “Surface” algorithm of Swiss Modeler with the following sigma contouring values: negative potentials were displayed as red (−1.80), neutral potentials were displayed as white, while molecular regions containing positive potentials were displayed as blue (+ 1.80). The physical dimensions (cubic Å and square Å) of each cavity for the catalytic groove was calculated using the “Calculate Cavities” function of SYBYL software and the volume of each cavity was rendered and displayed in Conolly mode using SYBYL software. The total polar and non-polar molecular surface areas for each molecular model was calculated by using Surface Racer 5.0 [40] while the number of cavities was calculated using the “cavities” function of Swiss Modeler [36–39].
In order to elucidate differences in long-range electrostatic interactions of each of the four groups of metalloproteinases in the absence of a docked substrate, field potential maps based on Coulombic interactions (electrostatic) in vacuum were generated using Poisson–Boltzmann calculations, with a dielectric constant (protein) of 70 and solvent ionic strength (total salt concentration in mol/l) based on the salt concentrations of PBS at pH 7.0 were performed using Swiss PDB Viewer and SYBYL software. These conditions are the same chemical environment for which the proteolytic metalloproteinase-dependent activities of C. s. scutulatus venom were analyzed [32]. The field and surface potential maps were superimposed and rendered in RGB as shown in Fig. 5.
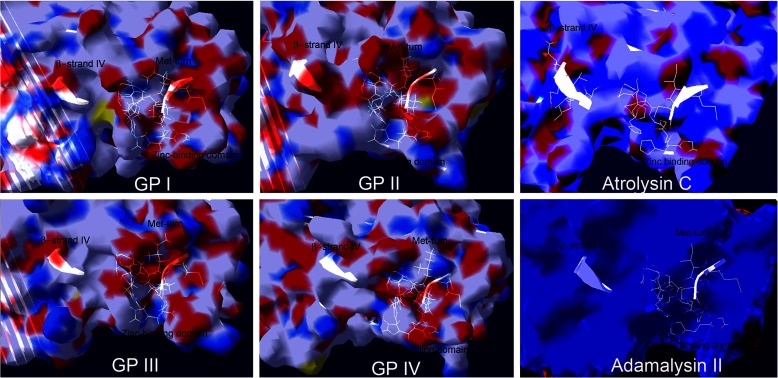
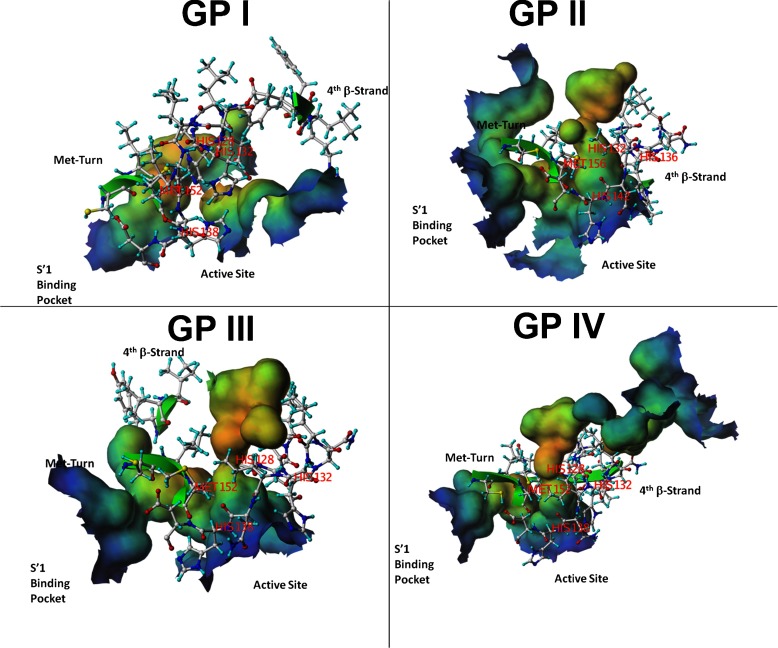
Fig. 5.
Molecular surfaces of the four groups of Mojave rattlesnake metalloproteinases and of atrolysin and adamalysin II crystal structures. Rendering of the surface potential maps for each molecular model of each C. s. scutulatus metalloproteinase group and the crystal structures of atrolysin C and adamalysin II. Positive surface potentials (Sigma: + 1.80) are shown as blue whereas negative surface potentials are shown in red (Sigma: −1.80) and neutral regions are rendered white. For convenience to the viewer, the side chains of the amino acid residues of the zinc binding domain, Met turn, and β-strand IV (top of the catalytic groove) have been displayed (white) in order to locate the catalytic groove
Molecular dynamic simulations
To assess the flexibility and dynamics of the interconnecting loops near the Met turn which form part of the catalytic site for each C. s. scutulatus metalloproteinase model, the AMBER99 force fields were assigned to each of the atoms of the models and a layer of water was added to generate PDBQT files. The shift in the central angle of the S’1-pocket for each model (includes residues 157–167) was analyzed for flexibility and mobility in solution for 10,000 femtoseconds at 300 Kelvins by using the Molecular Dynamics module of the SYBYL software. The flexibility of this region was plotted as the variation in angular displacement of this site over time.
Results
The complete amino acid sequences of the four C. s. scutulatus metalloproteinases were deduced from the open reading frames of the DNA sequences of the mature proteinase region of the metalloproteinases genes [32] (Fig. 1). Amino acid sequence alignments of the metalloproteinases showed 80.93 % homology to each other [32] (Fig. 1). The proteins consist of up to 192 amino acids in length, and the predicted molecular weights were between 21.7 and 22.2 kDa (http://expasy.org/). Hence, the four mature proteinases were classified as P-I type class of metalloproteinases based on the amino acid sequence alignments and molecular weight. GP2 showed the highest divergence among the four groups of metalloproteinases with 31 amino acid differences that were predominantly located within the N-terminal region of the proteinase domain. On the other hand, the C-terminal region of the four groups was highly conserved compared to the rest of the proteinase domain. GP1, GP3, and GP4 were highly similar to each other and had a 93.16 % amino acid sequence homology (Fig. 1). The amino acid sequences of GP3 and GP4 had a homology of 97.35 % (two amino acid substitutions). The proteinase domain of GP2 was 192 amino acids long or four amino acids longer compared to the three other groups of metalloproteinases due to an insertion of four consecutive amino acids located at amino acid positions 95-98 (TLEE). The predicted isoelectric focusing points for each of the four C. s. scutulatus metalloproteinase sequences were calculated as follows: GP1:6.17, GP2: 6.71, GP3:6.17, GP4:6.41.
Fig. 1.
Alignment of the amino acid sequences for the mature proteinase domains of C. s. scutulatus metalloproteinases. Positions for conserved cysteine (↑) and non-conserved cysteine (*) amino acid residues are indicated. Cysteine residues involved in the formation of disulfide bonds are colored red
All four metalloproteinases contained a conserved Zn2+ binding motif, HEXXHXXGXXHD (highly conserved amino acid residues indicated in bold, amino acids numbers 132–143, Fig. 1). The amino acid sequence in the methionine turn (Met-turn) [41], the region that forms the hydrophobic base of the Zn2+ binding site, was 100 % homologous (CIMASVL, residues 154 to 160) for all of the mature metalloproteinases and all four groups had four C-terminal cysteine residues at amino acid positions 147, 149, 154, and 187 that formed two disulfide bond bridges (Fig. 3). In addition to the two disulfide bonds, the GP1, GP3, and GP4 isoforms had a cysteine residue at amino acid position 106, whereas GP2 had a cysteine residue at amino acid position 171. All of the four groups contained an N-glycosylation site (defined by the sequence NXS/T) at position 170. However, unlike the three other groups of metalloproteinases, the glycosylation site for GP2 had a cysteine residue, which followed the asparagine residue rather than tyrosine (Y171 →171C, Fig. 1), and which was predicted to form a third disulfide bridge based on the molecular model of the proteinase domain (Figs. 2 and 3).
Fig. 2.
Secondary structural alignment for the molecular structures of the presumed Crotalus s. scutulatus metalloproteinases, atrolysins (Ht-a, Ht-c, Ht-d, and Ht-e,) and the crystal structures of atrolysin Ht-d and adamalysin II (1IAG). Gaps are introduced for a better alignment. Consensus amino acid residues (*) and chemically similar (.) amino acid residues are indicated. Residues that participate in the α-helix (h) and β-sheet (s) formation are indicated below the amino acid residues
Secondary structure analysis demonstrated that the four metalloproteinase groups were rich in α-helices and β-strands located within the N-terminal region. In summary, all four groups contained a series of highly conserved N-terminal α-helices and eight β-strands except for GP2, which lacked one N-terminal α-helix. The GP2 isoform also had a truncated N-terminal α-helix C, and a distorted α-helix D, which was predicted to split into two smaller segments (Fig. 2). All four groups showed a similar topology (arrangement of secondary structural elements) with five β-strands that formed the β-pleated sheet core of the proteinase domain that precedes the zinc binding domain. At the C-terminal domain, the four groups contained two short β-strands that harbor the Met turn. This structural region was arranged in the same order as reported for all RSVMPs [34, 35, 44, 45].
Molecular models for the four groups of Mojave metalloproteinase models were generated using the crystal structures of atrolysin C and adamalysin II as template structures and subjected to global energy minimization protocols, validated for quality, and subjected to molecular refinement protocols as explained in the Methods section of this manuscript. Each molecular model was thoroughly validated and analyzed for their overall quality compared to the crystal template structures by subjecting each model to several energy and structural quality control analyses as summarized in supplementary Table 1. Overall, the Mojave molecular models possessed either only one or no amino acids that produced undesirable clashes with other amino acids. In addition, all of the molecular models scored well for total solvent accessibility (73–75 % for GP1–GP4 vs. 85–86 % for template crystal structures) and for predicted tertiary structure analyses by comparing the predicted secondary structure for each metalloproteinase amino acid sequence to the molecular models (72–77 % for GP1–GP4 vs. 85–86 % for template structures). Furthermore, the mean force field energy for each of the four C. s. scutulatus molecular models was similar to that of the template crystal structures of adamalysin II and atrolysin C (supplementary Table 1). In addition, all three catalytic histidine residues bound successfully to zinc atoms at their catalytic sites (data imbedded in PDB files that can be downloaded in the electronic supplemental material section of this manuscript). Threading energy analyses, an energy metric used to study the chemical environment for each amino acid, demonstrated that all C. s. scutulatus metalloproteinase models contained similar threading energy profiles (data not shown) but only two cysteine amino acids (Cys 143 and Cys 145) contained high threading energies as these two residues were located in a surface loop that precedes the Met turn while the template structures possessed one amino acid each with high threading energies (Leu 2 for adamalysin II and Met 116). Solvent accessibility profiles demonstrated that all molecular models contain similar mean scores for predicted solvent accessibilities compared to the crystal template structures (Supplementary Table 1) and contained the expected stretches of hydrophobic amino acid residues buried within the zinc binding domains, in the methionine turns, and the central β-pleated sheet core while the C-terminal region is more solvent exposed (data not shown). The secondary structural prediction analysis (Fig. 2) showed that the four metalloproteinase groups, as displayed in Kabsch–Sander mode in Fig. 3, contained two major structural domains, an upper N-terminal domain and a lower C-terminal domain. All the molecular models have four conserved a-helices, termed A, B, C, and D, and five N-terminal β-strands (I–V) that form the central hydrophobic β-pleated sheet core region, which is arranged in the same topological conformation as published for the crystal structures of adamalysin II of C. adamanteus [34, 35, 43–45] and for atrolysin C (Ht-d) of C. atrox [34, 35]. The C-terminal domain included a downward loop preceding the Zn+2 binding site but located after α-helix C. All four models had the same conformation for the methionine-turn (Met turn) with the methionine residue located approximately 2 Å away from the zinc ion within the zinc binding pocket.
In order to analyze the spatial arrangement of structural domains among the four molecular models, we superimposed them using Swiss PDV Viewer. By using this method, we found that the Zn2+ binding site was highly conserved (RMS: <1.00 Å) and that the first six amino acid residues in the Zn2+ binding site superimposed very well, suggesting that this domain is an obligatory conserved region for snake venom metalloproteinases. The Met turn loop was stabilized by a disulfide bond (cysteine residues 147 and 154 for the GP2 and cysteine residues 143 and 150 for GP1, GP3, and GP4) in all four molecular models. GP1, GP2, and GP3 contained a second disulfide bond that linked the cysteine residues at positions 102 and 183, and for GP2, at amino acid positions 87 and 187. This second disulfide bond bridged the carboxyl terminus to the amino terminal end of the molecule in the same manner as shown by the crystal structures of rattlesnake metalloproteinases [34, 35, 43–45]. Unexpectedly, the GP2 model had an additional third disulfide bond (149–171) that pulled the α-helix D towards the methionine-turn loop and which caused α-helix D to be partitioned into two smaller segments (Fig. 3).
In order to compare the spatial arrangement of critical structural elements in the cleft of the active site, we superimposed the Ca-backbone traces for the whole protein, or for certain regions, using the “Magic” fit function of PDB Swiss Viewer. The spatial differences were expressed as the root mean square deviation (RMSD) values (measured in Å). These data are summarized in Table 1. Overall, with the exception of the Zn2+ binding domain and the Met turn, the molecular structures of the four groups showed significant structural deviations for other functional regions of the proteinase domains including the S’1-pockets and β-strand IV, two critical structural elements that form the base of the catalytic groove and the northern cleft wall, respectively (Table 1). The complete Cα-backbone traces of the molecular models of GP1 and GP3, two groups of enzymes with similar in vitro proteinase activities [32], superimposed well on each other (<1.00 Å) whereas GP4 and GP2, two metalloproteinase groups with different proteolytic activities, had a higher spatial deviation of their Cα-backbone traces (Table 1).
Table 1.
The root mean square deviation (RMSD) values of the superimposed Ca-backbone atoms of the complete proteinase domain, and the β-strand IV-linker, the S’1-pocket, and the zinc binding sites of the Crotalus s. scutulatus metalloproteinases, GP1, GP2, GP3, and GP4, with the metalloproteinase of Crotalus adamanteus adamalysin II (Adll), and Crotalus atrox atrolysins Ht-a, Ht-b, Ht-c, Ht-d, and Ht-e
| Proteinases | RMSDa | RMSDb | RMSDc | RMSDd |
|---|---|---|---|---|
| GP1 vs. GP2 | 1.52 Å | 1.69 Å | 1.00 Å | 0.50 Å |
| GP1 vs. GP3 | 0.56 Å | 0.49 Å | 0.31 Å | 0.55 Å |
| GP1 vs. GP4 | 1.25 Å | 1.11 Å | 1.00 Å | 0.80 Å |
| GP2 vs. GP3 | 1.60 Å | 1.73 Å | 1.02 Å | 0.65 Å |
| GP2 vs. GP4 | 1.82 Å | 2.10 Å | 1.40 Å | 0.50 Å |
| GP3 vs. GP4 | 1.31 Å | 1.07 Å | 1.00 Å | 1.00 Å |
| AdII vs. GP1 | 1.41 Å | 1.17 Å | 1.13 Å | 0.49 Å |
| AdII vs. GP2 | 1.62 Å | 2.20 Å | 1.30 Å | 0.55 Å |
| AdII vs. GP3 | 1.49 Å | 1.24 Å | 0.78 Å | 0.61 Å |
| AdII vs. GP4 | 1.77 Å | 1.21 Å | 1.41 Å | 0.87 Å |
| Ht-a vs. GP1 | 1.33 Å | 1.28 Å | 0.88 Å | 0.50 Å |
| Ht-b vs. GP1 | 1.41 Å | 1.24 Å | 1.09 Å | 0.52 Å |
| Ht-c vs. GP1 | 1.41 Å | 1.23 Å | 0.84 Å | 0.56 Å |
| Ht-d vs. GP1 | 1.42 Å | 1.32 Å | 0.93 Å | 0.48 Å |
| Ht-e vs. GP 1 | 1.45 Å | 1.01 Å | 1.23 Å | 0.43 Å |
| Ht-a vs. GP2 | 1.44 Å | 1.97 Å | 0.91 Å | 0.42 Å |
| Ht-b vs. GP2 | 1.58 Å | 2.03 Å | 1.50 Å | 0.53 Å |
| Ht-c vs. GP2 | 1.58 Å | 2.13 Å | 1.30 Å | 0.52 Å |
| Ht-dvs. GP2 | 1.58 Å | 2.01 Å | 1.34 Å | 0.45 Å |
| Ht-e vs. GP2 | 1.61 Å | 1.97 Å | 0.96 Å | 0.39 Å |
| Ht-a vs. GP3 | 1.40 Å | 1.37 Å | 1.05 Å | 0.40 Å |
| Ht-b vs. GP3 | 1.50 Å | 1.30 Å | 0.97 Å | 0.44 Å |
| Ht-c vs. GP3 | 1.52 Å | 1.55 Å | 0.73 Å | 0.61 Å |
| Ht-d vs. GP3 | 1.51 Å | 1.31 Å | 0.80 Å | 0.52 Å |
| Ht-e vs. GP3 | 1.55 Å | 1.06 Å | 1.17 Å | 0.42 Å |
| Ht-a vs. GP4 | 1.68 Å | 1.26 Å | 1.59 Å | 0.43 Å |
| Ht-b vs. GP4 | 1.74 Å | 1.43 Å | 1.50 Å | 0.43 Å |
| Ht-c vs. GP4 | 1.78 Å | 1.53 Å | 1.41 Å | 0.72 Å |
| Ht-d vs. GP4 | 1.73 Å | 1.42 Å | 1.42 Å | 0.44 Å |
| Ht-e vs. GP4 | 1.74 Å | 1.17 Å | 1.63 Å | 0.52 Å |
a Complete molecular structure RMSD values for Cα-backbone atoms
b β-strand IV-linker RMSD values for Cα-backbone atoms
c S′1-pocket RMSD values for Ca-backbone atoms
d Zinc binding site RMSD values for Ca -backbone atoms
To further understand a structural basis for the variation in proteinase activity in the venom of the Mojave rattlesnake, we generated four additional models of the mature proteinase domains from C. atrox(Western Diamondback rattlesnake), and searched for structural domains that are conserved between Mojave and Western Diamondback rattlesnake metalloproteinases. The amino acid sequences of C. atrox atrolysins Ht-a, Ht-b, Ht-c, and Ht-e were used to generate these models using the Homology Module of Insight 2000 software. The atrolysin molecular models were minimized and optimized for torsions, improper bonds, abnormal geometry, force-field energies, abnormal angles of amino acid side chains, and missing hydrogen atoms were added to incomplete amino acids. More importantly, all atrolysin models have high QMean scores (0.67–0.75), mean solvent accessibility (81–85 %), and predicted secondary structure scores (81–85 % as predicted by QMean analyses), possessed no amino acids making clashes to other amino acid side chains or carbon backbone atoms, and contained similar structural and energy profiles as the atrolysin C and adamalysin II template structures as validated by SYBYL (data not shown). The molecular models for atrolysins Ht-a, Ht-b, Ht-c, and Ht-eand the crystal structures for atrolysin C (Ht-d) and adamalysin II are illustrated in Fig. 4. Like the Mojave rattlesnake molecular models, the atrolysin molecular models and the crystal structures of atrolysin C and adamalysin II show a similar topology. The N-terminal domain, which includes the β-strands that form the central β-pleated sheet core, is highly conserved among all eight molecular models. Moreover, all molecular models contain three elongated N-terminal α-helices, and contain a highly conserved C-terminal domain. In a manner similar to the molecular model of the Mojave rattlesnake GP2 isoform, the molecular models for atrolysin A and E contained a third disulfide bond that bridges the downward loop of the Met-turn with the N-terminal portion of α-helix D.
Superimpositions of the Cα-carbon traces of the atrolysin models and the crystal structures onto the Cα-carbon traces of the Mojave rattlesnake metalloproteinase models were very similar except for some significant spatial deviations as described below (Table 1). As expected, the Zn2+ binding domains were well superimposed among the atrolysin and Mojave rattlesnake metalloproteinases (Table 1). This observation agrees with the view that the Zn2+ binding domain is an obligatory conserved region for snake venom metalloproteinases and further validates our molecular models. Interestingly, not only do all four Mojave rattlesnake metalloproteinase models have a lower amino acid homology compared to the mature proteinase domains of the atrolysin models, but all four Mojave rattlesnake metalloproteinase models showed a significant structural deviation RMSD range (RMSD: 1.33 ≤ 1.78 Å for C. s. scutulatus models) when superimposed onto each other while the atrolysins molecular models showed less deviation when superimposed among each other (RMSD: 0.48 ≤ 0.60 Å) or to adamalysin II (RMSD: 0.48 ≤ 0.60 Å) (Tables 1 and 2). In addition, the S’1-pocket of all four atrolysins molecular models superimposed well to each other (RMSD range, 0.33 ≤ 0.88 Å), suggesting that the atrolysins bind similar substrates that induce hemorrhage when cleaved. Interestingly, the S’1-pocket of the GP3 isoform of the Mojave rattlesnake, a non-hemorrhagic enzyme but containing a high proteolytic activity [32], superimposed well with the S’1-pockets of adamalysin II and atrolysins Ht-c and Ht-d (Table 1). In agreement with the proteolytic and hemorrhagic profiles for the C. s. scutulatus metalloproteinases, the S’1-pockets and Met turns of the GP1 Mojave metalloproteinase superimposed the best with atrolysins Ht-a and Ht-b (Table 1) compared to the other three C. s. scutulatus metalloproteinases.
Table 2.
The root mean square deviation (RMSD) values of superimposed Ca-backbone atoms of the complete proteinase domain, and the β-strand IV-linker, the S-pocket, and the zinc binding sites of and Crotalus atrox atrolysins Ht-a, Ht-b, Ht-c, Ht-d, and Ht-eas compared to each other
| Proteinases | RMSDa | RMSDb | RMSDc | RMSDd |
|---|---|---|---|---|
| Ht-a vs. Ht-b | 0.53 Å | 0.39 Å | 0.82 Å | 0.22 Å |
| Ht-a vs. Ht-c | 0.56 Å | 0.44 Å | 0.62 Å | 0.33 Å |
| Ht-a vs. Ht-d | 0.55 Å | 1.04 Å | 0.73 Å | 0.16 Å |
| Ht-a vs. Ht-e | 0.60 Å | 1.14 Å | 0.33 Å | 0.24 Å |
| Ht-b vs. Ht-c | 0.50 Å | 0.63 Å | 0.62 Å | 0.18 Å |
| Ht-b vs. Ht-d | 0.48 Å | 1.07 Å | 0.48 Å | 0.23 Å |
| Ht-b vs. Ht-e | 0.66 Å | 1.16 Å | 0.90 Å | 0.22 Å |
| Ht-c vs. Ht-d | 0.54 Å | 1.08 Å | 0.75 Å | 0.18 Å |
| Ht-c vs. Ht-e | 0.70 Å | 1.17 Å | 0.88 Å | 0.30 Å |
| Ht-d vs. Ht-e | 0.55 Å | 0.60 Å | 0.75 Å | 0.28 Å |
| Ht-a vs. AdII | 0.63 Å | 0.60 Å | 0.88 Å | 0.37 Å |
| Ht-b vs. AdII | 0.70 Å | 0.62 Å | 0.68 Å | 0.38 Å |
| Ht-c vs. AdII | 0.51 Å | 0.54 Å | 0.48 Å | 0.31 Å |
| Ht-d vs. AdII | 0.71 Å | 1.11 Å | 0.40 Å | 0.35 Å |
| Ht-e vs. AdII | 0.78 Å | 1.02 Å | 0.87 Å | 0.42 Å |
aComplete molecular structure RMSD values for Ca-backbone atoms
b β-strand IV-linker RMSD values for Ca-backbone atoms
c S-pocket RMSD values for Ca-backbone atoms
d Zinc binding site RMSD values for Ca-backbone atoms
The amino acid and structural alignments predict that the Mojave rattlesnake metalloproteinase groups contain catalytic grooves of variable sizes and contain different physico-chemical environments surrounding the catalytic groove. To address this question, we generated surface and field potential maps near the catalytic groove (Zn+2 binding domain, Met turn, and β-strand IV) for each molecular model. Although the Zn+2 binding sites of all metalloproteinase groups superimposed well to each other (Table 1), the electrostatic environment surrounding the Zn+2 binding domain and near the N-terminal portion of α-helix C was dramatically different for all Mojave rattlesnake molecular models. Interestingly, the GP2 model contained a highly basic molecular surface compared to the other molecular models (Fig. 5). Furthermore, a rendering of the field potential maps surrounding the catalytic sites of the metalloproteinase molecular models show that the field potentials near the N-terminal portion of α-helix C, which contains the Zn+2 binding site, has been reduced and disrupted for the GP2 model compared to the other three metalloproteinase models. This observation suggests that the ability of GP2 metalloproteinase to attract substrates in solution through long-range electrostatic interactions is limited (Fig. 6). Interestingly, the GP2 metalloproteinase field potential map resembles that of adamalysin II in that it contains a highly basic field potential surface whereas the GP1, GP3, and GP4 models contain a highly electronegative field potential surface near the catalytic cleft (Fig. 6). Interestingly, the field potential surface for atrolysin C, a hemorrhagic metalloproteinase, is extensive, wide, and predominantly acidic compared to all the other molecular models. These results suggest that a highly electronegative environment of GP1 and atrolysin C favorably attracts positively charged substrates to the active cleft. Furthermore, a thorough analysis of the molecular cavities showed that the four C. s. scutulatus metalloproteinase models have highly variable and amorphous catalytic binding grooves. The catalytic binding site of GP1 metalloproteinase is shallow and smaller, similar to atrolysin C (area: 277.5 Å2, volume: 263.81 Å3) hemorrhagic metalloproteinase, while the active site cavities were predicted to be wider and deeper for the other three C. s. scutulatus metalloproteinase models as shown in Fig. 7 (427 Å2 and 463 Å3 for GP2, 495 Å2 and 464 Å3 for GP3, 503 Å2 and 391.7 Å3 for GP4, respectively).
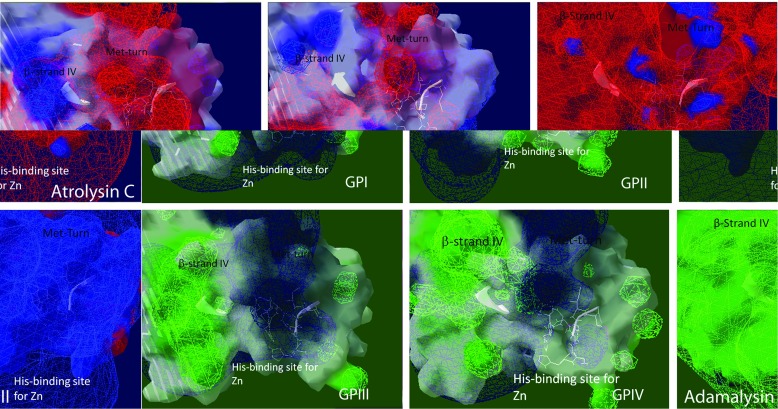
Fig. 6.
Rendering of field potential maps (Coulombic) for each molecular model of each C. s. scutulatus metalloproteinase group and for atrolysin C and adamalysin II crystal structures. Positive field potentials are shown as blue spheres whereas negative field potentials are shown as red spheres that conform the surface potential map and neutral regions are rendered white. For convenience to the viewer, the side chains of the amino acid residues of the zinc binding domain, Met turn, and β-strand IV (top of the catalytic groove) have been displayed (white) in order to locate the catalytic groove of each molecular model
Fig. 7.
Connolly surface rendering of the cavity that encompasses the catalytic active site and the S’1-pockets of the C. s. scutulatus molecular models. The cavity models have been color coded based on depth in which blue colored sections represent shallow portions of the cavity while red regions depict deeper areas of the cavity. For visual clarity, the amino acid residues of the three histidine residues of the active site and the methionine residue of the Met turn have been rendered in ball and stick mode. Note that all models have variable shape while the cavity size is shallow and smaller for GP1 metalloproteinase compared to the other three molecular models. The cavity area and volumes were calculated respectively for the four Mojave metalloproteinase models as follows: 277.5 Å2 and 263.81 Å3 for GP1, 427 Å2 and 463 Å3 for GP2, 495 Å2 and 464 Å3 for GP3, 503 Å2 and 391.7 Å3 for GP4, respectively
It has been suggested that the depth and accessibility of the S’1-pocket dictates hemorrhagic activity for metalloproteinases [35]. However, we did not find differences in the depth or a distance correlation between the center points of the catalytic and S’1-pockets (GPI: 8.31 Å, GPII: 8.54 Å, GPIII: 7.82 Å, and GPIV: 9.035 Å).
Discussion
Over the past decade, several high-resolution crystal structures of RSVMPs have been solved, and these data have enabled structural biologists to gain a deeper understanding of the substrate specificity and selectivity of these toxins [35, 44–47]. Given the limited number of available snake metalloproteinase crystal structures, predicting the molecular structures of additional RSVMPs by applying molecular modeling techniques is essential for designing catalytic inhibitors that block hemorrhage by these toxins. Indeed, molecular modeling and in silico affinity docking techniques have aided toxinologists in developing small molecular inhibitors of rattlesnake metalloproteinases and for understanding the molecular mechanisms of anti-venom compounds [48].
The P-I class of RSVMPs show variability in substrate selectivity, in binding affinities, and in their ability to degrade both fibrin clots and insoluble protein aggregates. Indeed, the high variation observed in rattlesnake venom composition—even within the same rattlesnake species—gives credence to the concept that pooling rattlesnake venom from individual rattlesnakes of the same species collected from different geographic locations is necessary to develop an effective “pan” anti-venom to efficiently treat bite victims [49]. Because of these biochemical properties, it is conceivable that rattlesnake recombinant metalloproteinases may find clinical applications such as treating humans affected by neurodegenerative disorders by degrading protein aggregates like α-synuclein and β-amyloid to stop or reverse mental degeneration. Recombinant forms of RSVMPs may also have application as anti-thrombolytic agents to treat deep vein thrombosis.
Although the mechanisms of substrate specificity of snake venom metalloproteinases are beginning to be elucidated, it is believed that small differences in amino acid sequences contribute to altering the physico-chemical landscape surrounding the catalytic region of the enzyme, which plays an important role in conferring substrate specificity. Indeed, several P-I metalloproteinases from the venom of the same snake species were found to show variation in proteolytic activities and substrate binding affinities. For example, two Akistrodon piscivorus leucostoma metalloproteinases that have high amino acid sequence homology and similar molecular weight sizes can degrade fibrinogen and fibrin to the same degree and do not produce hemorrhage in vivo [41]. In contrast, atrolysins from C. atrox, which have a much lower amino acid sequence homology to each other, degrade similar extracellular matrix substrates but produce very different digestion patterns and have different binding affinities to these substrates [50, 51].
The metalloproteinase composition of rattlesnake venom and their associated proteolytic activities may vary, even within the same species [5, 32, 33, 42, 43]. While the venom composition and hemorrhagic activities of C. atrox appears to be consistent among individual rattlesnakes [57], venom from individual C. s. scutulatushas different metalloproteinases whose activities vary [4, 32]. Previously, we clustered the venom metalloproteinases of C. s. scutulatus into four groups according to their hemorrhagic and protease activities [32]. The genomic DNA corresponding to these metalloproteinases was also sequenced [32]. Of the four groups, GP1 metalloproteinase was highly hemorrhagic and had high proteolytic activities, whereas the other three groups of metalloproteinases had modest or even lacked hemorrhagic activity and their protease activity varied [32].
Given the differences of the four groups of metalloproteinases, we sought to elucidate structural differences and molecular mechanisms that affect metalloproteinase activity. Using molecular modeling methodologies, the three-dimensional structures of the Mojave and atrolysin metalloproteinases were shown for the first time to be structurally homologous to the molecular structures of the metzincin-superfamily of zinc peptidases [41]. Primary, secondary, and structural superimpositions of the molecular models were done to identify specific structural elements that confer hemorrhagic and proteolytic activities to the C. s. scutulatus metalloproteinases. All eight molecular models that we studied, including atrolysin isoenzymes and the C. s. scutulatus metalloproteinases, had 3D structures similar to that of trypsin or chymotrypsin-like enzymes [41].
Overall, we found significant primary amino acid sequence differences among the four C. s. scutulatus metalloproteinase groups, which nevertheless contained highly conserved predicted tertiary structures. For instance, while GP2 had a lower amino acid homology compared to the other three C. s. scutulatus groups, this metalloproteinase contained minor differences in the length and alignment of certain secondary structural elements compared to the other three groups of metalloproteinases. These results suggest that the amino acid differences observed in GP2 may predict drastic differences in the spatial arrangement of structural elements among the four C. s. scutulatus metalloproteinase models. Indeed, by superimposing the complete Cα traces of the proteinase domains, RMSD analysis showed that the GP1 metalloproteinases had the highest structural homology to the atrolysins compared to the other three groups of C. s. scutulatus metalloproteinases. Even though the GP2 metalloproteinase superimposed better to the atrolysins compared to the GP4 proteinase, which is a weak hemorrhagic toxin (Table 1), it is conceivable that the lack of proteolytic and hemorrhagic activity of GP2 is explained by the presence of charge reversal amino acid substitutions surrounding the catalytic cleft compared to other metalloproteinase models that give rise to a different chemical landscape compared to other Mojave metalloproteinases (Fig. 6).
Moreover, prior to our studies, several studies showed that the hemorrhagic potency of rattlesnake venom metalloproteinases correlated with the degree of flexibility of the interconnecting loops that precede or follow the Met turn [52, 53]. For instance, it has been reported that the interconnecting loop that follows the Met turn, which also contains the S’1-specificity pocket, is remarkably more flexible in non-hemorrhagic metalloproteinases compared to hemorrhagic metalloproteinases [52]. In agreement with this observation [52, 53], the C. s. scutulatus metalloproteinases belonging to the non-hemorrhagic group of metalloproteinases (GP2) contained a much more dynamic S’1- pocket than the strongly or weakly hemorrhagic groups (GP1 and GP4) as measured by the standard deviations of the angular shift over time (GP1: ±4.04°, GP2: ±6.72°, GP3: ±6.73°, GP4: ±4.90°).
On the other hand, it is also plausible that GP2 is not expressed due to transcriptional repressive mechanisms as verified by a lack of immunoreactivity as determined by using anti-hemorrhagic metalloproteinase antibodies in rattlesnake venom [26]. In agreement with this concept, age-dependent microRNA-mediated suppression of RSVMPs has been suggested as an additional posttranslational mechanism for regulating rattlesnake venom composition and metalloproteinase-associated activities [54].
The N-terminal and C-terminal regions of the metalloproteinases delineate the active cleft, which is comprised of the Zn+2 binding site, the northern wall of the active site cleft (β-strand 4), and the S’1- pocket. The Zn+2 binding site in all four C. s. scutulatus metalloproteinases included six amino acid residues that formed part of the N-terminal portion of the α-helix C and six residues contained within the downward loop that follows α-helix C. The northern cleft wall (β-strand IV) of the active site cleft serves to orient the bound substrate towards non-primed amino acid residues that are C-terminally located towards the scissile peptide bond and determines the strength of enzyme-substrate interactions [41]. The northern cleft wall of the Mojave rattlesnake metalloproteinases superimposed well among each other based on their hemorrhagic potencies (GP1 vs. GP4, and GP3 vs. GP2). This finding suggests that a specific orientation of the northern cleft wall facilitates the access of specific extracellular matrix substrates to the catalytic site and induce hemorrhage when cleaved.
The S’1- pocket, which follows the Met-turn, harbors the P’1 side-chains of the bound substrate and is thought to determine the cleavage specificity of the enzymes [41]. Unlike the atrolysins, the 3D conformations of the S’1- pockets were not well conserved among the C. s. scutulatus metalloproteinases, suggesting that differences in substrate specificities for the C. s. scutulatus metalloproteinases is also conferred by spatial differences of the S’1 -pockets. Indeed, the GP2 and GP4 metalloproteinases, which are weakly hemorrhagic, showed a higher structural divergence for the S’1-pockets when superimposed to the atrolysin isoenzymes as shown in Table 1. The S’1-pocket of GP1 was more structurally homologous to the S’1-pockets of the atrolysins than of the other three C. s. scutulatus metalloproteinases, suggesting that GP1 and the atrolysins recognize similar substrates to produce hemorrhage. The depth and accessibility of the alternate binding cavity conformed by the S’1-pocket is thought to determine the ability of hemorrhagic metalloproteinases for binding bulky amino acid side chains (i.e., tyrosine and tryptophan) as found in collagen type IV (Gly-X), a critical factor that determines hemorrhagic activity of metalloproteinases [28]. In the present study, an analysis of the depth or the distance between the S’1-pocket and the catalytic site cavity revealed no significant differences or correlation to hemorrhagic activities for the C. s. scutulatus metalloproteinase models. While the atrolysins have been characterized as hemorrhagic toxins that cleave similar substrates in vitro [55, 56], the fact that their catalytic clefts superimpose well with each other but poorly to weakly hemorrhagic C. s. scutulatus metalloproteinases is consistent with this observation.
On the other hand, RMSD analyses of superimposed atrolysin metalloproteinase models showed that the spatial arrangement of specific molecular regions is not sufficient to explain their ability to induce hemorrhage since the catalytic binding clefts of the atrolysins unexpectedly superimposed well with that of adamalysin II, a non-hemorrhagic metalloproteinase (Table 2). Although RMSD analyses may explain conservation in catalytic activities for some metalloproteinases, these findings suggest that additional factors contribute to hemorrhagic activity of metalloproteinases and that slight deviations of specific structural elements or differences in amino acid sequences are not the factors that confer hemorrhagic and proteolytic activities of rattlesnake metalloproteinase in vivo [57]. Interestingly, we noticed that the catalytic clefts have similar sizes and depth for the GP2, GP3, and GP4 metalloproteinase, while that of GP1 metalloproteinase was shallower and smaller (Fig. 7), similar to that of the crystal structure of atrolysin C, a hemorrhagic metalloproteinase, suggesting that a narrow catalytic site of certain metalloproteinases is characteristic of hemorrhagic metalloproteinases.
Other than the disintegrin domain, some metalloproteinases contain disintegrin-like sequences imbedded within the proteinase domain which confer hemorrhagic activity. The proteinase domain of jararhagin, a Bothrops jararaca venom metalloproteinase that inhibits collagen-induced platelet aggregation, appears to interact with platelet integrin receptors independently of the disintegrin domain [3]. The RKKH segment, which is adjacent to the N-terminal β-strand 3 of the twisted β-pleated sheet, binds to the α2 I-domain of the integrin in such a way that the catalytic active site of jararhagin remains free to degrade the β1 subunit of the integrin [3]. None of the four groups of C. s. scutulatus metalloproteinases contain the RKKH segment and are not likely inhibitors of platelet aggregation.
The physico-chemical environment surrounding the catalytic cleft of RSVMPs is critical for dictating substrate specificity and proteolytic activities. A rendering of the surface and field potential maps of the metalloproteinase molecular models showed that the physico-chemical landscape surrounding catalytic cleft of GP2 metalloproteinase is significantly altered compared to the other three metalloproteinases. Specifically, GP2 contained a highly positively charged surface patch near the N-terminal portion of the Zn+2 binding domain and near the S’1-pocket, whereas the other three groups contained negatively charged surface landscapes. The positively charged field potential map surrounding the Met-turn of GP2 was severely contracted compared to the other metalloproteinases (Fig. 6). This observation suggests that the GP2 metalloproteinase may be impaired for attracting positively charged extracellular matrix substrates through long-range Coulombic interactions in the blood of an affected human individual. Likewise, atrolysin C and acutolysin A [35, 45], two strongly hemorrhagic metalloproteinases, also show very extensive and robust negative field potential maps compared to other metalloproteinases (data not shown). This result suggests that a series of acidic residues located around the catalytic cleft region is critical for attracting positively charged matrix substrates like collagen type IV, extracellular matrix components that form part of the basal lamina of capillaries or bloodstream. Conversely, adamalysin II, a non-hemorrhagic metalloproteinase, contained a robust basic surface landscape and an extensive positively charged field potential map near the catalytic cleft (Fig. 6). Similar observations were seen for other crystal structures of non-hemorrhagic metalloproteinases including the fibrinogenolases (TM-1, TM-2, and TM-3) from Taiwan Habu [58]. Indeed, these three non-hemorrhagic metalloproteinases contain a very robust basic field potential surface whereas acutolysin C and Bap1 from Bothrops asper, two weakly hemorrhagic metalloproteinases [53], show energy landscapes with mix polarities in that they contain a highly basic field potential landscape at the N-terminal region and an acidic field potential energy landscape near the catalytic site (data not shown). In further agreement with this view, the intensity of the negative field potential maps surrounding the Zn+2 binding domain and the Met turn for each of the C. s. scutulatus metalloproteinases correlated with hemorrhagic potency [32] in the following manner: atrolysin C > GP1 > GP4 > GP3 > GP2 > adamalysin II. A previous study successfully segregated 19 hemorrhagic and non-hemorrhagic PI metalloproteinases on the basis of their total polar molecular surface area. In brief, hemorrhagic metalloproteinases tended to contain a significantly higher polar molecular surface area compared to non-hemorrhagic metalloproteinase [59]. While our studies also support the notion that multiple factors contribute to hemorrhagic activity, including the physico-chemical landscape surrounding the catalytic cleft, we unexpectedly found an inverse relationship in the total polar surface area compared to the hemorrhagic activity of each C. scutulatus metalloproteinase (GP1: 3261 Å2, GP2: 3682 Å2, GP3: 3352 Å2, GP4: 3314 Å2, atrolysin C: 3440 Å2 and adamalysin II:3399 Å2), suggesting that other factors besides surface polarity play roles in conferring hemorrhagic activity for PI snake venom metalloproteinases. Hence, it is plausible that a combination of factors that include both the amount of surface polarity and the type of polarity (acidic vs. basic) near the catalytic sites of metalloproteinases may further explain differences in hemorrhagic activities. Interestingly, we also found a modest correlation in the number of cavities to hemorrhagic activity for each molecular model. The hemorrhagic metalloproteinase models and crystal structures (GP1, atrolysin C, acutolysin A, and GP4) contained fewer surface cavities (4, 2, 1, and 4 cavities respectively) while the non-hemorrhagic metalloproteinases GP2, GP3, and adamalysin II tended to contain a higher number of surface cavities (8, 5, and 5 respectively). Hence, future studies are required to further elucidate how the combination of all the aforementioned structural and physico-chemical factors lead to differences in hemorrhagic activities of rattlesnake venom metalloproteinases. Overall, these observations suggest that the molecular basis for differences in hemorrhagic activities for PI metalloproteinases is much more complex than anticipated.
Overall, our data suggest that the following factors confer metalloproteinases with differences in substrate selectivity and hemorrhagic activities: (a) differences in amino acid sequence within the N-terminal region of the mature proteinase domain, (b) spatial deviations of the northern cleft wall and S’1-pockets, (c) differences in the physico-chemical environment surrounding the catalytic groove (Figs. 5 and 6), (d) differences in the flexibility and size of the catalytic cleft, and the (e) molecular surface complexity (number of surface cavities). In aggregate, our results and other reports shed light on the molecular basis of substrate selectivity for hemorrhagic rattlesnake metalloproteinases and may aid in the design of basic peptide inhibitors that irreversibly anneal to the catalytic sites of hemorrhagic enzymes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported by NIH grants S06GM08012, RR08124, and P20 GM103554 (a COBRE grant in Cell Signaling Across Membranes). We also acknowledge technical assistance of Mr. Mukhiddin Ibodullaev and Mr. Noorzod Aripov of the Tashkent Ulugbek International School.
Conflict of interest
All authors that contributed to this study declare no conflicts of interest.
Footnotes
An erratum to this article is available at http://dx.doi.org/10.1007/s10867-014-9347-y.
Contributor Information
Ruben K. Dagda, Email: rdagda@medicine.nevada.edu
Sardar E. Gasanov, Email: sgasanov@yahoo.com
Boris Zhang, Email: bzhang249483@gmail.com.
William Welch, Email: welch@unr.edu.
Eppie D. Rael, Email: eppierael@yahoo.com
References
- 1.Markland FS. Rattlesnake venom enzymes that interact with components of the hemostatic system. J. Toxicol. Tox. Rev. 1983;2:119–160. doi: 10.3109/15569548309012695. [DOI] [Google Scholar]
- 2.Molina O, Seriel RK, Martinez M, Sierra ML, Varela-Ramirez A, Rael ED. Isolation of two hemorrhagic toxins from Crotalus basiliscus basiliscus (Mexican west coast rattlesnake) venom and their effect on blood clotting and complement. Intern. J. Biochem. 1990;22(3):253–261. doi: 10.1016/0020-711X(90)90337-3. [DOI] [PubMed] [Google Scholar]
- 3.Pentikainen O, Hoffren AM, Ivaska J, Kapyla J, Nyronen T, Heino J, Johnson MS. RKKH peptides from the snake venom metalloproteinase of Bothrops jararaca bind near the metal ion-dependent adhesion site of the human integrin alpha(2) I-domain. J. Biol. Chem. 1999;274(44):31493–31505. doi: 10.1074/jbc.274.44.31493. [DOI] [PubMed] [Google Scholar]
- 4.Rael ED, Lieb CS, Maddux N, Varela-Ramirez A, Perez J. Hemorrhagic and Mojave toxins in the venoms of the offspring of two Mojave rattlesnakes (Crotalus scutulatus scutulatus) Comp. Biochem. Physiol. 1993;106(3):595–600. doi: 10.1016/0305-0491(93)90136-s. [DOI] [PubMed] [Google Scholar]
- 5.Rael ED, Rivas JZ, Chen T, Maddux N, Huizar E, Lieb CS. Differences in fibrinolysis and complement inactivation by venom from different northern blacktailed rattlesnakes (Crotalus molossus molossus) Toxicon. 1997;35(4):505–513. doi: 10.1016/S0041-0101(96)00139-0. [DOI] [PubMed] [Google Scholar]
- 6.Hite LA, Jia LG, Bjarnason JB, Fox JW. cDNA sequences for four snake venom metalloproteinases: structure, classification, and their relationship to mammalian reproductive proteins. Arch. Biochem. Biophys. 1994;308(1):182–191. doi: 10.1006/abbi.1994.1026. [DOI] [PubMed] [Google Scholar]
- 7.Jia LG, Wang XM, Shannon JD, Bjarnason JB, Fox JW. Function of disintegrin-like/cysteine-rich domains of atrolysin A. Inhibition of platelet aggregation by recombinant protein and peptide antagonists. J. Biol. Chem. 1997;272(20):13094–13102. doi: 10.1074/jbc.272.20.13094. [DOI] [PubMed] [Google Scholar]
- 8.Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Semin. Cell. Dev. Biol. 2009;20(2):146–152. doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Fry BG. From genome to venome: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15(3):403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casewell NR. On the ancestral recruitment of metalloproteinases into the venom of snakes. Toxicon. 2012;60(4):449–454. doi: 10.1016/j.toxicon.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs’ MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25(11):2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda S, Takeya H, Iwanaga S. Snake venom metalloproteinases: structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta. 2012;1824(1):164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Serrano SM, Kim J, Wang D, Dragulev B, Shannon JD, Mann HH, Veit G, Wagener R, Koch M, Fox JW. The cysteine-rich domain of snake venom metalloproteinases is a ligand for von Willebrand factor A domains: role in substrate targeting. J. Biol. Chem. 2006;281(52):39746–39756. doi: 10.1074/jbc.M604855200. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007;581(13):2416–2422. doi: 10.1016/j.febslet.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 15.Takeda S, Igarashi T, Mori H. Crystal structure of RVV-X: an example of evolutionary gain of specificity by ADAM proteinases. FEBS Lett. 2007;581(30):5859–5864. doi: 10.1016/j.febslet.2007.11.062. [DOI] [PubMed] [Google Scholar]
- 16.Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, Mackie S, Olland S, Lin L, Zhong X, Kriz R, Reifenberg EL, Collins-Racie LA, Corcoran C, Freeman B, Zollner R, Marvell T, Vera M, Sum PE, Lavallie ER, Stahl M, Somers W. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17(1):16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275(12):3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez JM, Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000;82(9–10):841–850. doi: 10.1016/S0300-9084(00)01163-9. [DOI] [PubMed] [Google Scholar]
- 19.Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45(8):969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Jia LG, Wang XM, Shannon JD, Bjarnason JB, Fox JW. Inhibition of platelet aggregation by the recombinant cysteine-rich domain of the hemorrhagic snake venom metalloproteinase, atrolysin A. Arch. Biochem. Biophys. 2000;373(1):281–286. doi: 10.1006/abbi.1999.1517. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Q, Dangelmaier C, Smith JB. The hemorrhagin catrocollastatin inhibits collagen-induced platelet aggregation by binding to collagen via its disintegrin-like domain. Biochem. Biophys. Res. Commun. 1996;219(3):720–726. doi: 10.1006/bbrc.1996.0301. [DOI] [PubMed] [Google Scholar]
- 22.Rael ED, Martinez M, Molina O. Isolation of a fibrinolytic protease, M4, from venom of Crotalus molossus molossus (northern blacktail rattlesnake) Haemostasis. 1992;22(1):41–49. doi: 10.1159/000216290. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Hu P, Ritter MR, Swenson SD, Argounova S, Epstein AL, Markland FS. Molecular cloning and functional expression of contortrostatin, a homodimeric disintegrin from southern copperhead snake venom. Arch. Biochem. Biophys. 2000;375(2):278–288. doi: 10.1006/abbi.1999.1682. [DOI] [PubMed] [Google Scholar]
- 24.Takeda S, Takeya H, Iwanaga S. Snake venom metalloproteinases: structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta. 2012;1824(1):164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther. 1994;62(3):325–72. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 26.Shimokawa K, Jia LG, Wang XM, Fox JW. Expression, activation, and processing of the recombinant snake venom metalloproteinase, pro-atrolysin E. Arch. Biochem. Biophys. 1996;335(2):283–294. doi: 10.1006/abbi.1996.0509. [DOI] [PubMed] [Google Scholar]
- 27.Georgieva D, Ohler M, Seifert J, von Bergen M, Arni RK, Genov N, Betzel C. Snake venomic of Crotalus durissus terrificus—correlation with pharmacological activities. J. Proteome Res. 2010;9(5):2302–2316. doi: 10.1021/pr901042p. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Romero G, Rucavado A, Lazcano D, Gutierrez JM, Borja M, Lomonte B, Garza-Garcia Y, Zugasti-Cruz A. Comparison of venom composition and biological activities of the subspecies Crotalus lepidus lepidus, Crotalus lepidus klauberi and Crotalus lepidus morulus from Mexico. Toxicon. 2013;71:84–95. doi: 10.1016/j.toxicon.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Castro EN, Lomonte B, del Carmen Gutierrez M, Alagon A, Gutierrez JM. Intraspecies variation in the venom of the rattlesnake Crotalus simus from Mexico: different expression of crotoxin results in highly variable toxicity in the venoms of three subspecies. J. Proteomics. 2013;87:103–121. doi: 10.1016/j.jprot.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 30.Bjarnason JB, Tu AT. Hemorrhagic toxins from Western diamondback rattlesnake (Crotalus atrox) venom: isolation and characterization of five toxins and the role of zinc in hemorrhagic toxin e. Biochemistry. 1978;17(16):3395–3404. doi: 10.1021/bi00609a033. [DOI] [PubMed] [Google Scholar]
- 31.Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle SB. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57(5):657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Dagda, R.K., Gasanov, S., De La OIII, Y., Rael, E.D., Lieb, C.: Genetic basis for variation of metalloproteinase-associated biochemical activity in venom of the Mojave rattlesnake (Crotalus scutulatus scutulatus). Int. Res. Biochem. 2013, 251474 (2013). doi:10.1155/2013/251474 [DOI] [PMC free article] [PubMed]
- 33.Massey DJ, Calvete JJ, Sanchez EE, Sanz L, Richards K, Curtis R, Boesen K. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from southern Arizona. J. Proteomics. 2012;75(9):2576–2587. doi: 10.1016/j.jprot.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 34.Gomis-Ruth FX, Kress LF, Kellermann J, Mayr I, Lee X, Huber R, Bode W. Refined 2.0 An X-ray crystal structure of the snake venom zinc-endopeptidase adamalysin II. Primary and tertiary structure determination, refinement, molecular structure and comparison with astacin, collagenase and thermolysin. J. Mol. Biol. 1994;239(4):513–544. doi: 10.1006/jmbi.1994.1392. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Botos I, Gomis-Ruth FX, Doll R, Blood C, Njoroge FG, Fox JW, Bode W, Meyer EF. Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d) Proc. Natl. Acad. Sci. USA. 1994;91(18):8447–8451. doi: 10.1073/pnas.91.18.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peitsch MC. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 1996;24(1):274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 37.Peitsch MC, Wells TN, Stampf DR, Sussman JL. The Swiss-3DImage collection and PDB-Browser on the World-Wide Web. Trends Biochem. Sci. 1995;20(2):82–84. doi: 10.1016/S0968-0004(00)88963-X. [DOI] [PubMed] [Google Scholar]
- 38.Peitsch MC, Borner C, Tschopp J. Sequence similarity of phospholipase A2 activating protein and the G protein beta-subunits: a new concept of effector protein activation in signal transduction? Trends Biochem. Sci. 1993;18(8):292–293. doi: 10.1016/0968-0004(93)90038-O. [DOI] [PubMed] [Google Scholar]
- 39.Guex N, Diemand A, Peitsch MC. Protein modelling for all. Trends Biochem. Sci. 1999;24(9):364–367. doi: 10.1016/S0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]
- 40.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23(6):600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 41.Stocker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth FX, McKay DB, Bode W. The metzincins–topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 1995;4(5):823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wooldridge BJ, Pineda G, Banuelas-Ornelas JJ, Dagda RK, Gasanov SE, Rael ED, Lieb CS. Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Mol. Biol. 2001;130(2):169–179. doi: 10.1016/s1096-4959(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 43.French WJ, Hayes WK, Bush SP, Cardwell MD, Bader JO, Rael ED. Mojave toxin in venom of Crotalus helleri (Southern Pacific Rattlesnake): molecular and geographic characterization. Toxicon. 2004;44(7):781–791. doi: 10.1016/j.toxicon.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, Teng M, Niu L. Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases. Acta Crystallogr. 1999;55(Pt 11):1834–1841. doi: 10.1107/s0907444999010306. [DOI] [PubMed] [Google Scholar]
- 45.Gong W, Zhu X, Liu S, Teng M, Niu L. Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus. J. Mol. Biol. 1998;283(3):657–668. doi: 10.1006/jmbi.1998.2110. [DOI] [PubMed] [Google Scholar]
- 46.Bolger MB, Swenson S, Markland FS., Jr Three-dimensional structure of fibrolase, the fibrinolytic enzyme from southern copperhead venom, modeled from the X-ray structure of adamalysin II and atrolysin C. AAPS Pharm. Sci. 2001;3(2):E16. doi: 10.1208/ps030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomis-Ruth FX, Kress LF, Bode W. First structure of a snake venom metalloproteinase: A prototype for matrix metalloproteinases/collagenases. EMBO J. 1993;12(11):4151–4157. doi: 10.1002/j.1460-2075.1993.tb06099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sunitha K, Hemshekhar M, Gaonkar SL, Sebastin Santhosh M, Suresh Kumar M, Basappa Priya BS, Kemparaju K, Rangappa KS, Nanjunda Swamy S, Girish KS. Neutralization of haemorrhagic activity of viper venoms by 1-(3-dimethylaminopropyl)-1-(4-fluorophenyl)-3-oxo-1,3-dihydroisobenzofuran-5-car bonitrile. Basic Clin. Pharmacol. Toxicol. 2011;109(4):292–299. doi: 10.1111/j.1742-7843.2011.00725.x. [DOI] [PubMed] [Google Scholar]
- 49.Gibbs HL, Sanz L, Calvete JJ. Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol. 2009;68(2):113–125. doi: 10.1007/s00239-008-9186-1. [DOI] [PubMed] [Google Scholar]
- 50.Jia Y, Lucena S, Cantu E, Jr, Sanchez EE. Perez, J.C.: cDNA cloning, expression and fibrin(ogen)olytic activity of two low-molecular weight snake venom metalloproteinases. Toxicon. 2009;54(3):233–243. doi: 10.1016/j.toxicon.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baramova EN, Shannon JD, Bjarnason JB, Fox JW. Degradation of extracellular matrix proteins by hemorrhagic metalloproteinases. Arch. Biochem. Biophys. 1989;275(1):63–71. doi: 10.1016/0003-9861(89)90350-0. [DOI] [PubMed] [Google Scholar]
- 52.Wallnoefer HG, Lingott T, Gutierrez JM, Merfort I, Liedl KR. Backbone flexibility controls the activity and specificity of a protein–protein interface: specificity in snake venom metalloproteases. J. Am. Chem. Soc. 2010;132(30):10330–10337. doi: 10.1021/ja909908y. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe L, Shannon JD, Valente RH, Rucavado A, Alape-Giron A, Kamiguti AS, Theakston RD, Fox JW, Gutierrez JM, Arni RK. Amino acid sequence and crystal structure of BaP1, a metalloproteinase from Bothrops asper snake venom that exerts multiple tissue-damaging activities. Protein Sci. 2003;12(10):2273–2281. doi: 10.1110/ps.03102403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durban J, Perez A, Sanz L, Gomez A, Bonilla F, Rodriguez S, Chacon D, Sasa M, Angulo Y, Gutierrez JM, Calvete JJ. Integrated omics profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crolatus simus simus. BMC Genomics. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hite LA, Shannon JD, Bjarnason JB, Fox JW. Sequence of a cDNA clone encoding the zinc metalloproteinase hemorrhagic toxin e from Crotalus atrox: evidence for signal, zymogen, and disintegrin-like structures. Biochemistry. 1992;31(27):6203–6211. doi: 10.1021/bi00142a005. [DOI] [PubMed] [Google Scholar]
- 56.Hite LA, Fox JW, Bjarnason JB. A new family of proteinases is defined by several snake venom metalloproteinases. Biol. Chem. Hoppe Seyler. 1992;373(7):381–385. doi: 10.1515/bchm3.1992.373.2.381. [DOI] [PubMed] [Google Scholar]
- 57.Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther. 1994;62(3):325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 58.Huang KF, Hung CC, Chiou SH. Characterization of three fibrinogenolytic proteases isolated from the venom of Taiwan habu (Trimeresurus mucrosquamatus) Biochem. Mol. Biol. Int. 1993;31(6):1041–1050. [PubMed] [Google Scholar]
- 59.Ramos OH, Selistre-de-Araujo HS. Comparative analysis of the catalytic domain of hemorrhagic and non-hemorrhagic snake venom metallopeptidases using bioinformatic tools. Toxicon. 2004;44(5):529–538. doi: 10.1016/j.toxicon.2004.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.