Summary
Bacterial secretion systems often employ molecular chaperones to recognize and facilitate export of their substrates. Recent work demonstrated that a secreted component of the type VI secretion system (T6SS), hemolysin co-regulated protein (Hcp), binds directly to effectors, enhancing their stability in the bacterial cytoplasm. Herein, we describe a quantitative cellular proteomics screen for T6S substrates that exploits this chaperone-like quality of Hcp. Application of this approach to the Hcp secretion island I-encoded T6SS (H1-T6SS) of Pseudomonas aeruginosa led to the identification of a novel effector protein, termed Tse4 (type VI secretion exported 4), subsequently shown to act as a potent intra-specific H1-T6SS-delivered antibacterial toxin. Interestingly, our screen failed to identify two predicted H1-T6SS effectors, Tse5 and Tse6, which differ from Hcp-stabilized substrates by the presence of toxin-associated PAAR-repeat motifs and genetic linkage to members of the valine-glycine repeat protein G (vgrG) genes. Genetic studies further distinguished these two groups of effectors: Hcp-stabilized effectors were found to display redundancy in interbacterial competition with respect to the requirement for the two H1-T6SS-exported VgrG proteins, whereas Tse5 and Tse6 delivery strictly required a cognate VgrG. Together, we propose that interaction with either VgrG or Hcp defines distinct pathways for T6S effector export.
Introduction
Bacteria possess specialized secretion systems to export proteins to the extracellular milieu or directly into target cells. The type VI secretion system (T6SS) is a macromolecular assembly that spans the cell envelope of Gram-negative bacteria and functions to deliver toxic effector proteins into neighboring cells (Silverman et al., 2012). Some bacterial T6SSs appear to play a direct role in virulence through host cell targeting, while others have been shown to display potent antimicrobial activity (Hood et al., 2010, Murdoch et al., 2011, Pukatzki et al., 2006, Schwarz et al., 2010, Jani & Cotter, 2010). This diversity in targeting capability likely arises, in part, from the activity of effector proteins secreted by this system (Russell et al., 2014). For example, effectors that target peptidoglycan are secreted by antibacterial T6SSs, whereas the actin cross-linking activity exhibited by the Vibrio cholerae T6SS is responsible for its cytotoxic effects on eukaryotic cells (Russell et al., 2011, Ma et al., 2009, Srikannathasan et al., 2013). More recently, several diverse families of phospholipase effectors have been identified, with some characterized members having reported roles in both virulence and interbacterial interactions (Russell et al., 2013, Wilderman et al., 2001, Dong et al., 2013).
Given the significant structural and functional diversity among T6S effectors, a fundamental question that arises regarding this system is how substrates are specifically recognized for export among the diverse pool of proteins that exist in the bacterial cytosol. In other contact-dependent secretion systems, such as the well-characterized bacterial type III secretion system (T3SS), highly specific molecular chaperones have been shown to maintain substrates in a partially unfolded, secretion-competent state (Evdokimov et al., 2003, Rodgers et al., 2010, Stebbins & Galan, 2001). These effector-chaperone complexes then interact with a T3SS ATPase, which provides the necessary energy for chaperone dissociation and allows for subsequent effector translocation (Cooper et al., 2010, Gauthier & Finlay, 2003). Similarly, substrates of some type IV secretion systems (T4SSs) interact with so-called coupling proteins (CPs), which serve to recruit effectors to the base of the secretion apparatus. CPs not only bind to a signal sequence located at the C-terminus of their cognate substrates but also provide the energy required for their export via ATP hydrolysis (Beranek et al., 2004, Jurik et al., 2010, Buscher et al., 2005, Atmakuri et al., 2003, Sutherland et al., 2012). While a number of studies have contributed to our understanding of these substrate selection mechanisms employed by the T3 and T4SSs, comparatively less is known about this process in the T6SS.
Active T6SSs appear to universally export proteins belonging to the Hcp and VgrG families (Hachani et al., 2011, Hood et al., 2010, Pukatzki et al., 2007, Zheng & Leung, 2007, Rao et al., 2004). X-ray crystallographic and solution NMR studies have demonstrated that these proteins are structurally similar to the tail tube and spike proteins of contractile bacteriophage, respectively (Leiman et al., 2009, Pell et al., 2009, Mougous et al., 2006, Kanamaru et al., 2002). Moreover, Hcp is capable of forming cross-linked nanotubes in vitro and has been isolated in complex with VgrG in vivo (Ballister et al., 2008, Lin et al., 2013). It has been proposed that a phage tail-like structure containing Hcp and VgrG is ejected by contraction of a tubular sheath consisting of the conserved T6SS-associated cytoplasmic proteins, TssB and TssC (Lossi et al., 2013, Kapitein et al., 2013). A recent study demonstrating the physical association of Hcp with TssB lends biochemical credence to this model (Lin et al., 2013).
Beyond its basic structural role in the T6S apparatus, Hcp appears to function directly in effector recognition (Zheng & Leung, 2007, Silverman et al., 2013). The Hcp molecule assembles into a homohexameric ring that contains a large internal cavity measuring approximately 40 Å in diameter (Mougous et al., 2006, Osipiuk et al., 2011). Recent work by Silverman et al. has shown that effectors bind within the pore of this ring structure and that this binding is required for their stability in the cytoplasm. Each of the known H1-T6SS substrates demonstrated this property, including type VI secretion exported 1 (Tse1), a peptidoglycan amidase, Tse2, a cytoplasmic effector of unknown mechanism, and Tse3, a muramidase (Russell et al., 2011, Hood et al., 2010, Li et al., 2012). Therefore, Hcp binds effectors despite differences in their targeted cellular compartment, enzymatic activity and tertiary structure.
Like Hcp, VgrG proteins have been associated directly with effector delivery by the T6SS. In the simplest cases, VgrG proteins can bear C-terminal extensions that exhibit toxic functionality. For example, V. cholerae VgrG-1 and VgrG-3 contain C-terminal actin cross-linking and peptidoglycan hydrolase domains, respectively (Pukatzki et al., 2007, Dong et al., 2013, Brooks et al., 2013). The recent X-ray crystal structure of a VgrG-like chimera in complex with a PAAR-repeat domain suggests an additional mechanism for VgrG-associated effector delivery (Shneider et al., 2013). Not only does the PAAR-repeat domain appear to play a structural role at the tip of the VgrG spike, it also likely functions as a protein-protein interaction module that tethers effectors to VgrGs. Notably, PAAR repeats are found in many putative toxins including rearrangement hot-spot/tyr-asp (RHS/YD)-repeat containing proteins, which have recently been implicated in T6SS-dependent interbacterial growth inhibition (Zhang et al., 2012, Koskiniemi et al., 2013, Poole et al., 2011, Wenren et al., 2013).
Herein we use proteomic and bioinformatic methods to define three novel effectors of the H1-T6SS of P. aeruginosa. Genetic and phenotypic studies indicate that these effectors, along with the previously characterized Tse1-3 proteins, constitute two distinct classes based on their stabilization by Hcp and their linkage to the two VgrG proteins exported by the H1-T6SS. We find that the Hcp-stabilized effectors, Tse2 and Tse4, function independently of specific VgrG proteins, whereas our data suggest that the PAAR domain-containing effectors, Tse5 and Tse6, do not require Hcp for stability and display a strict functional requirement for their cognate VgrG proteins. This work significantly expands the number of known H1-T6SS substrates and suggests that effectors of a single T6SS can be recognized and transported via two distinct mechanisms.
Results
Identification of Hcp1 substrates by quantitative cellular proteomics
Our laboratory recently made the observation that Hcp1 is a secretion receptor that directly binds the known H1-T6SS substrates, Tse1-Tse3, and is required for their intracellular stability (Silverman et al., 2013). We hypothesized that this chaperone-like activity of Hcp1 could be exploited at a proteome-wide level to discover novel H1-T6SS effectors. Specifically, we reasoned that H1-T6SS effectors could be identified based on a marked decrease in cellular abundance in a Δhcp1 background relative to a parental strain. To test this, we first established a reference proteome based on a P. aeruginosa strain harboring in-frame deletions in retS and tssM1. The retS gene encodes a hybid sensor kinase/response regulator that when deleted results in increased expression of the H1-T6SS and its substrates (Goodman et al., 2004, Hood et al., 2010). Deletion of tssM1 was introduced to inactivate the H1-T6SS and remove the confounding factor of substrate secretion into the culture medium. Using conservative criteria for inclusion, which included a high abundance threshold (>8 spectral counts) for each protein in duplicate biological replicates, we arrived at a final reference proteome consisting of 550 proteins (Fig. 1A and Table S1).
Fig. 1. Quantitative cellular proteomics identifies a novel H1-T6SS effector candidate.
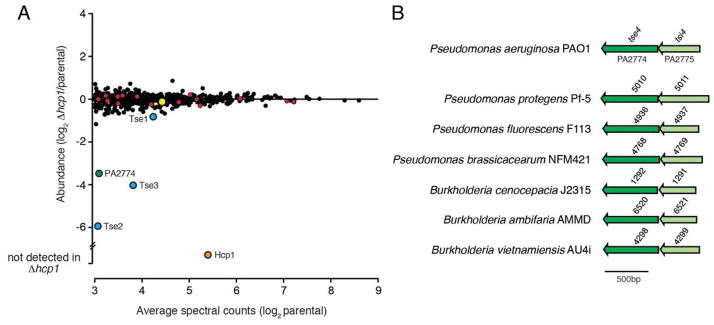
(A) Comparison of the cellular proteomes of a P. aeruginosa PAO1 parental strain (ΔretS ΔtssM1) and a derivative bearing an in-frame deletion of hcp1 (ΔretS ΔtssM1 Δhcp1) using quantitative mass spectrometry. Red, blue and orange circles indicate H1-T6SS apparatus components, previously identified H1-T6SS effector proteins (Tse1-3), and Hcp1, respectively. Green and yellow circles indicate the Hcp- and VgrG-associated effectors PA2774 and PA2684, respectively. (B) Genomic arrangement of PA2774–PA2775 (herein name tse4–tsi4) and homologous bicistrons from Pseudomonas spp. and Burkholderia spp. Shading is used to indicate candidate effector (dark) and immunity (light) open reading frames.
Next we compared our reference proteome to that of the parental strain harboring the Δhcp1 allele. Outside of Hcp1, no proteins found in the reference proteome were undetected in in this strain. Moreover, 546 of the 549 proteins detected in the Δhcp1 background were within 0.5–2.3-fold abundance relative to the reference, indicating that deletion of hcp1 did not result in gross changes to the cellular proteome. Among those proteins not significantly impacted by the absence of Hcp1 was each of the 15 detected components of the H1-T6SS apparatus (Fig. 1A, red circles). The three proteins whose abundance was significantly altered were identified as Tse2, Tse3 and a hypothetical protein encoded by the PA2774 locus, which were 61, 16 and 11-fold decreased in abundance relative to the reference proteome, respectively. Tse1 exhibited only a modest decrease in abundance (1.8-fold) compared to Tse2 and Tse3. Nonetheless, Tse1 was the fifth-most destabilized protein in the Δhcp1 background. The observation that both Tse2 and Tse3 are highly destabilized corroborates recent work from our lab that identified these proteins, along with Tse1, as substrates of the Hcp1 chaperone (Silverman et al., 2013). To our knowledge, the remaining protein, PA2774, had not previously associated with T6S nor studied in any capacity. Based on our findings, we postulated the PA2774 protein represents a previously unidentified effector of the H1-T6SS.
PA2774-PA2775 function as an antibacterial effector–immunity pair
T6S effector genes are invariably located adjacent to loci encoding specific immunity determinants. In P. aeruginosa, the tse genes are found in bicistrons with cognate immunity genes, termed type VI secretion immunity 1-3 (tsi1-3). Examination of the genomic context of PA2774 indicated that like tse1-3, this gene is encoded within a predicted bicistron. Furthermore, we found this bicistronic arrangement conserved in PA2774 homologs identified in other Pseudomonads and members of the Burkholderia (Fig. 1B). Based on these observations, we hypothesized that the open reading frame (ORF), PA2775, encodes a PA2774-specific immunity determinant.
To test whether PA2774–PA2775 constitute an effector–immunity (E–I) pair, we generated an in-frame deletion of the PA2774–PA2775 bicistron and assessed the ability of this strain to compete against assorted donors strains under contact-promoting growth conditions. We performed these and subsequent experiments involving P. aeruginosa in the ΔretS background, as constitutive activation of the H1-T6SS in this strain permits robust intraspecific competition phenotypes independent of yet unknown physiological stimulators (Hood et al., 2010, Basler & Mekalanos, 2012). We found that donor strains possessing PA2774 and a functional T6SS display a significant fitness advantage against the ΔPA2774 ΔPA2775 recipient (Fig. 2A). In agreement with the hypothesis that PA2774 functions as an antibacterial effector delivered by the H1-T6SS, deletion of PA2774 or inactivation of the T6SS in the donor restored fitness to the recipient. The fitness defect in the ΔPA2774 ΔPA2775 strain could also be restored by plasmid-borne expression of PA2775, consistent with this protein acting as a PA2774-specific immunity determinant. Together, these data suggest PA2775-PA2775 (hereafter referred to as Tse4–Tsi4) function as an H1-T6SS effector–immunity pair. Tse4 escaped detection in our prior secretome-based T6S effector discovery study and we have been unable to detect Tse4 in P. aeruginosa supernatants by immunoblotting (data not shown). With clear phenotypic data indicating Tse4 is a T6S-delivered toxin, these observations underscore the utility of our cellular proteomics approach for defining T6S effectors.
Fig. 2. Tse4–Tsi4 are an H1-T6SS effector–immunity pair.

(A) Growth competition assays between the indicated P. aeruginosa donor and recipient strains. PAO1 ΔretS is the parental strain in this and all subsequent interbacterial competition studies involving P. aeruginosa. A deletion of clpV1 inactivates the H1-T6SS (Mougous et al., 2006). Donor and recipient strains were mixed 1:1, grown for 20 hrs on solid media, and differentiated using blue/white screening. (B and C) Viability of E. coli cells grown on solid media harboring inducible plasmids expressing the indicated proteins. Empty vector controls are indicated by a dash. Error bars represent ± SD (n=3).
Examination of Tse4 using sequence or structure-based functional prediction algorithms yielded no substantial insights into its activity. As a first step toward understanding the mechanism of this effector, we asked whether it directs toxicity from the cytoplasmic or periplasmic compartment of bacteria. Often, the site of activity of a T6S effector can be deduced from the predicted localization of its cognate immunity protein. However, in the case of Tse4, the predicted two-pass inner membrane topology of Tsi4 complicates this inference. Consistent with Tse4 exerting toxicity from the periplasm, we observed significant reduction of viability in E. coli cells expressing Tse4 targeted to the periplasm, while E. coli producing cytoplasmic Tse4 retained viability similar to a vector control (Fig. 2B). To determine if periplasmic Tse4-induced toxicity against E. coli is physiologically relevant, we next measured the impact of Tsi4 co-expression on viability. Expression of Tsi4, but not non-cognate periplasmic immunity proteins, fully rescued E. coli viability, indicating that the Tse4-dependent growth inhibition observed is a consequence of specific activity elicited by the protein (Fig. 2C). Together, these data indicate that Tse4 is an H1-T6SS effector that exerts its toxicity in the periplasm of bacterial cells.
Identification of two VgrG-associated H1-T6SS effector proteins
A recent comprehensive bioinformatic analysis of polymorphic toxin systems by Zhang et al. suggests that there exist additional families of T6S substrates that have yet to be functionally characterized (Zhang et al., 2012). Specifically, these authors predict that proteins containing RHS/YD- and/or PAAR-repeat motifs function as toxins secreted by the T6SS. In support of this supposition, it was recently shown in Dickeya dadantii 3937 that deletion of vgrGA and vgrGB abrogates the growth advantage conferred by the RHS/YD- and PAAR repeat-containing protein encoded by the rhsB gene (Koskiniemi et al., 2013). PAAR repeat-containing proteins physically interact with VgrG molecules and are often fused to predicted toxins (Shneider et al., 2013). The genome of P. aeruginosa PAO1 contains a single predicted RHS/YD-repeat containing protein encoded by the PA2684 locus. Intriguingly, this gene is immediately adjacent to PA2685, which encodes VgrG4, a protein our group previously showed is exported by the H1-T6SS (Fig. 3A)(Hood et al., 2010). PA2684 contains a cryptic PAAR domain at its N-terminus that is predicted by the Phyre2 tertiary structure prediction algorithm to adopt the same fold as other PAAR domains, but lacks the signature pro–ala–ala–arg motif (Kelley & Sternberg, 2009). Another toxin module predicted by Zhang and colleagues that is found in the P. aeruginosa genome is the Toxin_61/NTox46 family (Zhang et al., 2012). The protein containing this domain, PA0093, also possesses an N-terminal PAAR motif. Moreover, the PA0093 locus is in close proximity to the H1-T6SS core gene cluster and the vgrG1 gene, which encodes a protein previously demonstrated to transit the H1-T6S pathway (Fig. 3A). Taken together, we postulated that PA2684 and PA0093 represent previously undiscovered H1-T6SS substrates.
Fig. 3. Two PAAR repeat containing proteins are H1-T6SS effectors with cognate immunity.
(A) Genomic context of the PA2684–PA2684.1 (herein named tse5–tsi5) and PA0092-PA0093 (herein named tse6–tsi6). Putative effector and immunity open reading frames are colored dark and light blue, respectively. For previously unannotated genes, locus tags are provided below the corresponding locus. (B and D) Growth competition assays between the indicated P. aeruginosa donor and recipient strains. Donor and recipient strains were mixed 1:1 and grown for 20 hrs on solid media. Populations were differentiated using antibiotic sensitivity (B) or blue/white screening (D). (C and E) Viability of E. coli cells grown on solid media harboring inducible plasmids expressing the indicated proteins. Empty vector controls are indicated by a dash. Error bars represent ± SD (n=3).
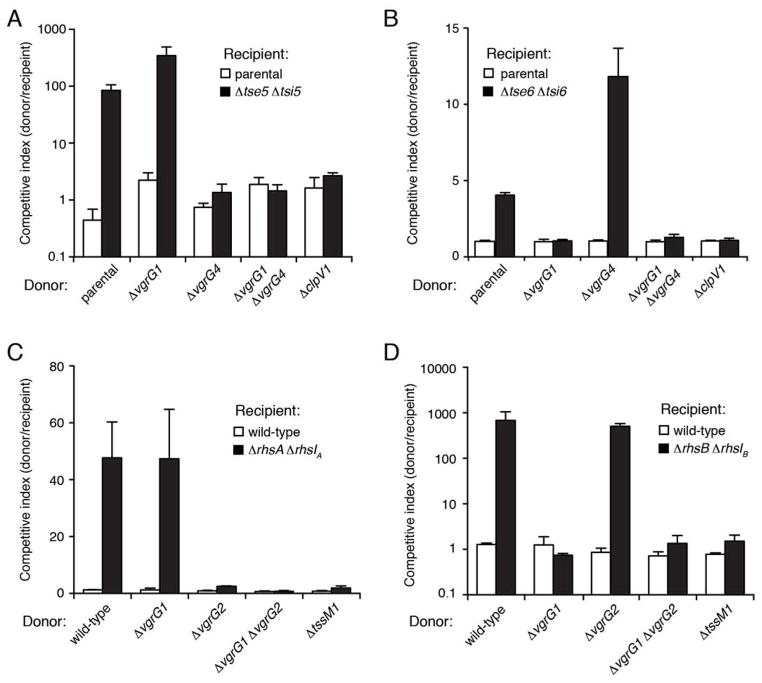
As all T6 antibacterial effectors identified to-date are co-expressed with cognate immunity proteins, we were surprised by the lack of an annotated ORF immediately adjacent to PA2684 that would encode a candidate immunity protein. However, analysis of DNA sequences flanking PA2684 led to the identification of a downstream ORF with a predicted ribosomal binding site, herein termed PA2684.1. The product of PA2684.1 is a short, 86 residue protein with two predicted transmembrane helices. To test if PA2684–PA2684.1 constitute an E–I pair, we constructed an in-frame deletion of the genes and measured the capacity of this strain to compete with its parental strain. Consistent with our hypothesis that PA2684 is an effector protein and PA2684.1 its cognate immunity protein, the fitness of the double deletion strain was significantly reduced when placed in competition with parental strains containing PA2684 and a functional H1-T6SS (Fig. 3B). Furthermore, its fitness was restored by expression of PA2684.1. Hereafter we refer to PA2684 and PA2684.1 as tse5 and tsi5, respectively.
Informatic and experimental studies on Rhs/YD-repeat containing proteins have shown that the C-terminus of these proteins harbors their toxic activity (Poole et al., 2011, Koskiniemi et al., 2013). This region is demarcated from the Rhs core structural domain by a conserved DPXGL-(18)-DPXGL motif, which forms the active site of an aspartyl protease that releases the the C-terminal toxin domain (Jackson et al., 2009, Busby et al., 2013). Bioinformatic analysis of the apparent C-terminal toxin domain of Tse5 (Tse5-CT) suggests this domain is unique to P. aeruginosa and failed to provide functional insights. As a first step towards understanding how this domain exerts its toxicity in cells, we expressed Tse5-CT (residues 1168-1317) in both the cytoplasm and periplasm of E. coli. Consistent with functioning as a toxin that acts from the periplasm, expression of Tse5-CT reduced E. coli viability only when targeted to the periplasmic compartment of the cell (Fig. 3C). However, co-expression of Tsi5 was not able to rescue this toxicity despite its capacity to abrogate Tse5-based intoxication in P. aeruginosa (Fig. 3B). Since Tsi5 is predicted to exist in the inner membrane, we speculate the inability of Tsi5 to rescue Tse5-mediated toxicity in E. coli is due to difficulties associated with membrane protein overexpression in a heterologous host (Wagner et al., 2006).
Bioinformatic analysis of PA0093 identified several homologous sequences (35–40%identical) in Pseudomonas spp.; all located upstream of a small ORF that encodes a protein with predicted cytoplasmic localization (Fig. S1). We postulated that this ORF in P. aeruginosa, PA0092, encodes a PA0093-specific immunity determinant. To investigate whether PA0092–PA0093 function as an H1-T6SS E–I pair, we measured the impact of PA0093 and H1-T6SS function in donor strains grown in competition under contact-promoting conditions against a P. aeruginosa ΔPA0092 ΔPA0093 recipient. Consistent with PA0093 serving as an H1-T6SS antibacterial toxin, the ΔPA0092 ΔPA0093 strain was four-fold less fit than its parent, and this defect depended on PA0093 and clpV1 in the donor strain (Fig. 3D). The PA0093-dependent donor fitness advantage was abrogated by PA0092 expression in the ΔPA0092 ΔPA0093 background. Taken together, these data indicate that PA0093–PA0092 are H1-T6SS E–I pair that participate in interbacterial interactions. Hereafter we refer to PA0093 and PA0092 as Tse6 and Tsi6, respectively.
Sequence analysis of Tse6 using the PFAM database suggests that its toxic activity lies in a putative Toxin_61 domain located at its C-terminus (Tse6-CT) (Punta et al., 2012, Zhang et al., 2012). To test this prediction, we examined the ability of Tse6-CT (residues 265–430) to exhibit toxicity when expressed in E. coli. As expected for a cytotoxic protein, Tse6-CT expression resulted in a significant decrease in E. coli viability compared to the control strain (Fig. 3E). Furthermore, the observed toxicity was specific to Tse6-CT as co-expression of Tsi6, but not the non-cognate cytoplasmic immunity protein Tsi2, provided full rescue of E. coli viability. These results demonstrate that the Toxin_61 domain of Tse6 is a potent cytoplasmic toxin whose activity can be specifically neutralized by Tsi6.
Elucidation of distinct H1-T6SS effector export pathways
Above we show that Tse5 and Tse6, like Tse1-Tse4, function as antibacterial effectors secreted by the H1-T6SS. Despite this, Tse5 and Tse6 were not identified in our quantitative cellular proteomic screen for proteins that rely on Hcp1 for cytoplasmic stability. Tse6 was not detected; however, Tse5 was present in high abundance in the P. aeruginosa PAO1 ΔretS ΔtssM reference proteome but exhibited no significant change in abundance compared to the P. aeruginosa PAO1 ΔretS ΔtssM Δhcp1 strain (Fig. 1A, yellow circle). This observation led us to conclude that Tse5, and by interference the other H1-T6SS PAAR repeat-containing effector, Tse6, do not use Hcp1 as a chaperone to transit the T6SS apparatus. Instead, given the genetic linkage found between genes encoding PAAR domain proteins and vgrG, we surmised that Tse5 and Tse6 transit the H1-T6SS in a manner requiring a cognate VgrG, thereby defining a second export pathway distinct from Hcp association. Hcp and VgrG export is co-dependent; however, in instances of multiple VgrG export by a single T6SS, VgrG proteins support Hcp export in a redundant fashion. We reasoned that if these two pathways function in a single T6SS, effectors transiting the pathways should be distinguishable by their behavior in strains bearing single versus double vgrG deletions. In agreement with this concept, it was previously shown that the Hcp1-interacting effector Tse3 requires Hcp1 for export, but is secreted independently of either of the VgrG proteins exported by the H1-T6SS (Hachani et al., 2011).
To examine the VgrG-dependency of the PAAR domain-containing effectors Tse5 and Tse6, we measured the fitness advantage of strains lacking vgrG1, vgrG4, or both vgrG genes relative to the parental strain in growth competition assays against sensitive recipients. These experiments showed that inactivation of vgrG4 blocks Tse5 intercellular delivery whereas vgrG1 inactivation blocks Tse6 delivery (Fig. 4A and B). Neither of these Tse proteins showed diminished activity in strains lacking non-cognate vgrG. On the contrary, we observed an approximate two-fold increase in Tse6-dependent fitness in strains lacking vgrG4, suggesting that VgrG–effector complexes might compete for docking to the apparatus.
Fig. 4. PAAR repeating containing effectors require cognate VgrG proteins for intercellular delivery.
(A–D) Donor strains bearing deletions of individual vgrG genes inhibit T6S-dependent intercellular transfer of PAAR repeat-containing effectors in two bacteria. Growth competition assays between the indicated P. aeruginosa (A and B) or E. cloacae (C and D) donor and recipient strains. P. aeruginosa or E. cloacae donor and recipient strain were mixed 1:1 and grown for 20 hrs or 4 hrs on solid media, respectively. Populations were differentiated using antibiotic sensitivity (A, C, and D) or blue/white screening (B). Error bars represent ± SD (n=3).
While the P. aeruginosa vgrG genes are located in close proximity to their cognate effectors, PAAR-domain containing proteins in other organisms can be found unlinked to other T6S genes. To test whether PAAR domain-containing proteins in these organisms also require a cognate VgrG protein, we investigated the two Rhs/YD-repeat effectors, rhsA and rhsB, encoded by the Enterobacter cloacae genome. The rhsA gene and the two vgrG genes of the organism, vgrG1 and vgrG2, reside in a T6S gene cluster, while rhsB is located at a distant site unlinked to a vgrG gene or other readily identifiable T6SS apparatus components (Fig. S2). Both rhsA and rhsB reside immediately upstream of genes encoding predicted cognate immunity determinants, rhsIA and rhsIB, respectively. Importantly, both RhsA and RhsB confer significant T6S-dependent interbacterial fitness in donor strains grown in competition with recipient strains lacking cognate immunity genes (Fig. S3). Mirroring our result with P. aeruginosa, mutational inactivation of vgrG1 specifically abrogated RhsB-mediated growth inhibition, whereas inactivation of vgrG2 exclusively prevented RhsA delivery (Fig. 4C and D). Taken together, these data show that T6S-exported PAAR-repeat toxins require specific cognate VgrG proteins to exert growth inhibition on target cells.
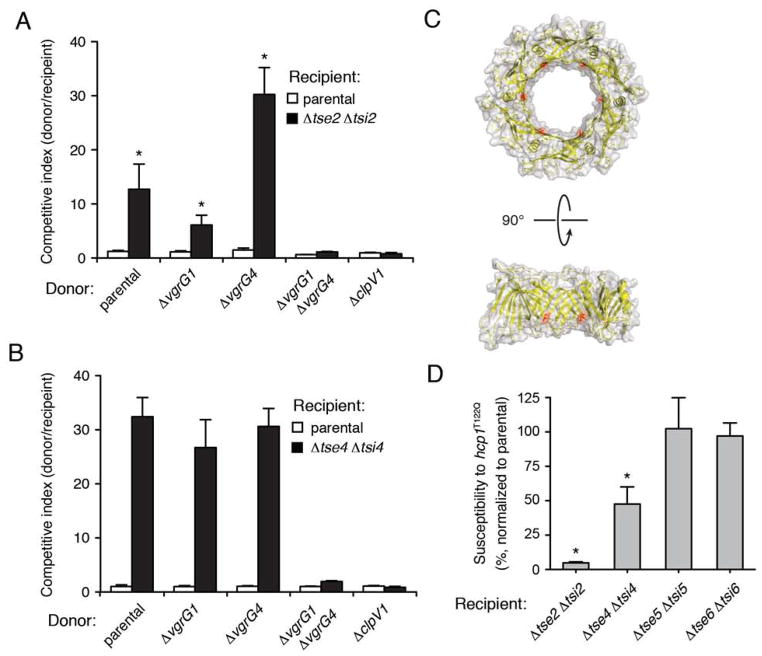
Next we turned our attention to the Hcp-associated H1-T6SS substrates. As tse2 and tse4 both confer strong intraspecific growth advantage against susceptible recipients, we used these substrates to examine the dependence of Hcp-associated effectors on VgrG proteins. Our experiments showed Tse2 and Tse4 retain the capacity to target recipient cells via the H1-T6SS in strains lacking either vgrG1 or vgrG4 (Fig. 5A and B). Interestingly, we observed an approximate 2.5-fold increase in Tse2-dependent fitness in strains lacking vgrG4 suggesting that VgrG1 may facilitate more efficient delivery of Tse2–Hcp1 complexes. As expected, deletion of both vgrG1 and vgrG4 – previously shown to abrogate Hcp1 secretion – blocked the ability of these effectors to target recipient cells (Hood et al., 2010). Thus, unlike PAAR repeat-containing effectors, Tse2- and Tse4-dependent delivery occurs independent of a specific VgrG protein.
Fig. 5. Hcp-associated effectors are delivered independently of a specific VgrG protein.
(A and B) Intercellular transfer of Tse2 and Tse4 by the H1-T6SS occurs in donor strains bearing individual, but not double vgrG deletions. (C) X-ray crystal structure of the Hcp1 hexamer from P. aeruginosa shown as a space filling representation. The location of threonine 122 is highlighted in red. (D) A donor strain expressing a chromosomal Hcp1T122Q point mutant exhibits reduced fitness relative to its parental strain towards recipients sensitive to Hcp1-associated effectors, Tse2 and Tse4. (A, B and D) Growth competition assays between the indicated P. aeruginosa donor and recipient strains. Donor and recipient strains were mixed 1:1 and grown for 20 hrs on solid media. Populations were differentiated using antibiotic sensitivity (Δtse5 Δtsi5) or blue/white screening (Δtse2 Δtsi2, Δtse4 Δtsi4, and Δtse6 Δtsi6). Asterisks indicate competition outcomes significantly different from (A) the ΔvgrG1 ΔvgrG4 donor competed against the Δtse2 Δtsi2 recipient or (D) the parental donor competed against the Δtse2 Δtsi2 or the Δtse4 Δtsi4 recipient (P < 0.05). Error bars represent ± SD (n=3).
The pore of the Hcp protein can interact with T6S effectors; however, the protein is also generally required for T6 apparatus assembly and for the export of VgrG proteins (Mougous et al., 2006, Hood et al., 2010, Zheng & Leung, 2007, Lin et al., 2013). Indeed, previous work in our laboratory has shown that hcp1 deletion blocks VgrG1 and VgrG4 secretion. The multiple roles that Hcp plays in the T6SS necessitates an alternative to hcp1 knockout in order to specifically evaluate its role in the export of the Tse proteins. Notably, Hcp1 variants bearing amino acid substitutions located on the inner ring surface have been found to reduce stability and export of Hcp1-associated effectors without compromising other functions of the T6S apparatus (Fig. 5C)(Silverman et al., 2013). Among a panel of hcp1 alleles examined previously, a mutant encoding a glutamine substitution at position 122 (hcp1T122Q) resulted in significantly decreased Tse1-3 export, indicating that this substitution compromises the ability of Hcp1 to translocate its substrates. We found that P. aeruginosa encoding Hcp1T122Q from the native hcp1 locus exhibited reduced capacity to deliver Tse2 and Tse4, while Tse5 and Tse6 delivery was unaffected (Fig. 5D). These data suggest that Tse5 and Tse6 do not require interaction with the Hcp1 pore for intercellular transit. In total, our data support a model in which effectors transit the T6SS by two genetically distinct pathways (Fig. 6).
Fig. 6. Model summarizing P. aeruginosa H1-T6SS effectors and their export pathways.
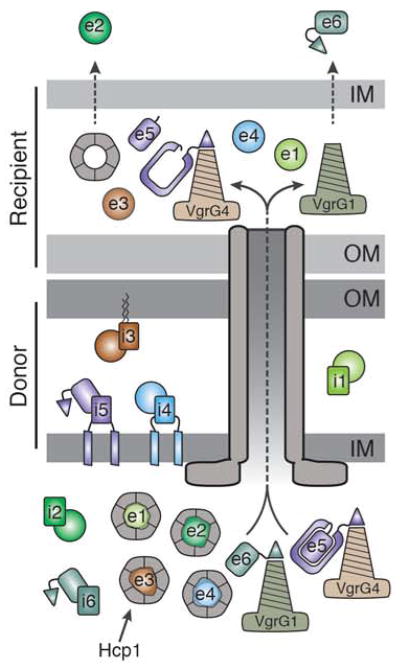
The schematic depicts a donor P. aeruginosa cell delivering its effector payload to a recipient Gram-negative bacterium. The inner (IM) and outer membranes (OM) of the donor and recipient cells are indicated. The H1-T6SS apparatus is depicted as a grey tube, as its organization was not the focus of this work (for a more detailed structural description see). Data from this and previous studies suggest that Tse1-4 (e1–e4) interact with Hcp1 hexamers while PAAR domain (triangles)-containing effectors, Tse5 and Tse6 (e5 and e6), associate with VgrG4 and VgrG1, respectively. Based on recent structural insights, the C-terminal toxin domain of Tse5 is depicted within a shell corresponding to the N-terminal Rhs domain of the protein (Busby et al., 2013, Gatsogiannis et al., 2013). The toxin domain is likely released by intramolecular proteolysis in the recipient cell as observed in the Rhs-containing insecticidal toxins of Y. entomophaga and Photorhabdus luminescens. The dashed arrows in the recipient cell inner membrane emphasize that Tse2 and Tse6 reach the cytoplasm of the recipient cell by an unknown mechanism. Immunity neutralization of cognate effectors is demonstrated in the donor cell (i1–i6). The subcellular localization of each immunity protein is based on experimental evidence (i1–i3) or bioinformatic predictions (i4–i6). A similar schematic depicting E. cloacae delivering effectors into a neighboring Gram-negative cell can be found in the supplement (Figure S4).
Discussion
We have developed and successfully implemented a quantitative cellular proteomic screen for the identification of T6S substrates. This approach presents several advantages over previously reported experimental methods for T6 effector identification. Transposon mutagenesis coupled with next generation sequencing (Tn-seq) was recently employed for the identification of T6S immunity genes and, by inference, effector loci, in Vibrio cholerae (Dong et al., 2013). While this is a high-resolution genome-wide approach, its utility is inherently restricted to the identification of effectors that are toxic in the recipient cell periplasm. Immunity determinants of effectors that act in the cytoplasm are essential and thus cannot be readily distinguished from other essential genes by transposon mutagenesis. The screen developed in this work does not discriminate between peri- and cytotoxic effectors, as effectors destined for both compartments associate with Hcp. One frequently utilized, and perhaps more intuitive, method for discovering T6 substrates is to define candidates based on their decreased abundance in bacterial secretomes prepared from strains bearing T6S-inactivating mutations relative to wild-type (Fritsch et al., 2013, Hood et al., 2010). A limitation of this method is that like other contact-dependent intercellular delivery systems, T6 is generally under the control of specialized posttranslational mechanisms that repress effector export under conditions suitable for secretome preparation. The best characterized of these is the threonine phosphorylation pathway (TPP) of the P. aeruginosa H1-T6SS, which was overridden by genetic manipulation in order to facilitate the discovery of Tse1-3 (Mougous et al., 2007, Hood et al., 2010). As only one-third of T6SSs possess clear functional equivalents of TPP components, the precise genetic perturbations necessary for achieving activation of the majority of T6SSs remains unknown. A second benefit of identifying T6S effectors within cellular proteomes is that it avoids the highly proteolytic extracellular milieu (Scott et al., 2013). Under physiological conditions, T6S substrates are delivered directly between donor and recipient cells, and therefore have not evolved to persist extracellularly. On the contrary, instability in the absence of Hcp favors effector identification using our cellular proteomics method. Consistent with this, Tse4 was not detected in prior P. aeruginosa secretome studies and it is a predicted unstructured protein with 20% glycine content.
An interesting observation that arose from our cellular proteomics screen is that the PAAR repeat-containing toxin, Tse5, does not require Hcp1 for stability. The Tse6 protein was not detected in our analysis; however, this suggests that a subset of T6S effectors do not directly contact Hcp. Taken together with our data showing that P. aeruginosa Tse5/6 and E. cloacae RhsA/B require cognate VgrG proteins to exert growth inhibition on susceptible recipients, we postulate that export of these effectors is dependent on specific VgrG contacts (Fig. 6 and Fig. S4). This hypothesis is supported by the recently determined crystal structure of a chimera composed of a bacteriophage T4 gp5 fragment fused to C-terminal VgrG residues in complex with a PAAR-repeat domain (Shneider et al., 2013). The C-terminus of the PAAR domain extends away from the structure in such a way that may accommodate fusion to toxin domains.
The finding that T6 effectors can associate with Hcp or VgrG proteins raises the question of what, if any, properties of effectors necessitate their export by either of these two pathways. Peptidoglycan glycoside hydrolase activity has been ascribed to both P. aeruginosa Tse3 and V. cholerae VgrG-3, indicating that the activity of an effector is unlikely to dictate a corresponding export mechanism (Russell et al., 2011, Brooks et al., 2013). Protein size could provide an explanation for the requirement of multiple effector export mechanisms. Hcp-associated effectors bind the internal cavity of the hexameric Hcp ring, limiting the diameter of their smallest dimension. Indeed, with the exception of Tse3, all effectors shown to interact with Hcp are under 20 kDa and thus near the expected molecular weight cutoff for a globular protein bound to the ~40 Å internal pore of the Hcp hexamer (Silverman et al., 2013, Chou et al., 2012, Zheng & Leung, 2007, Lin et al., 2013). At 44 kDa, Tse3 is considerably above this cutoff; however, the recent determination of its crystal structure reveals an elongated fold that could also be accommodated within Hcp ring(s) (Li et al., 2013, Wang et al., 2013). In contrast, bioinformatic data suggest that most VgrG-associated effectors are in excess of 60 kDa (Russell et al., 2013). Structures of RHS/YD-repeat containing proteins indicate this group of VgrG-associated proteins adopts a cocoon-like structure that incases the toxin domain. The narrowest constriction of these structures is over 50 Å, well beyond the diameter of the Hcp pore (Busby et al., 2013, Gatsogiannis et al., 2013). Another factor to consider is that the folded state of effectors interacting with the Hcp pore remains unknown, but would clearly impact the molecular weight cutoff of this pathway. If effectors do interact with Hcp prior to attaining their active conformation, it might be that the capacity to adopt a quasi-folded or alternative-folded state – rather than molecular weight per se – that dictates export through the two pathways.
The current study doubles the number of known effectors of the H1-T6SS, which raises the question of why one T6SS would release such a complex payload of effectors? Among the P. aeruginosa H1-T6SS substrates, Tse1 and Tse3 function as a peptidoglycan amidase and muramidase, respectively, while the activity of the remaining effectors remains unknown and is a current focus of investigation. Given the complete lack of sequence homology between Tse proteins, it is unlikely that these proteins have overlapping function. One explanation for a diverse T6S payload is that each might function optimally under distinct and rapidly changing environmental conditions. For example, cell wall-degrading effectors would be expected to be highly effective in hypotonic conditions but less so when the surrounding environment is iso- or hyperosmotic. Perhaps a more parsimonious explanation is that by the simultaneous deployment of multiple T6S effectors with distinct activities, bacteria minimize the ability of a target population to evolve resistance.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
A detailed list of all strains and plasmids generated in this study can be found in the supplement (Tables S2 and S3). All P. aeruginosa strains generated were derived from the sequenced strain PAO1 (Stover et al., 2000). P. aeruginosa strains were grown in Luria-Bertani (LB) media or LB agar at 37 °C supplemented with 30 μg ml−1 gentamycin, 25 μg ml−1 irgasan, 5% (w/v) sucrose and 75 μg ml−1 tetracycline where appropriate. The pEXG2 suicide vector was used for in-frame chromosomal deletions as described previously (Mougous et al., 2006). The E. cloacae strains generated in this study were derived from the sequenced strain Enterobacter cloacae subspecies cloacae ATCC 13047 (Ren et al., 2010). E. cloacae strains were grown in LB media or LB agar at 37 °C supplemented with 150 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 200 μg ml−1 rifampicin, 100 μg ml−1 spectinomycin and 10 μg ml−1 tetracycline where appropriate. Gene-deletion constructs were generated using plasmids pKAN or pSPM as described previously (Hayes et al., 2002, Koskiniemi et al., 2013). Briefly, DNA fragments from upstream and downstream of the target gene were amplified and ligated to pKAN or pSPM to flank the antibiotic-resistance cassette. The resulting constructs were linearized by restriction digestion and electroporated into E. cloacae cells expressing phage λ Red recombinase proteins as described (Chaveroche et al., 2000). Transformants were selected on LB-agar supplemented with either kanamycin or spectinomycin. All chromosomal deletions were confirmed by whole-cell PCR analysis. E. coli strains used included DH5α for cloning, SM10 for conjugal transfer of plasmids into P. aeruginosa and BL21 (DE3) pLysS for toxicity assays. E. coli strains were either grown in LB or LB agar at 37 °C supplemented with 50 μg ml−1 kanamycin, 150 μg ml−1 carbenicillin, 30 μg ml−1 chloramphenicol, 200 μg ml−1 trimethoprim, 0.1% (w/v) L-rhamnose and the indicated concentrations of IPTG as required.
Preparation of samples for cellular proteomic analysis
Overnight cultures of P. aeruginosa PAO1 ΔretS ΔtssM1 and P. aeruginosa PAO1 ΔretS ΔtssM1 Δhcp1 were diluted to an OD600 of 0.1 and 100 μL of each strain was plated on LB agar. After incubation at 37 °C for 8 hours, cells were harvested from the plates and frozen at −80 °C. The cell pellets were then thawed in lysis buffer containing 100 mM ammonium bicarbonate, 8 M urea and 0.1% (w/v) RapiGest™ SF surfactant (Waters). The cell suspensions were then sonicated six times with 10 second pulse lengths followed by centrifugation to remove cellular debris. 200 μg of each sample was then reduced with 5 mM TCEP for 1 hour at 37 °C followed by alkylation using 10 mM iodoacetamide for 30 min in the dark at room temperature. Alkylation reactions were quenched using 12 mM N-acetyl-cysteine and subsequently diluted with 100 mM ammonium bicarbonate to reduce the urea concentration to 1.5M. Samples were then treated with 10 μg of sequencing grade trypsin (Promega) overnight at 37 °C. The RapiGest™ SF surfactant was then precipitated by acidification of the samples to pH 2–3 by the addition of 2M HCl followed by incubation at 37 °C for 15 min. Precipitated detergent was removed by centrifugation at 10,000 g for 5 min. Samples were then diluted with 100% acetonitrile and 10% (w/v) trifluoroacetic acid (TFA) to a final concentration of 5% (v/v) and 0.1% (w/v) and applied to MacroSpin™ C18 columns (The Nest Group, Inc.) that had been charged with two washes of 100% acetonitrile followed by one wash with ddH2O. Bound tryptic peptides were then washed twice in 5% (v/v) acetonitrile, 0.1% (w/v) TFA before elution with 80% (v/v) acetonitrile, 25 mM formic acid.
MS analysis of tryptic peptides
Peptide digests were analyzed by electrospray ionization in the positive ion mode on a hybrid quadrupole-orbitrap mass spectrometer (Q Exactive™, Thermo Fisher, San Jose, CA). The Q Exactive was equipped with a nanoflow HPLC system (NanoAcquity; Waters Corporation, Milford, MA) fitted with a home-built helium-degasser. Peptides were trapped on a homemade 100 μm i.d. × 20 mm long pre-column packed with 200 Ǻ (5μm, C18AQ; Michrom BioResources Inc., Auburn, CA, USA). Subsequent peptide separation was on an in-house constructed 75 μm i.d. × 180 mm long analytical column pulled using a Sutter Instruments P-2000 CO2 laser puller (Sutter Instrument Company, Novato, CA) and packed with 100 Å (5 μm, C18AQ: Michrom) particle. For each injection, an estimated amount of 1 μg of peptide mixture was loaded onto the pre-column at 4 μl/min in water/acetonitrile (95/5) with 0.1% (v/v) formic acid. Peptides were eluted using an acetonitrile gradient flowing at 250 nl/min using mobile phase consisting of: A, water, 0.1% formic acid; B, acetonitrile, 0.1% formic acid. Peptides were eluted using an acetonitrile gradient flowing at 250 nL/min using mobile phase gradient of 5–35% acetonitrile over 60 min. with a total gradient time of 95 min. Ion source conditions were optimized using the tuning and calibration solution recommended by the instrument provider. Data-dependent analyses were acquired using MS survey scans in the Orbitrap followed by data dependent selection of the 20 most abundant precursors for tandem mass spectrometry. Singly charged ions were excluded from data-dependent analysis. Data redundancy was minimized by excluding previously selected precursor ions for 60 seconds following their selection for tandem mass spectrometry. Data was acquired using Xcalibur, version 2.2 (Thermo Fisher). Samples were analyzed in triplicate.
Tandem mass spectra were searched for sequence matches against the UniProt P. aeruginosa PAO1 database using MaxQuant v1.4.1.2. The following modifications were set as search parameters: peptide mass tolerance at 6 ppm, trypsin digestion cleavage after K or R (except when followed by P), 2 allowed missed cleavage site, carbamidomethylated cysteine (static modification), and oxidized methionine, protein N-term acetylation (variable modification/differential search option). Search results were validated with peptide and protein FDR both at 0.01. Differences in relative expression of proteins were calculated using peptide spectral counting algorithm followed by QSpec statistical analysis (http://www.nesvilab.org/qspec.php/).
The semi-quantitative technique of spectral counting was used to determine the relative abundance of identified proteins in each sample. Spectral counting utilizes the total number of MS/MS spectra identified for a particular protein as a measure of protein abundance (Liu et al., 2004). A normalization criterion was applied to normalize the spectral counts so that the values of the total spectral counts per sample were similar. An average of the spectral counts was generated for each sample (based on the triplicate samples) and variably present proteins between biological replicates were omitted from further analysis. As the premise of the screen was to identify proteins whose abundance decreased significantly in the absence of Hcp1, only proteins with an average spectral count of eight or greater were considered in our analyses of the P. aeruginosa PAO1 ΔretS ΔtssM1 reference proteome. Ratios of the average spectral counts between the reference and the P. aeruginosa PAO1 ΔretS ΔtssM1 Δhcp1 proteome were calculated to determine the relative abundance of all identified proteins between the two strains.
Growth competition assays
For P. aeruginosa competitions, the recipient strain contained lacZ inserted at the neutral phage attachment site to enable its differentiation from the unlabeled donor. For E. cloacae competitions, recipient strain CFU enumeration was performed by plating on LB agar containing the appropriate antibiotic (Table S2). Overnight cultures of donor and recipient strains were mixed in a 1:1 (v/v) ratio and diluted 1:2 (v/v) in LB. Starting ratios of donor and recipient were enumerated by plating on LB agar containing either 40 μg/ml X-gal or the appropriate antibiotic. 10 μl of each competition mixture was then spotted in triplicate on a 0.2 μm nitrocellulose membrane overlaid on a 3% LB agar plate and incubated face up at 37 °C for 20 h (P. aeruginosa) or 4 h (E. cloacae). Competitions were then harvested by resuspending cells in LB and enumerating CFUs by plating on LB agar containing either 40 μg/ml X-gal or the appropriate antibiotic. The final donor/recipient CFUs were normalized to the starting ratio of donor and recipient strains.
E. coli toxicity assays
Overnight cultures of E. coli BL21 pLysS cultures harboring the appropriate plasmids (Table S3) were diluted 106 in 10-fold increments and each dilution was spotted onto 3% LB agar plates containing the appropriate antibiotics. For comparison of cytoplasmic versus periplasmic toxicity of Tse4 and Tse5, cells were induced with 100 μM IPTG. For Tse4/Tsi4 co-expression experiments, Tse4 and Tsi4/Tsi1/Tsi3 expression were induced with 40 μM IPTG and with 0.1% (w/v) L-rhamnose, respectively. For Tse6/Tsi6 co-expression experiments, Tse6 and Tsi6/Tsi2 expression was induced with 0.1% (w/v) L-rhamnose and 100 μM IPTG, respectively.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Mougous lab for helpful discussions. This work was supported by grants from the National Institutes of Health to J.D.M. (AI080609), C.S.H. (GM078634) and D.A.L (GM102318), and by the University of Maryland Baltimore, School of Pharmacy Mass Spectrometry Center to D.R.G. (SOP1841-IQB2014). J.C.W. was supported by a postdoctoral research fellowship from the Canadian Institutes of Health Research, A.B.R. was supported by a graduate research fellowship from the National Science Foundation (DGE-0718124), J.A.D. was supported by a Mary Gates Research Scholarship, and J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
References
- Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Molecular microbiology. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. Journal of bacteriology. 2004;186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. The Journal of biological chemistry. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 2013;501:547–550. doi: 10.1038/nature12465. [DOI] [PubMed] [Google Scholar]
- Buscher BA, Conover GM, Miller JL, Vogel SA, Meyers SN, Isberg RR, Vogel JP. The DotL protein, a member of the TraG-coupling protein family, is essential for Viability of Legionella pneumophila strain Lp02. Journal of bacteriology. 2005;187:2927–2938. doi: 10.1128/JB.187.9.2927-2938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroche MK, Ghigo JM, d’Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic acids research. 2000;28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Bui NK, Russell AB, Lexa KW, Gardiner TE, LeRoux M, Vollmer W, Mougous JD. Structure of a peptidoglycan amidase effector targeted to Gram-negative bacteria by the type VI secretion system. Cell reports. 2012;1:656–664. doi: 10.1016/j.celrep.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CA, Zhang K, Andres SN, Fang Y, Kaniuk NA, Hannemann M, Brumell JH, Foster LJ, Junop MS, Coombes BK. Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLoS pathogens. 2010;6:e1000751. doi: 10.1371/journal.ppat.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov AG, Phan J, Tropea JE, Routzahn KM, Peters HK, Pokross M, Waugh DS. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nature structural biology. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- Fritsch MJ, Trunk K, Alcoforado Diniz J, Guo M, Trost M, Coulthurst SJ. Proteomic identification of novel secreted anti-bacterial toxins of the Serratia marcescens Type VI secretion system. Molecular & cellular proteomics: MCP. 2013 doi: 10.1074/mcp.M113.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsogiannis C, Lang AE, Meusch D, Pfaumann V, Hofnagel O, Benz R, Aktories K, Raunser S. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature. 2013;495:520–523. doi: 10.1038/nature11987. [DOI] [PubMed] [Google Scholar]
- Gauthier A, Finlay BB. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. Journal of bacteriology. 2003;185:6747–6755. doi: 10.1128/JB.185.23.6747-6755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Developmental cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. The Journal of biological chemistry. 2011;286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. The Journal of biological chemistry. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Thomas GH, Parkhill J, Thomson NR. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC genomics. 2009;10:584. doi: 10.1186/1471-2164-10-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell host & microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurik A, Hausser E, Kutter S, Pattis I, Prassl S, Weiss E, Fischer W. The coupling protein Cagbeta and its interaction partner CagZ are required for type IV secretion of the Helicobacter pylori CagA protein. Infection and immunity. 2010;78:5244–5251. doi: 10.1128/IAI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Molecular microbiology. 2013;87:1013–1028. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang W, Liu Q, Gao Y, Gao Y, Wang Y, Wang DZ, Li Z, Wang T. Structural Insights on the bacteriolytic and self-protection mechanism of muramidase effector Tse3 in Pseudomonas aeruginosa. The Journal of biological chemistry. 2013;288:30607–30613. doi: 10.1074/jbc.C113.506097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Le Trong I, Carl MA, Larson ET, Chou S, De Leon JA, Dove SL, Stenkamp RE, Mougous JD. Structural basis for type VI secretion effector recognition by a cognate immunity protein. PLoS pathogens. 2012;8:e1002613. doi: 10.1371/journal.ppat.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Ma LS, Lai EM. Systematic dissection of the agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PloS one. 2013;8:e67647. doi: 10.1371/journal.pone.0067647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. The Journal of biological chemistry. 2013;288:7536–7548. doi: 10.1074/jbc.M112.439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell host & microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nature cell biology. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. Journal of bacteriology. 2011;193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J, Xu X, Cui H, Savchenko A, Edwards A, Joachimiak A. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. Journal of structural and functional genomics. 2011;12:21–26. doi: 10.1007/s10969-011-9107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole SJ, Diner EJ, Aoki SK, Braaten BA, t’Kint de Roodenbeke C, Low DA, Hayes CS. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS genetics. 2011;7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic acids research. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PS, Yamada Y, Tan YP, Leung KY. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Molecular microbiology. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- Ren Y, Ren Y, Zhou Z, Guo X, Li Y, Feng L, Wang L. Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. Journal of bacteriology. 2010;192:2463–2464. doi: 10.1128/JB.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers L, Mukerjea R, Birtalan S, Friedberg D, Ghosh P. A solvent-exposed patch in chaperone-bound YopE is required for translocation by the type III secretion system. Journal of bacteriology. 2010;192:3114–3122. doi: 10.1128/JB.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nature reviews Microbiology. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends in microbiology. 2010;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott NE, Hare NJ, White MY, Manos J, Cordwell SJ. Secretome of Transmissible Pseudomonas aeruginosa AES-1R Grown in a Cystic Fibrosis Lung-Like Environment. Journal of proteome research. 2013;12:5357–5369. doi: 10.1021/pr4007365. [DOI] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Molecular cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annual review of microbiology. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikannathasan V, English G, Bui NK, Trunk K, O’Rourke PE, Rao VA, Vollmer W, Coulthurst SJ, Hunter WN. Structural basis for type VI secreted peptidoglycan DL-endopeptidase function, specificity and neutralization in Serratia marcescens. Acta crystallographica Section D, Biological crystallography. 2013;69:2468–2482. doi: 10.1107/S0907444913022725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS pathogens. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends in biotechnology. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wang T, Ding J, Zhang Y, Wang DC, Liu W. Complex structure of type VI peptidoglycan muramidase effector and a cognate immunity protein. Acta crystallographica Section D, Biological crystallography. 2013;69:1889–1900. doi: 10.1107/S090744491301576X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. mBio. 2013;4 doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PJ, Vasil AI, Johnson Z, Vasil ML. Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Molecular microbiology. 2001;39:291–303. doi: 10.1046/j.1365-2958.2001.02282.x. [DOI] [PubMed] [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biology direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Molecular microbiology. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.