Abstract
Dilated cardiomyopathy (DCM) is a highly heterogeneous trait with sarcomeric gene mutations predominating. The cause of a significant percentage of DCM remains unknown and no gene-specific therapy is available. Based on resequencing with 513 DCM cases and 1,150 matched controls from various ethnically distinct cohorts, we discovered rare, functional RAF1 mutations in three of them (South India, North India and Japan). The prevalence of RAF1 mutations was ~9% in childhood-onset DCM cases in those three cohorts. Biochemical studies showed that DCM-associated RAF1 mutants had altered kinase activity, resulting in largely unaltered ERK activation but AKT that was hyperactivated in a BRAF-dependent manner. Constitutive expression of these mutants in zebrafish embryos resulted in a heart failure phenotype with AKT hyperactivation that was rescued by rapamycin treatment. These findings provide new mechanistic insights and potential therapeutic targets for RAF1-associated DCM and further expand the clinical spectrum of RAF1-related human disorders.
Dilated cardiomyopathy (DCM) is characterized by left ventricular dilation with systolic dysfunction, affecting approximately one person in 250. This disorder is highly genetically heterogeneous with mutations in 40 different genes, including many encoding sarcomeric and other structural proteins1-5. The underlying genetic causes of roughly 50%-60% of DCM cases remain unknown. Of the known genes mutated in DCM, many are also implicated in hypertrophic cardiomyopathy (HCM), including those genes encoding titin, cardiac actin, β-myosin heavy chain, cardiac troponin T and α-tropomyosin3-6.
Signaling through mitogen-activated protein kinases (MAPKs) plays crucial roles in myocardial biology7. Germ-line mutations in genes encoding RAS-MAPK pathway members lead to several overlapping developmental syndromes termed the RASopathies, which include a high prevalence of HCM8-12. RASopathy genes do not appear to play a significant causal role in non-syndromic HCM13. Based on mouse genetic models, altered activities of certain proteins relevant for signaling through MAPKs such as loss of SHP-2, RAF1 or increased p38 activation can induce DCM7. However, inherited abnormalities in RAS-MAPK signaling have not been previously implicated in human patients with DCM.
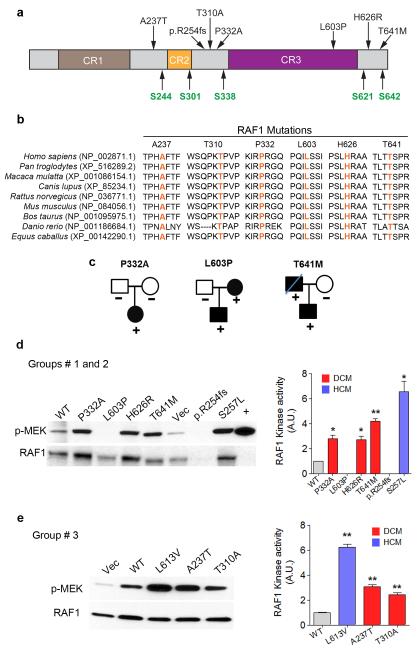
To explore whether mutations in genes encoding proteins in the RAS-MAPK pathway contribute to non-syndromic DCM, we first screened DNAs from 218 individuals with isolated DCM from South India (Group 1) for mutations in nine RAS-MAPK genes (PTPN11, HRAS, KRAS, RAF1, BRAF, SOS1, MEK1, MEK2 and SHOC2) and identified five novel RAF1 missense variants with the following predicted amino acid substitutions: p.Pro332Ala, p.Leu603Pro (2), p.His626Arg, and p.Thr641Met (Fig. 1a and Supplementary Fig. 1). No change was observed in the other eight genes. Each RAF1 variant altered a residue that was evolutionarily conserved among vertebrate RAF1 orthologs (Fig.1b) and was absent in 500 ethnically matched normal South Indian individuals. We also sequenced all RAF1 coding exons in 420 South Indians (100 of the 500 controls, 190 individuals with solid cancers, 100 individuals with coronary artery disease and 30 with atrial septal defects), finding only two synonymous alleles (T543T (n=1) and T638T (n=9)) in those exons and their splice sites. Missense RAF1 mutation frequency was significantly higher among the South Indian DCM cohort than those controls (5/436 vs. 0/840, p=0.0046). None of the RAF1 non-synonymous variants had been observed previously in NS with or without HCM, as a somatic change associated with cancer, or among the 13,600 CEU and African-American alleles in the Exome Sequencing Project (ESP)10,11. The RAF1-positive individuals screened negatively mutations in 12 known cardiomyopathy genes (MYH7, MYH6, MYBPC3, TNNT2, TPM1, MYL3, MYL2, TNNI3, PRKAG2, LMNA, PLN and TTN (Supplementary Table 1). In silico functional analyses revealed that the DCM cases (Supplementary Table 2) had significantly more variants predicted to alter RAF1 function than the population-matched controls (Polyphen-2: probably damaging 4/436 vs. 0/840; p=0.0135; pMUT: pathological 5/436 vs. 0/840; p=0.0046). We were able to analyze the families of three probands with DCM harboring a RAF1 missense mutation. For one affected individual harboring the RAF1 p.Pro332Ala allele who had a family history negative for DCM, analysis of the parental DNAs confirmed paternity and showed an absence of the RAF1 variant in both parents, consistent with a de novo change. The two other probands’ family histories suggested additional affected relatives, and the relevant RAF1 variants were observed in another symptomatic individual (Fig. 1c).
Figure 1. RAF1 mutants observed in dilated cardiomyopathy.
a. Schematic representation of RAF1 structure and location of residues altered in DCM patients. CR1-CR3 represents conserved regions of RAF1. Regulatory serine residues are shown in green. b. Alignment of RAF1 protein sequences from different species with the amino acid residues altered in DCM shown in orange. c. Pedigrees of DCM families with their RAF1 amino acid change indicated. d and e. RAF1 kinase assays. Vector alone (Vec), full-length wild-type (WT), HCM-associated (p.Leu613Val and p.Ser257Leu), and DCM-associated (p.Ala237Thr, p.Thr310Ala, p.Pro332Ala, p.Leu603Pro, p.His626Arg, p.Thr641Met and p.R254fs) RAF1 proteins were expressed in HEK293 cells as indicated. RAF1 was immunoprecipitated from EGF-stimulated cells at 15 min along with a positive control (+). Linked kinase assays were performed using inactive MEK1. RAF1 and phosphorylated MEK1 (p-MEK) were detected with anti-RAF1 (lower row) and anti-p-MEK (upper row) antibodies. Activation (p-MEK/RAF1) is expressed as Relative Expression compared to level in the WT cells. Data are mean values ± SD of two independent experiments. *p <0.05 or **p <0.01 vs. WT.
Next, we screened 200 North Indian (genetically distinct from South Indians) and 35 Japanese probands with DCM (Groups 2 and 3, respectively) for RAF1 mutations. In Group 2, two additional RAF1 sequence variants were identified: a single base-pair deletion leading to a protein truncation (p.R254fs) and a missense mutation predicting a p.Thr641Met substitution. Neither variant was identified in 350 ethnically matched North Indian controls (Figs. 1a, 1b and Supplementary Fig. 1). In Group 3, two additional novel RAF1 missense mutations were identified (p.Ala237Thr and p.Thr310Ala). Neither variant was detected among 300 Japanese controls. Both of the altered residues are evolutionary conserved (Figs. 1a and b). Although both variants were predicted to be tolerated (Supplementary Table 2), each had functional consequences when assessed in vitro (see below). None of these missense variants was observed in public databases (dbSNP, the 1000 Genomes Project and the ESP), and no other damaging allele (nonsense or frameshift) was described. Including Group 4, which is described below, the frequency of RAF1 missense or damaging mutations was significantly higher among DCM subjects than in the ESP cohorts (9/1026 vs. 29/13006; p=0.0004). Thus, we concluded that RAF1 variants were strongly associated with non-syndromic DCM (all DCM vs. population unmatched ESP: OR (Odds Ratio) =3.96, 95% CI=1.87-8.39; South Indian DCM vs. population matched controls: OR=21.42, 95% CI = 1.18-388.41).
The clinical features of RAF1-associated DCM are notable (Table 1). Of the ten subjects with a RAF1 mutation and known age of onset, eight presented in childhood or adolescence. The average age at presentation was 12.6 years, less than the approximate average age of 20 years associated with DCM caused by sarcomeric genetic mutations. Consistent with this, screening of DNAs from 60 Italian DCM patients with age at diagnosis > 18 years who were negative for mutations in nine known DCM genes (Group 4) revealed no disease-associated RAF1 mutation. Among the 218 South Indian cohort, 33 had childhood-onset disease with age at diagnosis <18 years, which included all five with RAF1 mutations. Similarly, 30 of the 200 North Indian patients had childhood-onset DCM and included both individuals with RAF1 mutations. RAF1 mutations in Indian subjects presented during childhood significantly more frequently than expected (7 of 63 total Indian childhood-onset cases (11%) compared to 0 of 355 adult-onset cases, p<0.0001). Including the one RAF1 mutation from among 25 Japanese childhood-onset cases, 8 of 88 (9%) of individuals with childhood-onset DCM harbored RAF1 mutations (p<0.0001). Thus, RAF1 is the first gene strongly associated with isolated DCM to be predominantly associated with pediatric-onset disease. Of note, the HCM associated with the RASopathies also presents early in life as have the rare cases of apparently isolated HCM with RAS/MAPK mutations identified so far10,11,13.
Table 1. Clinical features of RAF1 mutation-positive subjects*.
| P1 | P2A‡ | P2B‡ | P3 | P4 | P5 | P6§ | P7 | P8 | P9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid change |
p.Pro 332Ala |
p.Leu 603Pro |
p.Leu 603Pro |
p.His 626Arg |
p.Thr 641Met |
p.Leu 603Pro |
p.R254fs | p.Thr 641Met |
p.Ala 237Thr |
p.Thr 310Ala |
| Age, yr | 21 | 40 | 4 | 20 | 15 | 21 | 3 | 21 | 44 | 2 |
| Age of onset, yr |
13 | 24 | 3 | 10 | 7 | 10 | 1 | 16 | 40 | 2 |
| Gender | F | F | M | M | F | F | M | M | M | F |
| Origin | South India | North India | Japan | |||||||
| NYHA III or IV |
YES | YES | YES | YES | YES | YES | YES | YES | YES | YES |
| ST-T change | YES | YES | YES | NO | YES | YES | YES | YES | NA | YES |
| Ventricular arrhythmia |
YES | NO | YES | NO | YES | NO | YES | YES | NO | NO |
| LVIDd, mm | 52 | 55 | 74 | 70 | 72 | 68 | 71 | 65 | 85 | 63 |
| LVEF,% | 31 | 27 | 17 | 22 | 24 | 20 | 18 | 25 | 18 | 12 |
| Mitral regurgitation |
Mod | Mild | Sev | Mod | Mod | Mod | Sev | Mod | Mild | Mod |
Clinical features refer to time of presentation. Of note, no person had other features of a RASopathy (facial dysmorphism, short stature, webbed neck, chest deformity or mental retardation)
P2A is the father of P2B (Fig.1C) and
P6 is deceased. Abbreviations: LVEF, left ventricular ejection fraction; LVIDd, left ventricular internal diastolic dimension; NYHA, New York Heart Association; Mod, moderate; Sev, severe.
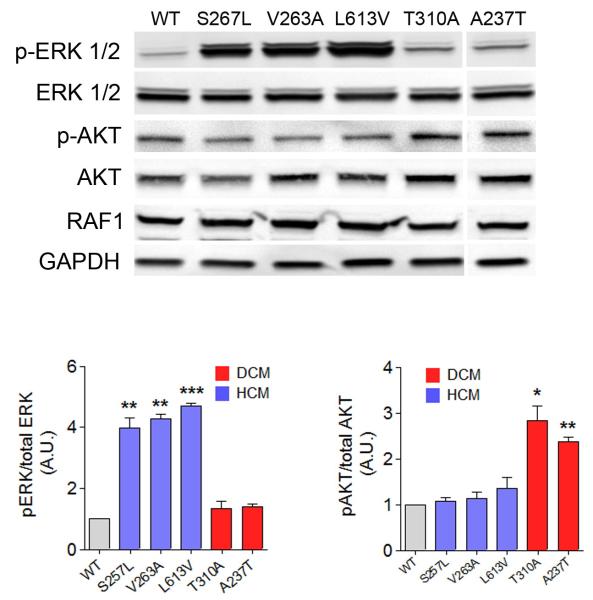
To understand the functional consequences of the DCM-associated RAF1 mutations and how they differ from HCM-associated RAF1 mutations observed in RASopathies, we transiently expressed several RAF1 mutations in human embryonic kidney (HEK293) cells and assessed their kinase activity and ERK activation. Five DCM-associated RAF1 missense mutant proteins (p.Ala237Thr, p.Thr310Ala, p.Pro332Ala, p.His626Arg, and p.Thr641Met) showed kinase activities that were mildly increased compared to wild type but less augmented in comparison to the HCM-associated RAF1 mutants (p.Leu613Val and p.Ser257Leu) tested (Figs. 1d and e). ERK activation engendered by the DCM RAF1 mutants was similar to that of the wild-type protein and significantly less than from the HCM mutants (Fig. 2 and Supplementary Fig. 2).
Figure 2. DCM-associated RAF1 mutants activate AKT.
Representative immunoblot with total lysates from HEK293 cells expressing wild-type (WT), HCM-associated RAF1 mutant proteins (p.Ser257Leu, p.Val263Ala and p.Leu613Val) and DCM-associated RAF1 mutant proteins (p.Thr310Ala and p.Ala237Thr) that were stimulated with EGF for 15 min and were probed with anti-phospho-ERK1/2, anti-ERK1/2, anti-phospho-AKT and anti-AKT antibodies. Expression levels were normalized to respective total proteins and expressed as Relative Expression compared to level in the WT cells. GAPDH levels were used as loading control. Data are mean values ± SD of four independent experiments assayed in duplicates. *p <0.05, **p <0.01 or ***p <0.001 vs WT.
One DCM-associated missense mutant (p.Leu603Pro) showed impaired kinase activity and reduced ERK activation as did the truncated RAF1 protein (p.R254fs). Of note, Leu603 is located in the kinase domain, so its substitution with a Pro could plausibly render RAF1 non-functional (Fig. 1d and Supplementary Fig. 2). Thus, DCM-associated RAF1 mutants had biochemical profiles that were distinct from those observed with RASopathy-associated mutants causing HCM.
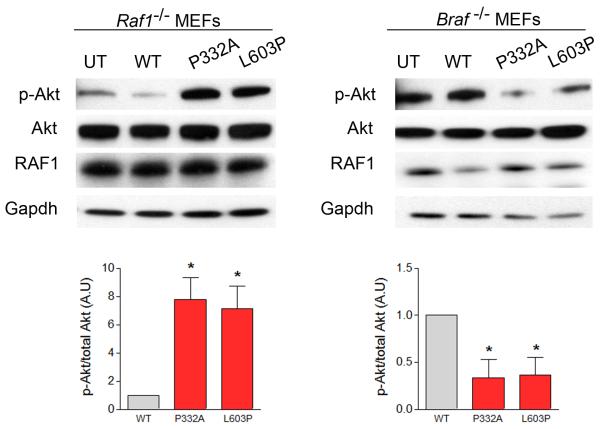
Furthermore, we found evidence supporting the hypothesis that DCM-associated RAF1 mutants signal through the AKT/mTOR pathway. Overexpression of DCM-associated RAF1 mutants in HEK293 cells resulted in excessive activation of AKT and tuberin, a downstream target of the AKT/mTOR pathway, following EGF or IGF-1 stimulation, compared to wild type. In contrast, expression of HCM-related RAF1 mutant proteins (p.Leu613Val, p.Val263Ala and p.Ser257Leu) did not excessively activate AKT or tuberin (Fig. 2, Supplementary Figs. 2 and 3). To determine whether DCM-associated RAF1 mutants depend upon BRAF for downstream signaling as has been documented for the HCM-associated RAF1 mutants14, we transiently expressed two representative DCM mutants, p.Pro332Ala (kinase active) and p.Leu603Pro (kinase impaired), in Raf1−/− and Braf −/− mouse embryonic fibroblasts (MEFs). After EGF stimulation, Erk activation in cells expressing those DCM mutants was similar to wild type with both types of MEFs (Supplementary Figs. 4 and 5). Akt hyperactivation was still observed when the DCM mutants were expressed in the Raf1−/− MEFs. In contrast, expression of DCM mutants in Braf −/− MEFs resulted in markedly reduced Akt activation (Fig. 3). Collectively, these data suggest that DCM-associated RAF1 mutants selectively induce AKT hyperactivation that is dependent upon BRAF, possibly through heterodimerization.
Figure 3. DCM-associated RAF1 mutants require Braf for Akt activation.
Representative immunoblots with total lysates from mouse embryonic fibroblasts (MEFs) from Raf1 and Braf knockout mice (Raf1−/− and Braf−/−, respectively) expressing wild-type (WT) and two representative DCM mutants (p.Pro332Ala and p.Leu603Pro) that were probed with anti-phospho-Akt and anti-Akt antibodies. Expression levels were normalized to respective total proteins and expressed as Relative Expression compared to level in the WT cells. Gapdh levels were used as loading control. Data are mean values ± SD of four independent experiments assayed in duplicates. *p <0.05 vs WT.
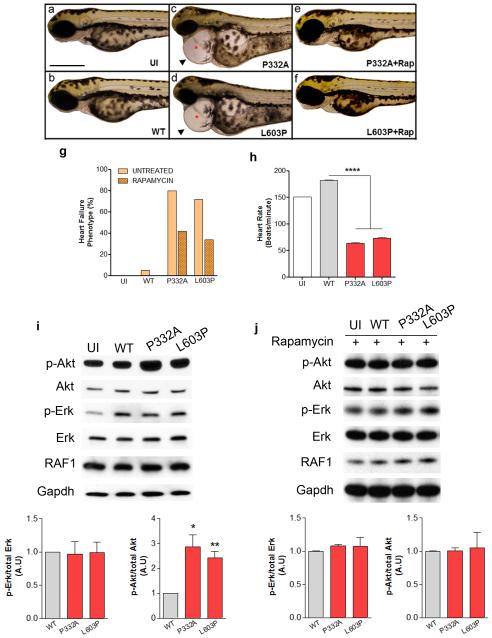
To determine whether RAF1-associated DCM mutations were sufficient to impair cardiac structure or function, we expressed wild-type RAF1 and two representative DCM mutants, p.Pro332Ala (kinase active) and p.Leu603Pro (kinase impaired), in zebrafish embryos via mRNA injection at the one-cell stage. Three days after injection, embryos expressing wild-type RAF1 had a cardiac status that was indistinguishable from uninjected embryos. In contrast, expression of both RAF1 mutants engendered heart defects mimicking a heart failure phenotype that included elongated ventricular and atrial chambers, profound pericardial edema, blood congestion at the cardiac inflow tract and impaired cardiac contractions (Figs. 4a-h). Immunoblotting revealed that the zebrafish hearts expressing the DCM-associated RAF1 proteins had ERK activation that was similar to uninjected or wild-type RAF1-expressing hearts but significantly hyperactivated Akt (Fig. 4i). After treatment with rapamycin (an AKT/mTOR inhibitor), the heart defects in the embryos expressing the DCM-associated RAF1 mutant proteins were partially rescued and AKT activation was normalized (Figs. 4e-g, i and j). These results suggest that the cardiac failure induced by DCM-associated mutant RAF1 is mediated by increased AKT signaling. Taken together with the genetic and biochemical data, the results provide compelling evidence that RAF1 mutations play a critical role in DCM. While the role of RAS/MAPK signaling in myocardial biology is well established7,12, this is the first demonstration that its alteration can contribute to isolated DCM in humans.
Figure 4. DCM-associated RAF1 mutants induce heart defects mimicking heart failure phenotype in zebrafish.
Lateral view of zebrafish embryos at 72 hours post fertilization (hpf) that were uninjected (a), injected with WT (b), p.Pro332Ala (c) or p. Leu603Pro (d) RAF1 mRNA. The two representative DCM mutants (p.Pro332Ala and p.Leu603Pro) showing string-like cardiac chambers (asterisk) with pericardial edema (arrow). Treatment with rapamycin rescued the heart failure phenotypes in the p.Pro332Ala and p.Leu603Pro RAF1 mRNA-injected embryos (e and f, respectively). Scale bar, 500 μm. g. Percentage of zebrafish embryos at 72 hpf after injection of the indicated RAF1 mRNA exhibiting heart defects with and without rapamycin treatment (n=150 in each group). h. Measurements of mutant heart rate showing severe bradycardia at 72 hpf. Data represent means ± SD of 30 embryos. i and j. Erk and Akt activation assays. Representative immunoblots with total lysates from untreated (i) and rapamycin-treated (j) zebrafish heart tissues at 72 hpf without injection (UI), injected with wild-type (WT), p.Pro332Ala and p.Leu603Pro RAF1 mRNA that were probed with anti-phospho-Erk, anti-Erk, anti-phospho-Akt and anti-Akt antibodies. Expression levels were normalized to respective total proteins and expressed as Relative Expression compared to level in the WT cells. Gapdh levels were used as loading control. Data are mean values ± SD of four independent experiments assayed in duplicates. *p <0.05, **p <0.01 or ****p <0.0001 vs WT.
Notably, RASopathy-associated RAF1 mutations as a cause of HCM appear to be functionally distinct from the newly identified DCM-associated RAF1 alleles. The HCM- and DCM-associated RAF1 mutations are mutually exclusive. Whereas HCM mutations cluster in two hot spots at Ser259 and Ser612, DCM RAF1 mutants are more widely distributed and do not alter residues critical to the regulation of RAF1 (Ser642 being an exception). DCM-associated RAF1 mutants exhibit modestly increased or impaired kinase activity, hyperactivate AKT, but not ERK, and are partially rescued by an AKT/mTOR inhibitor in zebrafish model of disease. In contrast, HCM-associated RAF1 mutants display greatly enhanced kinase activity, leading to robust ERK activation exclusively. Moreover, HCM in a mouse model of a RASopathy-associated RAF1 mutation (p.Leu613Val) was rescued using a MEK inhibitor, which prevents signaling to ERK1/215. Thus, the cardiomyopathies associated with RAF1 mutations are allelic but biologically distinct.
Finally, the complexities of signal transduction in myocardial biology are highlighted by the finding that loss-of-function PTPN11 alleles underlying another RASopathy, Noonan syndrome with multiple lentigines (NSML; formerly, LEOPARD syndrome), potentially with dominant negative properties, result in HCM. Based on a knock-in mouse model, an NSML-associated Ptpn11 allele results in hyperactivation of Akt but not Erk1/2 in the myocardium, leading to HCM earlier in life that transforms to DCM at older ages16. Of note, an mTOR inhibitor prevented or rescued the HCM phenotype in those mice, leading to the suggestion that a clinical trial of a rapamycin analog is indicated for patients with LEOPARD syndrome and HCM. If subsequent pre-clinical studies, most likely with a mouse model of a DCM-associated RAF1 mutation, recapitulate the cardiac disease and drug efficacy, a similar route to therapy can be envisioned for this genetic form of DCM.
GenBank accession numbers: RAF1 (NM_002880) and TTN (NM_001267550.1)
Methods
Clinical evaluations
A total of 513 hospitalized, unrelated DCM patients (Group 1, 2, 3 and 4) from (1) Madurai Rajaji Hospital, Madurai; (2) Sri Chitra Tirunal Institute of Medical Sciences and Technology, Trivandrum; Government Medical College Hospital, Kozhikode (representing South India as group 1) (3) Post Graduate Institute of Medical Education and Research, Chandigarh; and (4) Seth GS Medical College and KEM Hospital, Mumbai (representing North India as group 2) (5) Tokyo Women’s Medical University, Tokyo, Japan (representing Japan as group 3) (6) Monaldi Hospital, Second University of Naples (SUN), Naples, Italy (representing Italy as group 4) were recruited with informed written consent. In addition, a total of 320 registered South Indian cases with various other diseases (190 individuals with solid cancers, 100 individuals with coronary artery disease and 30 with atrial septal defects) were obtained from the referral centers outlined above. The institutional review boards of the study centers approved the protocol.
Diagnostic criteria of the index patients
DCM: A standard international protocol was followed in diagnosing the DCM cases based on published criteria5,6,17. Accordingly, individuals were diagnosed with DCM when their echocardiogram revealed depressed left ventricular (LV) systolic function (LV ejection fraction (LVEF) <0.45 and/or fractional shortening <0.25) and a dilated LV (LV end-diastolic dimension >117% of the predicted value corrected for age and body surface area in the absence of other cardiac or systemic causes including coronary and valvular diseases.
Details of the subject and control cohorts
In Group 1, the patients and controls were matched with respect to geographical region, ethnicity (self reported), sex (67% males vs 64% males) and age (40.02 ± 22 yrs vs. 45 ± 17.2 yrs). In Group 2, the patients and controls were again matched for geographical region (as outlined in group 1), ethnicity (self reported), sex (64% males vs 68% males) and age (47.22 ± 22 yrs vs 49 ± 17.2 yrs). In Group 3, the patients and controls were again matched for geographical region (as outlined in group 1), ethnicity (self reported), sex (60% males vs 62% males), age (40 ± 19yrs vs 40 ± 11 yrs). The percentage of LVEF of the patients in the group 1, 2 and 3 at the time of enrollment ranged from 29 ±11%, 32 ± 10% and 25 ± 14% with the average age of onset being 37 ± 12yrs, 40 ± 13yrs and 39 ± 11yrs respectively. The controls were apparently healthy volunteers with no familial history or symptoms of cardiovascular diseases with normal ECG and ECHO parameters from the hospitals mentioned above and were unrelated to the cardiomyopathy patients. Population stratification analysis was performed earlier using 50 ancestry-informative markers (AIMs) as described17.
Assessment of family members
The family members of the three unrelated index patients with the RAF1 mutations from South India were invited to participate in the present genetic study. The family members who participated in the present study were assessed with ECG and echocardiography. For two of the three families, additional affected members were identified. Further details about the affected family members are given in Table 1. Apart from these families, family members of the other 510 unrelated index cases were not assessed.
Sequencing and mutation analysis
Genomic DNA of individuals with DCM and controls were isolated from peripheral blood lymphocytes. Exons and their flanking intronic boundaries of PTPN11, HRAS, KRAS, BRAF, SOS1, MEK1, MEK2, SHOC2 and RAF1 were amplified from genomic DNA. Amplified PCR products were gel-isolated with a QiaxII Gel Extraction Kit (Qiagen) and sequenced on an ABI3730 DNA analyzer (Perkin-Elmer Corp., Applied Biosystems, Hitachi, Japan), using the BigDye terminator kit (Applied Biosystems, Foster City, CA, USA) and analyzed on an ABI3730 DNA Analyzer.
Cloning
RAF1 mutations were introduced into a Flag-tagged human RAF1 construct by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene). For zebrafish studies, pcDNA3 encoding human wild type and two DCM-related RAF1 mutations expressing p.Pro332Ala and p.Leu603Pro proteins were amplified and subcloned into the pCR8/GW/TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA) in combination with Gateway compatible vectors to create an mRNA expression vector appropriate for zebrafish embryos encoding six copies of a Myc-tag at the N-terminus of RAF1. The sequences of all of the mutant constructs were verified.
Expression and assessments of the mutant RAF1 proteins
Routinely used HEK293 cells as well as Raf1−/− and Braf−/− MEFs (obtained from Dr. Maneula Baccarini) were transfected with wild-type or mutant RAF1 plasmids using Lipofectamine (Invitrogen). Forty-eight hours after transfection, cells were switched to serum-starvation medium for 16 h. After stimulation with EGF (10 ng/ml) for the indicated intervals at 37 °C, the cells were lysed in RIPA buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 10 mM EDTA, 10% glycerol, 1% Triton X-100, 0.1% SDS, 1 protease inhibitor cocktail). Cell lysates were measured using the Bradford method and approximately 30 μg of total protein was loaded, separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The membranes were blocked in 5% nonfat milk and incubated with primary antibodies overnight at 4 °C. The membranes were incubated with appropriate secondary antibodies conjugated to horseradish peroxidase (Pierce) and signal intensities were visualized by chemiluminescence (Pierce).The following primary antibodies were from Cell Signaling Technology: p-Erk1/2 (Cat. No. 4370), Erk1/2 (Cat. No. 4695), p-Akt (Cat. No. 4060), p-Tsc2 (Cat. No. 3617) and β-actin (Cat. No. 4967). Total Raf1 (Cat. No. 610152), Akt (Cat. No. ab64148) and Gapdh (Cat. No. SAB2701826) antibodies were obtained from BD Biosciences, Abcam and Sigma respectively. All the cell lines were tested negative for mycoplasma contamination.
RAF1 kinase activity assay
Protein samples were prepared from HEK293 cells transfected with wild-type or mutant RAF1 expression constructs as detailed above. Lysates (containing 800 μg to 1 mg protein) were incubated with 4 μg of antibody to Flag (anti-Flag) overnight at 4 °C. Lysates were further incubated with 40 μl protein G-Sepharose beads (Roche) for 2 h at 4 °C. Bead-immune complexes were washed three times with chilled immunoprecipitation wash buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 0.2% Triton, 1X protease inhibitor) and once with the RAF1 assay reaction buffer. Beads were incubated with inactive MEK1 (RAF1 kinase assay kit, Upstate, Cat. No.17-357) at 30 °C for 1 h with shaking. Reactions were stopped by adding SDS loading buffer, boiled for 5 min, separated on SDS-PAGE and transferred to PVDF membrane. Products were detected on a protein blot using anti-phosphorylated MEK (phospho-MEK) (Upstate; 1:2,000) and goat anti-rabbit secondary antibody. RAF1 was detected with anti-Flag antibody (Sigma; 1:2,000).
In silico analysis
Analysis of the likelihood of pathogenic effect of each RAF1 variants was carried out using three bioinformatics tools: PolyPhen2, SIFT and PMut. The amino acid conservation across species was analyzed by comparing the protein sequences of various vertebrate species using ClustalW2 software.
Zebrafish maintenance and embryo injection
Adult male and female wild-type (TAB14, AB and TAB5) zebrafish (Danio rerio) were maintained on a 14:10 hour light:dark cycle at 28 °C. Transgenic fish expressing GFP in cardiomyocytes under the cmlc2 promoter (Tg(cmlc2:EGFP)) were generated by constructs contained in the tol2 kit18. Fertilized embryos collected following natural spawning were cultured at 28 °C in fish water (0.6 g/l Crystal Sea Marinemix; Marine Enterprises International, Baltimore, MD) containing methylene blue (0.002 g/l). Needles were calibrated to inject 4 nl per embryo using a Narishige IM-300 microinjector. mRNA encoding Myc-tagged human RAF1 was obtained using mMessage mMachine (Ambion, USA). By titrating mRNA concentrations, 75 ng of RNA injected per embryo was identified as the maximal tolerable and minimal effective concentration. The injections with wild-type and two mutant (p.Pro332Ala and p.Leu603Pro) RAF1 mRNAs were carried out in one-to four-cell-stage embryos. Subsequent experiments were neither randomized nor blinded. The Icahn School of Medicine at Mount Sinai’s Institutional Animal Care and Use Committee approved the necessary ethical protocols regarding zebrafish maintenance, handling and care.
Statistical analysis
Data were expressed as mean ± standard error of the mean. Differences between experimental groups were evaluated for statistical significance using Student’s t testing. P values ≤0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Heart, Lung, and Blood Institute to B.D.G. (HL071207) and grants from Telethon-Italy to M.T. (GGP10020, GGP13107). J.E. was a research Fellow supported by the Sarnoff Cardiovascular Research Foundation. Md.A.R is a postdoctoral fellow of the American Heart Association (Great Rivers Affiliate). K.T was supported by Network project grant (CARDIOMED-BSC0122) of Council of Scientific and Industrial Research (CSIR), Government of India. The authors thank Alexander Mir and Closser Evan for their technical support for the zebrafish studies, Poulikos Poulikakos and Maneula Baccarini for the Braf and Raf1 knockout MEFs and Pradeep Vaideeswar for initial collection of cardiomyopathy patient samples.
Footnotes
Author contributions
P.S.D. conceived the project, with input from B.D.G. P.S.D., M.A.R., U.M., S.K., D.S.R., G.L., A.B., M.K., A.R., E.F., M.T., T.Y., M.F., T.N., R.M. and K.T. collected and screened the various cardiomyopathy cases and controls. P.S.D., U.M., S.K., M.A.R., B.R., P.G., J.J.E. and I.R. performed the major experiments including sequencing and functional studies with the help of J.S., S.P. and S.M-N. J.R., R.J.H., D.L., K.C.S., and B.D.G. provided reagents for the study. P.S.D. drafted the manuscript, with input from B.D.G.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Hershberger RE, Siegfried JD. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2011;57:1641–1649. doi: 10.1016/j.jacc.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Curr. Opin. Cardiol. 2010;25:198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins H. Genetic clues to disease pathways in hypertrophic and dilated cardiomyopathies. Circulation. 2003;107:1344–1346. doi: 10.1161/01.cir.0000057860.52586.9c. [DOI] [PubMed] [Google Scholar]
- 4.Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies. N. Engl. J. Med. 2011;364:1643–1656. doi: 10.1056/NEJMra0902923. [DOI] [PubMed] [Google Scholar]
- 5.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2005;45:969–981. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 7.Sala V, et al. Signaling to cardiac hypertrophy: insights from human and mouse RASopathies. Mol. Med. 2012;18:938–947. doi: 10.2119/molmed.2011.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin AE, et al. Clinical, pathological, and molecular analyses of cardiovascular abnormalities in Costello syndrome: a Ras/MAPK pathway syndrome. Am. J. Med. Genet. A. 2011;155A:486–507. doi: 10.1002/ajmg.a.33857. [DOI] [PubMed] [Google Scholar]
- 10.Pandit B, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 11.Razzaque MA, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 12.Gelb BD, Tartaglia M. RAS signaling pathway mutations and hypertrophic cardiomyopathy: getting into and out of the thick of it. J. Clin. Invest. 2011;121:844–847. doi: 10.1172/JCI46399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaski JP, et al. Prevalence of sequence variants in the RAS-mitogen activated protein kinase signaling pathway in pre-adolescent children with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 2012;5:317–326. doi: 10.1161/CIRCGENETICS.111.960468. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, et al. Increased BRAF heterodimerization is the common pathogenic mechanism for noonan syndrome-associated RAF1 mutants. Mol. Cell. Biol. 2012;32:3872–3890. doi: 10.1128/MCB.00751-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, et al. MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the Raf1(L613V) mutation. J. Clin. Invest. 2011;121:1009–1025. doi: 10.1172/JCI44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marin TM, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhandapany PS, et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat. Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan KM, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.